Abstract
Despite expressing stem cell self-renewal factors, intermediate progenitor cells possess restricted developmental potential, which allows them to give rise exclusively to differentiated progeny rather than stem cell progeny. Failure to restrict the developmental potential can allow intermediate progenitor cells to revert into aberrant stem cells that might contribute to tumorigenesis. Insight into stable restriction of the developmental potential in intermediate progenitor cells could improve our understanding of the development and growth of tumors, but the mechanisms involved remain largely unknown. Intermediate neural progenitors (INPs), generated by type II neural stem cells (neuroblasts) in fly larval brains, provide an in vivo model for investigating the mechanisms that stably restrict the developmental potential of intermediate progenitor cells. Here, we report that the transcriptional repressor protein Earmuff (Erm) functions temporally after Brain tumor (Brat) and Numb to restrict the developmental potential of uncommitted (immature) INPs. Consistently, endogenous Erm is detected in immature INPs but undetectable in INPs. Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to all known neuroblast self-renewal factors in INPs. We also identified that the BAP chromatin-remodeling complex probably functions cooperatively with Erm to restrict the developmental potential of immature INPs. Together, these data led us to conclude that the Erm-BAP-dependent mechanism stably restricts the developmental potential of immature INPs by attenuating their genomic responses to stem cell self-renewal factors. We propose that restriction of developmental potential by the Erm-BAP-dependent mechanism functionally distinguishes intermediate progenitor cells from stem cells, ensuring the generation of differentiated cells and preventing the formation of progenitor cell-derived tumor-initiating stem cells.
Keywords: Earmuff, Brm, Developmental potential, Intermediate neural progenitor, Neuroblast, Self-renewal factors
INTRODUCTION
Tissue-specific stem cells often generate differentiated cell types indirectly through intermediate progenitor cells during normal development and the maintenance of homeostasis (Lui et al., 2011; Ming and Song, 2011; Weng and Lee, 2011; Chang et al., 2012; Homem and Knoblich, 2012; Franco and Müller, 2013). Intermediate progenitor cells possess restricted developmental potential, which allows them to give rise to exclusively differentiated progeny, thereby amplifying the output of stem cells. Accumulating evidence suggests that the acquisition of aberrant stem cell properties by intermediate progenitor cells might be an underlying mechanism that leads to the initiation of tumorigenesis (Weng et al., 2010; Liu et al., 2011; Haenfler et al., 2012; Xiao et al., 2012; Schwitalla et al., 2013; Komori et al., 2014). Thus, understanding the mechanisms that restrict the developmental potential of intermediate progenitor cells might lead to the discovery of novel strategies to attenuate tumor growth.
The type II neuroblast lineage in the fly larval brain provides an excellent genetic model in which to investigate the mechanisms that restrict the developmental potential of intermediate progenitor cells in vivo (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Weng et al., 2010; Xiao et al., 2012; Komori et al., 2014). A type II neuroblast can be unambiguously identified by the expression of Deadpan (Dpn+) and lack of Asense (Ase-), and divides asymmetrically to self-renew and to generate a newly born immature intermediate neural progenitor (INP) (Fig. 1A). Although the expression of self-renewal factors is maintained in the type II neuroblast, their expression becomes rapidly extinguished in the newly born immature INP (Xiao et al., 2012). This newly born INP undergoes a stereotypical maturation process during which its developmental potential becomes stably restricted and the expression of Ase is activated. Upon completing maturation, an INP divides only five or six times to generate exclusively differentiated progeny despite reactivating the expression of all known neuroblast self-renewal factors. Thus, it is likely that the restriction of developmental potential during the maturation of an immature INP results in attenuated competence to respond to the neuroblast self-renewal factors in an INP, but the mechanisms are not understood.
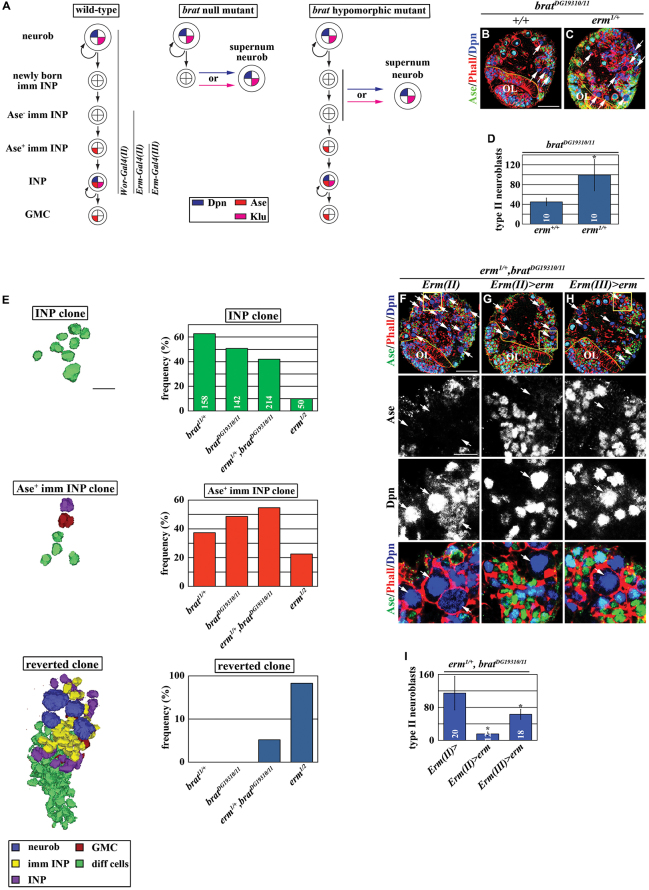
Fig. 1.
Erm functions in immature INPs to suppress supernumerary type II neuroblast formation. (A) A summary of the brat mutant phenotype and the expression patterns of the Gal4 drivers used in this study. Neurob, neuroblast; imm INP, immature INP; GMC, ganglion mother cell. (B-D) The heterozygosity of erm enhances the supernumerary neuroblast phenotype in bratDG19310/11 brains (as seen in B,C). Phalloidin (Phall) marks the cell cortex. Scale bar: 40 μm. (D) Quantification of the average number of type II neuroblasts (Dpn+Ase-) per brain lobe in larvae of the indicated genotypes. Error bars indicate s.d. (E) Reduction in erm function increases the frequency of formation of supernumerary neuroblasts originating from the Ase- immature INPs or INPs. Lineage clones marked with β-gal were induced and analyzed following the scheme shown in supplementary material Fig. S2. INP clone: a clone derived from a single INP; Ase+ imm INP clone: a clone derived from a single Ase+ immature INP; reverted clone: a clone containing supernumerary neuroblasts. The bar graphs show the frequency of clones observed in larval brains of the indicated genotype, and the total number of clones used to determine the frequency of the clones is shown in the bar graph for the INP clone. (F-I) Restoring erm function in the Ase- immature INPs or Ase+ immature INPs rescues the enhancement of the supernumerary neuroblast phenotype in bratDG19310/11 brains induced by the heterozygosity of erm. High magnification images of the boxed areas in the low magnification image are shown below. Scale bars: 40 μm (low magnification images); 10 μm (high magnification images). (I) The quantification of the average number of type II neuroblasts per brain lobe of the indicated genotypes. Key for this and all subsequent figures: dotted yellow line separates the brain from the optic lobe (OL); white arrow, type II neuroblast; white arrowhead, newly born immature INP and Ase- immature INP; yellow arrow, Ase+ immature INP; yellow arrowhead, INP. *P<0.05 between the marked genotype and the control genotype in the same bar graph, as determined by the Student’s t-test. n.s., not statistically significant.
The neuroblast self-renewal factors include Dpn, Klumpfuss (Klu), Enhancer of split mγ [E(spl)mγ] and Notch (Weng et al., 2010; San-Juán and Baonza, 2011; Xiao et al., 2012; Zacharioudaki et al., 2012; Zhu et al., 2012). Removal of Notch function alone or dpn and E(spl)mγ function simultaneously leads to premature neuroblast differentiation, whereas overexpression of any of the neuroblast self-renewal factors in type II neuroblasts leads to massive formation of supernumerary neuroblasts. Unexpectedly, whereas overexpression of klu in Ase- immature INPs driven by the Erm-Gal4(II) driver induces a robust supernumerary neuroblast phenotype, overexpression of klu in Ase+ immature INPs driven by the Erm-Gal4(III) failed to induce supernumerary neuroblast formation (Xiao et al., 2012). The expression level of Erm-Gal4(III) is ∼50% of Erm-Gal(II) (D.H.J. and C.-Y.L., unpublished observation). However, overexpression of two copies of the UAS-klu transgenes driven by two copies of the Erm-Gal4(III) driver was not sufficient to induce a supernumerary neuroblast phenotype remotely comparable to overexpression of one copy of the UAS-klu transgene driven by one copy of the Erm-Gal4(II) driver (Xiao et al., 2012). Although we cannot quantitatively control the exact expression level of the UAS-klu transgenes driven by Erm-Gal4(II) versus Erm-Gal4(III) in these experiments, these results suggest that Ase+ immature INPs are significantly less responsive to the expression of neuroblast self-renewal factors than Ase- immature INPs. Understanding the mechanisms that alter the responsiveness to neuroblast self-renewal factors in Ase+ immature INPs will provide important insight into the restriction of developmental potential.
The transcription factor Erm (also known as dFezf) functionally distinguishes an INP from a neuroblast (Weng et al., 2010). erm encodes an evolutionarily conserved C2H2 zinc-finger transcription factor, and the vertebrate orthologs of Erm can activate or repress gene expression in a context-dependent manner (Hirata et al., 2006; Weng et al., 2010; Yang et al., 2012). Erm is dispensable for the formation of INPs, but INPs in erm-null brains spontaneously revert into supernumerary type II neuroblasts. Importantly, restoring erm function by overexpressing erm or the vertebrate ortholog of erm (fez or fezl) rescued the supernumerary neuroblast phenotype, strongly suggesting that the function of Erm is evolutionarily conserved. Erm suppresses the reversion of INPs by antagonizing Notch signaling (Weng et al., 2010). However, the mechanisms by which Erm restricts the functional output of Notch signaling in INPs are completely unknown. In addition, understanding whether Erm exerts a similar regulatory effect on other neuroblast self-renewal factors will provide important insight into the mechanisms that functionally distinguish an INP from a neuroblast.
In this study, we show that Erm functions as a transcriptional repressor to stably restrict the developmental potential in immature INPs. Erm functions temporally after Brat and Numb in immature INPs to suppress the formation of supernumerary neuroblasts, and endogenous Erm protein is exclusively expressed in immature INPs. Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to all known neuroblast self-renewal factors in INPs. Thus, the Erm-dependent mechanism stably and globally restricts the genomic response to neuroblast self-renewal factors. We identified that the BAP chromatin-remodeling complex also functions temporally after Brat and Numb to restrict the developmental potential of immature INPs. Importantly, overexpression of a dominant-negative form of Brm strongly enhanced the supernumerary neuroblast phenotype in erm hypomorphic brains. Thus, we propose that Erm and the BAP complex function cooperatively to stably restrict the developmental potential of immature INPs and to functionally distinguish an INP from a neuroblast.
RESULTS
Erm functions in immature INPs to suppress the formation of supernumerary type II neuroblasts
We hypothesized that restriction of the developmental potential in immature INPs alters the responsiveness to neuroblast self-renewal factors, preventing INPs from aberrantly reverting into supernumerary neuroblasts in response to the re-activation of neuroblast self-renewal factors. We used the bratDG19310/11 hypomorphic genetic background to investigate the mechanisms that restrict the developmental potential of immature INPs (Xiao et al., 2012; Komori et al., 2014). Briefly, Brat prevents the formation of supernumerary neuroblasts by acting at two temporally distinct stages during maturation (Fig. 1A). First, Brat prevents a newly born immature INP, which lacks the expression of both Erm-Gal4(II) and Ase (Fig. 1A; supplementary material Fig. S1D), from reverting into a supernumerary neuroblast by rapidly extinguishing the function of neuroblast self-renewal factors. Newly born immature INPs mutant for brat rapidly revert into supernumerary neuroblasts instead of progressing through maturation, and removing the function of the neuroblast self-renewal gene klu or dpn suppressed the supernumerary neuroblast phenotype (supplementary material Fig. S1A-C) (Xiao et al., 2012). Second, Brat continues to play an important role in the Ase- immature INP, which shows the expression of Erm-Gal4(II) (Fig. 1A), to promote the maturation of immature INPs. We confirmed that the temporal expression pattern of Erm-Gal4(II) and Erm-Gal4(III) is not altered in bratDG19310/11 brains (supplementary material Fig. S1D-G). Restoring brat function in Ase- immature INPs suppressed the supernumerary neuroblast phenotype in bratDG19310/11 brains, whereas restoring brat function in Ase+ immature INPs had no effects (supplementary material Fig. S1H-L). Thus, Brat functions in the newly born immature INP and the Ase- immature INP to prevent supernumerary neuroblast formation.
The unstable nature of Ase- immature INPs provides an excellent in vivo system to elucidate the mechanisms that restrict developmental potential during the maturation of immature INPs. We identified the erm gene as a genetic enhancer of brat by screening for haploinsufficient loci that further exacerbate the supernumerary neuroblast phenotype in bratDG19310/11 brains. Although the heterozygosity of erm alone did not lead to a supernumerary neuroblast phenotype, it doubled the number of supernumerary neuroblasts in bratDG19310/11 brains (Fig. 1B,C). To examine whether the enhancement of the supernumerary neuroblast phenotype in bratDG19310/11 brains by the heterozygosity of erm originated from Ase- or Ase+ immature INPs, we induced β-gal-marked lineage clones derived from either a single Ase+ immature INP or an INP by FRT-mediated recombination (supplementary material Fig. S2A). Briefly, first instar larvae carrying a UAS-flipase transgene and a flip-out reporter transgene under the control of Erm-Gal4(III) and Tub-Gal80ts were heat-shocked at 30°C for 0-12 hours (supplementary material Fig. S2B-D). We determined that in the absence of heat shocking, the leaky basal level of Erm-Gal4(III) expression was sufficient to induce an average of three clones per lobe in the control brain (supplementary material Fig. S2E). The low frequency of clone induction under this condition is ideal for analyzing the identity of cells in an individual clone. We only recovered Ase+ immature INP clones, which contain one INP per clone, GMCs and differentiated cells, and INP clones, which contain only differentiated cells, in control bratDG19310/+ or erm1/+ brains (Fig. 1E). This is consistent with a wild-type INP maintaining restricted developmental potential. In bratDG19310/11 brains, more than 99% of the clones were either Ase+ immature INP clones or INP clones, and only 0.7% of the clones contained supernumerary neuroblasts (the reverted clone) (Fig. 1E). This result is consistent with Brat mainly functioning in the newly born immature INP and the Ase- immature INP to prevent the formation of supernumerary neuroblasts. Importantly, 3.5% of the clones in bratDG19310/11 brains heterozygous for erm were the reverted clones, and these clones consistently possessed more supernumerary neuroblasts than the reverted clones in bratDG19310/11 brains (Fig. 1E; supplementary material Fig. S2F). Thus, heterozygosity of erm increases the frequency of Ase+ immature INPs or INPs reverting to neuroblasts in bratDG19310/11 brains. The occurrence of the reverted clones increased dramatically in erm-null brains, consistent with erm playing a crucial role suppressing supernumerary neuroblast formation (Fig. 1E). These data strongly suggest that erm functions temporally after brat in immature INPs to prevent supernumerary neuroblast formation.
We tested whether erm indeed functions temporally after brat in immature INPs by restoring erm function in either Ase- or Ase+ immature INPs in bratDG19310/11 brains heterozygous for erm. Consistent with our hypothesis, restoring erm function in either Ase- or Ase+ immature INPs rescued the enhancement of the supernumerary neuroblast phenotype in bratDG19310/11 brains induced by the heterozygosity of erm (Fig. 1F-I). Thus, we conclude that erm functions temporally after brat in immature INPs to suppress the formation of supernumerary neuroblasts.
Erm functions temporally after Brat and Numb in immature INPs
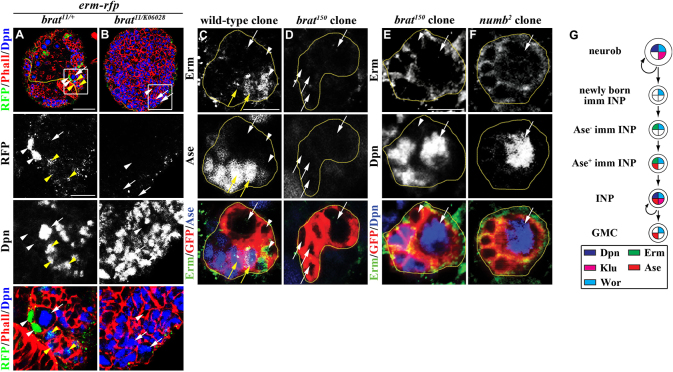
We assessed the expression pattern of endogenous Erm to confirm that erm indeed functions in Ase- and Ase+ immature INPs. We generated a transgenic fly line carrying a BAC clone containing the entire erm genomic locus fused in frame to a RFP epitope (erm-rfp). In the type II neuroblast lineage, the expression of erm-RFP was detectable in immature INPs located immediately adjacent to the type II neuroblast but became rapidly downregulated in INPs (Fig. 2A). The relative position of these Erm-expressing immature INPs to the type II neuroblast strongly suggests that endogenous Erm is expressed in Ase- immature INPs. To unambiguously verify the identity of cells in which endogenous Erm is expressed, we generated a specific antibody against the Erm protein. Co-immunolocalization using specific antibodies against Ase and Erm confirmed that endogenous Erm was absent from the newly born immature INP but became detectable in both Ase- and Ase+ immature INPs (Fig. 2C). These data are consistent with erm functioning in Ase- and Ase+ immature INPs to restrict developmental potential.
Fig. 2.
Erm is exclusively expressed in Ase- and Ase+ immature INPs. (A,B) The expression of erm-rfp is detected in immature INPs in wild-type brains but undetectable in brat-null brains. High magnification images of the boxed area are shown below. Scale bars: 40 μm (low magnification images); 10 μm (high magnification images). (C,D) Endogenous Erm is undetectable in the newly born immature INP. The GFP-marked lineage clones are outlined in yellow. Scale bar: 10 μm. (E,F) Erm is undetectable in brat- or numb-null mutant clones. The GFP-marked lineage clones are outlined in yellow. Scale bar: 10 μm. (G) A cartoon summarizing the expression pattern of Erm in the type II neuroblast lineage.
We net examined the expression pattern of endogenous Erm in brat-null brains to confirm that erm indeed functions temporally after brat in immature INPs. Indeed, the expression of erm-RFP was completely absent from the brat-null brain, and endogenous Erm was undetectable in the newly born immature INP in brat-null type II neuroblast clones (Fig. 2B,D,E). Thus, these results confirm that Erm functions temporally after Brat in immature INPs.
Numb functions in parallel to Brat to prevent the formation of supernumerary neuroblasts (Xiao et al., 2012). A numb-null type II neuroblast clone contained many supernumerary neuroblasts and showed an aberrant accumulation of immature INPs lacking Ase expression. Importantly, we never detected the expression of Erm in immature INPs lacking Ase expression in numb-null clones, indicating that these cells are newly born immature INPs (Fig. 2F). Thus, a numb-null clone aberrantly accumulates newly born immature INPs. Taken together, these data indicate that Erm functions temporally after Brat and Numb to restrict the developmental potential of immature INPs.
Erm restricts the developmental potential of immature INPs by repressing gene transcription
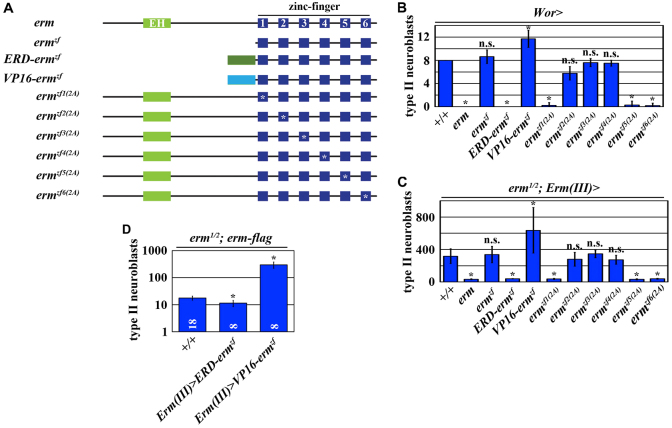
Because the vertebrate orthologs of Erm can activate or repress gene expression in a context-dependent manner (Hirata et al., 2006; Yang et al., 2012), we investigated whether Erm restricts developmental potential by activating or repressing gene expression. Although mis-expression of wild-type Erm in neuroblasts induced premature differentiation of all eight type II neuroblasts in a larval brain lobe, mis-expression of Ermzf (containing only the zinc-fingers) had no effect (Fig. 3A,B). Thus, the N-terminus of the Erm protein is essential for its function in restricting developmental potential. We fused the Engrailed repressor domain (ERD) to Ermzf, converting it to act solely as a transcriptional repressor (ERD-Ermzf), or the VP16 transactivation domain to Ermzf, converting it to act solely as a transcriptional activator (VP16-Ermzf) (Fig. 3A). Mis-expression of ERD-Ermzf in neuroblasts also led to premature differentiation of type II neuroblasts whereas mis-expression of VP16-Ermzf resulted in a mild increase in type II neuroblasts (Fig. 3B). These data strongly suggest that Erm restricts developmental potential by acting as a transcriptional repressor. We tested this hypothesis by taking two complementary approaches. First, we tested whether overexpression of the erm transgene can rescue the supernumerary neuroblast phenotype in erm-null brains. Restoring wild-type Erm function driven by the Erm-Gal4(III) driver rescued the supernumerary neuroblast phenotype in erm-null brains (Fig. 3C) (Weng et al., 2010). Importantly, overexpression of ERD-Ermzf under the identical conditions also rescued the supernumerary neuroblast phenotype in erm-null brains, but overexpression of VP16-Ermzf enhanced the supernumerary neuroblast phenotype (Fig. 3C). Second, we tested whether overexpression of the erm transgene can rescue the supernumerary neuroblast phenotype in an erm hypomorphic genetic background (erm1/2; erm-flag) (Fig. 3D). We confirmed that the temporal expression pattern of Erm-Gal4(III) is not altered in erm hypomorphic brains (supplementary material Fig. S3). Consistently, overexpression of ERD-Ermzf driven by the Erm-Gal4(III) driver rescued the supernumerary neuroblast phenotype in erm hypomorphic brains, but overexpression of VP16-Ermzf enhanced the supernumerary neuroblast phenotype (Fig. 3D). Together, these data led us to conclude that Erm restricts the developmental potential in immature INPs by acting as a transcriptional repressor, and that VP16-Ermzf can exert a dominant-negative effect on the restriction of developmental potential.
Fig. 3.
Erm restricts the developmental potential in immature INPs by repressing gene transcription. (A) Schematics of the UAS-erm transgenes used in this series of experiments. ERD, engrailed repressor domain; VP16, transactivation domain. (B) Overexpression of erm induces premature neuroblast differentiation by repressing gene transcription. Third instar larval brains of the indicated genotype were stained for Dpn, Ase and Phall, and the total number of type II neuroblasts (Dpn+Ase-) per brain lobe was quantified. (C) Restoring erm expression rescues the supernumerary neuroblast phenotype in erm-null brains by repressing gene transcription. Third instar larval brains of the indicated genotypes were treated and analyzed as in B. (D) Overexpression of VP16-ermzf exerts a dominant-negative effect and further exacerbates the supernumerary neuroblast phenotype in erm hypomorphic brains. Third instar larval brains of the indicated genotypes were treated and analyzed as in B.
We next examined which C2H2 zinc-finger elicits the function of Erm in restricting developmental potential. We generated UAS-ermzf(2A) transgenes that encode Erm transgenic proteins containing substitutions of alanine for cysteine in individual zinc-fingers (Fig. 3A). Mis-expression of ermzf2(2A), ermzf3(2A) or ermzf4(2A) failed to induce premature differentiation of type II neuroblasts in wild-type brains and failed to rescue the supernumerary neuroblast phenotype in erm-null brains (Fig. 3B,C). By contrast, mis-expression of ermzf1(2A), ermzf5(2A) or ermzf6(2A) induced premature differentiation of type II neuroblasts and rescued the supernumerary neuroblast phenotype in erm null brains (Fig. 3B,C). Thus, the zinc-finger 2-4 are essential to confer Erm function. Together, we conclude that Erm restricts the developmental potential in immature INPs by repressing gene transcription through the zinc-finger 2-4.
Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to Klu in INPs
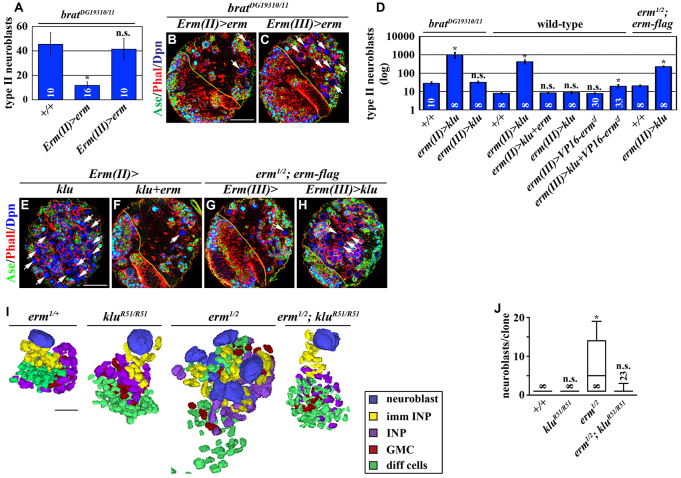
To begin investigating the mechanisms by which Erm restricts the developmental potential of immature INPs, we examined whether overexpression of erm can substitute for the function of brat and suppress the supernumerary neuroblast phenotype in bratDG19310/11 brains. Indeed, overexpression of erm in Ase- immature INPs efficiently suppressed the supernumerary neuroblast phenotype in bratDG19310/11 brains, but overexpression of erm in Ase+ immature INPs could not (Fig. 4A-C). These results strongly suggest that brat and erm suppress supernumerary neuroblast formation by regulating similar downstream mechanisms. Brat suppresses supernumerary neuroblast formation by antagonizing Klu (Xiao et al., 2012). Similar to overexpression of klu in wild-type brains, mis-expression of klu in Ase- immature INPs in bratDG19310/11 brains led to supernumerary neuroblast formation, but mis-expression of klu in Ase+ immature INPs had no effect (Fig. 4D). The inefficiency in inducing supernumerary neuroblasts by mis-expression of klu in Ase+ immature INPs correlates with Erm mainly functioning in Ase+ immature INPs to restrict developmental potential. Thus, we hypothesized that Erm restricts developmental potential by antagonizing Klu function. Consistently, co-expression of erm completely suppressed supernumerary neuroblast formation induced by mis-expression of klu in Ase- immature INPs in wild-type brains (Fig. 4D-F). Furthermore, although mis-expression of VP16-ermzf or klu alone in Ase+ immature INPs did not have any effect, co-expression of VP16-ermzf and klu induced supernumerary neuroblast formation (Fig. 4D). This result indicates that the downstream mechanisms regulated by Erm can act cooperatively with Klu to induce supernumerary neuroblast formation. Finally, mis-expression of klu in Ase+ immature INPs also enhanced supernumerary neuroblast formation in erm hypomorphic brains (Fig. 4D,G,H). Together, these data strongly suggest that Erm restricts developmental potential by antagonizing Klu function.
Fig. 4.
Erm-dependent restriction of developmental potential in immature INPs leads to attenuated competence to respond to Klu in INPs. (A-C) Overexpression of erm in Ase- immature INPs can suppress the supernumerary neuroblast phenotype in brat hypomorphic brains as seen in B and C. Scale bar: 40 μm. (A) Quantification of total type II neuroblasts (Dpn+Ase-) per brain lobe of the indicated genotypes. (D-H) Co-expression of erm can suppress the supernumerary neuroblast phenotype induced by mis-expression of klu, as seen in E-H. Scale bar: 40 μm. (D) The quantification of total type II neuroblasts (Dpn+Ase-) per brain lobe of the indicated genotypes. (I,J) Removing klu function suppresses supernumerary neuroblast formation in erm-null brains. (I) Three-dimensional reconstructed images of clones of the genotype indicated. Third instar larval brains carrying GFP-marked mosaic clones derived from single neuroblasts of the genotype indicated were stained for GFP, Dpn, Ase, Pros and Elav. Scale bar: 10 μm. (J) Quantification of total type II neuroblasts (Dpn+Ase-) per clone for the indicated genotypes.
We directly tested whether removing klu function can suppress supernumerary neuroblast formation in erm-null brains. In erm-null brains, GFP-marked mosaic clones derived from single type II neuroblasts contained multiple neuroblasts per clone (Fig. 4I,J). Clonally removing klu function for 72 hours was not sufficient to induce premature differentiation of type II neuroblasts (Berger et al., 2012; Xiao et al., 2012). By contrast, clonally removing klu function strongly suppressed supernumerary neuroblasts in erm-null brains (Fig. 4I,J). Thus, klu is required for supernumerary neuroblast formation in erm-null brains. Because Klu expression is extinguished in immature INPs but becomes re-activated in INPs (Xiao et al., 2012), INPs are most likely the cells of origin of supernumerary neuroblasts in erm-null brains. Therefore, Erm probably restricts developmental potential by indirectly antagonizing Klu function. We conclude that Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to Klu in INPs.
Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to Dpn and E(spl)mγ
Similar to klu, dpn is also required for supernumerary neuroblast formation in brat-null brains (supplementary material Fig. S1A-C). In addition, mis-expression of dpn in Ase+ immature INPs also led to a significantly milder supernumerary neuroblast phenotype in wild-type as well as in bratDG19310/11 brains compared with mis-expression in Ase- immature INPs (Fig. 5A). These results prompted us to test whether Erm restricts developmental potential by antagonizing Dpn function. Consistent with our hypothesis, co-expression of erm completely suppressed supernumerary formation induced by mis-expression of dpn in Ase- immature INPs in wild-type brains (Fig. 5A-C). Furthermore, co-expression of VP16-ermzf and dpn in Ase+ immature INPs led to a significant increase in supernumerary neuroblasts compared with mis-expression of dpn alone under the identical conditions (Fig. 5A). Finally, mis-expression of dpn in Ase+ immature INPs enhanced supernumerary neuroblast formation in erm hypomorphic brains (Fig. 5A,D,E). Together, these data strongly suggest that Erm restricts developmental potential by antagonizing Dpn function. Importantly, although not affecting the maintenance of type II neuroblasts (Zacharioudaki et al., 2012; Zhu et al., 2012) (Fig. 5F), clonally removing the function of dpn strongly suppressed the supernumerary neuroblast phenotype in erm-null brains (Fig. 5F). Because Dpn expression is extinguished in immature INPs but becomes re-activated in INPs (Xiao et al., 2012), INPs are most likely the cells of origin for supernumerary neuroblasts in erm-null brains. Thus, Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to Dpn in INPs.
Fig. 5.
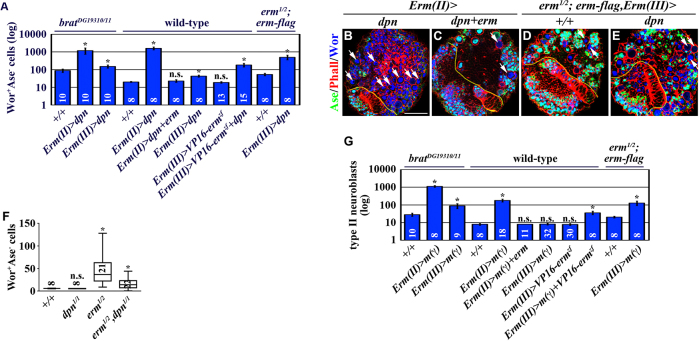
Erm-dependent restriction of developmental potential in immature INPs leads to attenuated competence to respond to Dpn and E(spl)mγ in INPs. (A-E) Co-expression of erm can suppress the supernumerary neuroblast phenotype induced by mis-expression of dpn, as seen in B-E. Scale bar: 40 μm. (A) Quantification of total Wor+Ase- cells (including type II neuroblasts and Ase- immature INPs) per brain lobe of the indicated genotypes. (F) Removing dpn function suppresses supernumerary neuroblast formation in erm-null brains. Quantification of total Wor+Ase- cells (including type II neuroblasts and Ase- immature INPs) per clone for the indicated genotypes. (F) Third instar larval brains carrying GFP-marked mosaic clones derived from single neuroblasts of the indicated genotypes were stained for GFP, Wor, Ase, Pros and Elav. (G) Co-expression of erm can suppress the supernumerary neuroblast phenotype induced by mis-expression of E(spl)mγ. Quantification of total type II neuroblasts (Dpn+Ase-) per clone for the indicated genotypes.
A recent study showed that E(spl)mγ also functions as a neuroblast self-renewal factor (Zacharioudaki et al., 2012). Similar to klu and dpn, co-expression of erm strongly suppressed supernumerary formation induced by mis-expression of E(spl)mγ in Ase- immature INPs in wild-type brains (Fig. 5G). In addition, co-expression of VP16-ermzf and E(spl)mγ in Ase+ immature INPs induced supernumerary neuroblasts, and mis-expression of E(spl)mγ in Ase+ immature INPs enhanced supernumerary neuroblast formation in erm hypomorphic brains (Fig. 5G). Because E(spl)mγ expression is extinguished in immature INPs but becomes re-activated in INPs (L. Anhezini and C.-Y.L., unpublished observation), E(spl)mγ also probably contributes to the reversion of INPs into supernumerary neuroblasts in erm-null brains. These results show that Erm-dependent restriction of the developmental potential in immature INPs leads to attenuated competence to respond to all known neuroblast self-renewal factors in INPs.
The BAP complex suppresses supernumerary neuroblast formation by restricting the developmental potential of immature INPs
To elucidate the mechanisms by which Erm restricts the developmental potential of immature INPs, we characterized additional haploinsufficient loci that enhanced the supernumerary neuroblast phenotype in bratDG19310/11 brains. We identified that the brm, mor and osa genes act as genetic enhancers of brat. Specifically, although the heterozygosity of brm, mor or osa did not have effects on the type II neuroblast lineage in wild-type brains, the heterozygosity of any of these three genes enhanced the supernumerary neuroblast phenotype in bratDG19310/11 brains (Fig. 6A-E). Because brm, mor and osa encode the core components of the BAP chromatin-remodeling complex (Mohrmann et al., 2004; Carrera et al., 2008), we hypothesize that the BAP complex functions in immature INPs to suppress supernumerary neuroblast formation. Consistently, overexpression of a UAS-brmDN transgene, which encodes a dominant-negative form of Brm, specifically in Ase- immature INPs enhanced the supernumerary neuroblast phenotype in bratDG19310/11 brains (Fig. 6F-I). These data support our hypothesis that the BAP complex functions temporally after Brat in immature INPs to suppress supernumerary neuroblast formation. Consistent with the observations from a previous study (Neumüller et al., 2011), we confirmed that knocking down or removing the function of brm, osa or mor led to supernumerary neuroblast formation (data not shown). We ruled out the possibility that supernumerary neuroblasts induced by the loss of the BAP complex function arise from symmetric neuroblast division because a mitotic osa mutant type II neuroblast displayed normal establishment and maintenance of the apical-basal cortical polarity (data not presented). Together, these results strongly suggest that the BAP complex functions temporally after Brat in immature INPs to prevent supernumerary neuroblast formation.
Fig. 6.
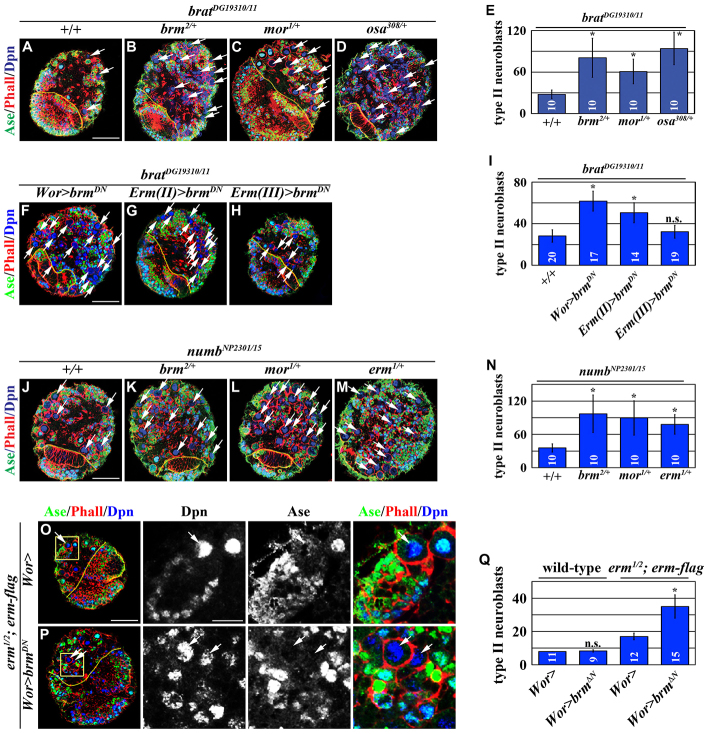
The BAP complex functions cooperatively with Erm to restrict the developmental potential in immature INPs. (A-E) Reduced function of the BAP complex enhances the supernumerary neuroblast phenotype in brat hypomorphic brains, as seen in A-D. Scale bar: 40 μm. (E) Quantification of total type II neuroblasts per lobe for the indicated genotypes. (F-I) Reducing brm function in Ase- immature INPs enhances the supernumerary neuroblast phenotype in brat hypomorphic brains (F-H). Scale bar: 40 μm. (I) The quantification of total type II neuroblasts per lobe for the indicated genotypes. (J-N) Reducing the function of the BAP complex or erm enhances the supernumerary neuroblast phenotype in numb hypomorphic brains (J-M). Scale bar: 40 μm. (N) Quantification of total type II neuroblasts per lobe for the indicated genotypes. (O-Q) Reducing brm function enhances the supernumerary neuroblast phenotype in erm hypomorphic brains (O,P). High magnification images of the boxed areas are shown on the right. Scale bars: 40 μm in the low magnification images; 10 μm in the high magnification images. (Q) Quantification of total type II neuroblasts per lobe for the indicated genotypes.
We next tested whether the BAP complex also functions temporally after Numb to suppress supernumerary neuroblast formation. Similar to Brat, Numb also functions to prevent the newly born immature INP from aberrantly reverting into a supernumerary neuroblast (Fig. 2F). However, the supernumerary neuroblast phenotype displayed by a numb-null type II neuroblast clone is too severe to test gene function in immature INPs (Xiao et al., 2012). By contrast, a numbNP2301/15 hypomorphic brain lobe, which contained 35.7±7.5 type II neuroblasts and many INPs, provides a sensitized genetic background for testing gene function in immature INPs (Fig. 6J,N). Consistent with our hypothesis, the heterozygosity of brm or mor enhanced the supernumerary neuroblast phenotype in numbNP2301/15 brains (Fig. 6K,L). These results strongly suggest that the BAP complex also functions temporally after Numb in immature INPs to suppress supernumerary neuroblast formation.
Both Erm and the BAP complex function temporally after Brat and Numb in immature INPs to suppress supernumerary neuroblast formation. Thus, we tested whether Erm and the BAP complex might function cooperatively to restrict the developmental potential of immature INPs. Similar to the BAP complex, the heterozygosity of erm also enhanced the supernumerary neuroblast phenotype in numbNP2301/15 brains (Fig. 6M,N). Most importantly, although overexpression of the UAS-brmDN transgene alone did not have any effect on the type II neuroblast lineage, overexpression of brmDN significantly increased the formation of supernumerary neuroblasts in erm hypomorphic brains (Fig. 6O-Q). Taken together, these data strongly suggest that Erm and the BAP complex function cooperatively to restrict the developmental potential of immature INPs.
DISCUSSION
Understanding the molecular mechanisms that stably restrict the developmental potential of progenitor cells may lead to the identification of novel therapeutic targets to selectively target progenitor cell-derived tumor-initiating stem cells. However, restriction of developmental potential may occur while intermediate progenitor cells acquire their functional identity. Thus, well-established stem cell lineage information is essential for investigating the molecular mechanisms that restrict developmental potential. The type II neuroblast lineage in fly larval brains offers a unique model system to investigate the restriction of the developmental potential in uncommitted intermediate progenitor cells (Weng et al., 2010; Xiao et al., 2012; Komori et al., 2014). In this study, we show that stable restriction of developmental potential in immature INPs required a temporally coordinated effort of the asymmetrically inherited proteins Brat and Numb and the transcriptional repressor protein Erm. Brat and Numb function in the newly born immature INP where they prevent the reversion into a supernumerary neuroblast induced by the activities of self-renewal factors (Fig. 7). Erm functions downstream of Brat and Numb in the Ase- and Ase+ immature INPs to restrict their genomic response to neuroblast self-renewal factors, leading to permanently attenuated competence to respond to these factors in INPs (Fig. 7). Furthermore, we also identified that the BAP chromatin-remodeling complex functions synergistically with Erm in immature INPs to suppress the formation of supernumerary neuroblasts. Taken together, we propose that Erm functions cooperatively with the BAP complex to implement a stable restriction of the developmental potential in immature INPs by modifying their genome, which leads to an attenuated response to all neuroblast self-renewal factors in INPs (Fig. 7).
Fig. 7.
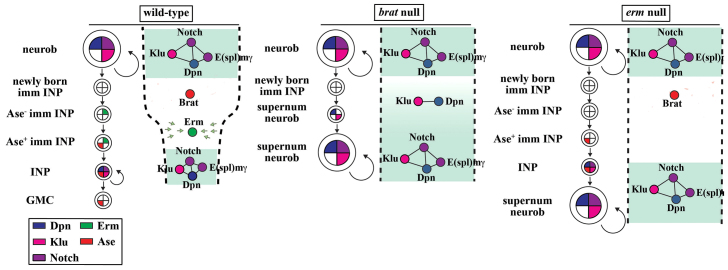
Schematic models depicting the role of Brat and Erm in the regulation of immature INPs. In wild-type brains, Erm functions temporally after Brat to restrict the competence of immature INPs to respond to the self-renewal network (indicated by the width between dotted lines). The self-renewal network is expressed in type II neuroblasts (light cyan area), but is deactivated during maturation by Brat (white area). An Erm-dependent restriction of developmental potential (green arrows) in immature INPs, leads to an attenuated competence to respond to the re-activation of neuroblast self-renewal factors in INPs (light cyan area). In brat amorphic brains, the newly born immature INPs fail to undergo maturation and rapidly revert into supernumerary neuroblasts. In erm amorphic brains, immature INPs undergo successful maturation, but their competence to respond to neuroblast self-renewal factors is not attenuated. Thus, upon re-expression of neuroblast self-renewal factors, INPs revert to form supernumerary neuroblasts.
Erm-dependent restriction of the developmental potential in immature INPs leads to an attenuated competence to respond to all self-renewal transcription factors in INPs
Several pieces of evidence led us to conclude that Erm mainly restricts the developmental potential in the Ase+ immature INPs. Erm is primarily detected in the Ase- and Ase+ immature INPs but becomes undetectable in the INP, strongly suggesting that Erm functions in the Ase- and Ase+ immature INPs (Fig. 2). Consistently, heterozygosity of erm further exacerbated supernumerary neuroblast formation from the Ase+ immature INPs in brat hypomorphic brains, and restoring erm function in the Ase+ immature INPs rescued the enhancement of the supernumerary neuroblast phenotype (Fig. 1E-I). Furthermore, restoring erm function in the Ase+ immature INPs also rescued the supernumerary neuroblast phenotype in erm-null brains (Weng et al., 2010). Finally, overexpression of VP16-Ermzf, a dominant-negative form of Erm, in the Ase+ immature INPs significantly increased supernumerary neuroblasts in various genetic backgrounds (Fig. 3). Although these data do not exclude the possibility that erm might still function in the INPs, erm most likely functions mainly in immature INPs to restrict developmental potential and suppress supernumerary neuroblast formation.
Erm restricts the developmental potential of immature INPs by acting as a transcriptional repressor, and removing the function of klu or dpn completely suppressed the supernumerary neuroblast phenotype in erm-null brains (Figs 3, 4, 5). Thus, Erm might restrict the developmental potential of immature INPs by directly repressing the transcription of the genes encoding neuroblast self-renewal factors or indirectly attenuating the competence to respond to these factors. However, Ase+ immature INPs in erm-null type II neuroblast clones never showed a premature onset of Dpn expression, and overexpression of erm in neuroblasts did not affect Dpn expression (Weng et al., 2010). In addition, transcriptome analyses indicated that dpn and klu are upregulated to a similar level in brat- and erm-null brains as in control brains (H.K. and C.-Y.L., unpublished data). Thus, it is unlikely that Erm directly regulates the transcription of neuroblast self-renewal genes. We favor the mechanism that Erm indirectly attenuates the competence to respond to these neuroblast self-renewal factors in INPs by restricting the developmental potential in immature INPs. Consistently, although overexpression of the dominant-negative VP16-Ermzf or a single neuroblast self-renewal factor alone induced a very weak supernumerary neuroblast phenotype, co-expression of VP16-Ermzf and a single neuroblast self-renewal factor led to a very robust supernumerary neuroblast phenotype (Figs 4, 5). Taken together, these data strongly suggest that stable restriction of developmental potential by the Erm-dependent mechanism in immature INPs leads to attenuated competence to respond to the re-activation of neuroblast self-renewal factors in INPs.
Erm restricts the developmental potential in immature INPs by repressing gene transcription
The vertebrate orthologs of Erm, Fezf1 and Fezf2, regulate cortical development either by activating or repressing gene expression (Hirata et al., 2006; Yang et al., 2012). As overexpression of either fezf1 or fezf2 can functionally substitute for the role of Erm in the Ase+ immature INPs (Weng et al., 2010), the results from the vertebrate studies prompted us to investigate the molecular mechanism by which Erm restricts the developmental potential in immature INPs. Overexpression of ERD-Ermzf, which functions solely as a transcriptional repressor protein, efficiently rescued the supernumerary neuroblast phenotype in the Ase+ immature INPs (Fig. 3). By contrast, overexpression of VP16-Ermzf exerted a dominant-negative effect and led to a further increase in supernumerary neuroblasts in brat or erm hypomorphic brains (Fig. 3). Taken together, these data strongly suggest that Erm restricts developmental potential in immature INPs by repressing gene transcription.
We previously mapped the molecular lesion induced by the erm1-null allele to a single amino acid substitution in the third C2H2 zinc finger of Erm, indicating that the third zinc finger is essential for the function of Erm. The C2H2 zinc finger transcription factor typically binds to DNA with two or three zinc fingers (Brayer and Segal, 2008). Consistently, perturbing the folding of zinc-finger 2 (Ermzf2(2A)) or 4 (Ermzf4(2A)) also renders the transgenic protein unable to restrict the developmental potential in the Ase+ immature INPs whereas perturbing the folding of zinc-finger 1 (Ermzf1(2A)), 5 (Ermzf5(2A)) or 6 (Ermzf6(2A)) had no effects (Fig. 3). These data indicate that the zinc-finger 2, 3 and 4 most likely mediate the binding of Erm to DNA.
Erm might function cooperatively with the BAP chromatin-remodeling complex to modify the genomic response to neuroblast self-renewal factors
A genome-wide RNAi study showed that knocking down the function of several subunits in the BAP complex results in supernumerary neuroblast formation in fly larval brains (Neumüller et al., 2011). We independently identified that brm, mor and osa, which encode the core components of the BAP complex, probably function temporally after Brat and Numb to restrict the developmental potential in the Ase- immature INPs (Fig. 6). Because Brm and Osa are expressed ubiquitously in all cells in larval brains (H.K. and C.-Y.L., unpublished data), the BAP complex probably functions cooperatively with a transcription factor that is uniquely expressed in the immature INPs to restrict developmental potential. Erm is the only known transcriptional factor that is uniquely expressed in the immature INPs, and is an excellent candidate for functioning cooperatively with the BAP complex to restrict the developmental potential (Fig. 2G). Consistently, reducing the function of Brm enhanced the supernumerary neuroblast phenotype in erm hypomorphic brains (Fig. 6O-Q). Thus, we propose that Erm restricts the developmental potential in the immature INPs by recruiting the BAP complex to specific genomic loci where the BAP complex alters the nucleosome structures, leading to attenuated competence to respond to the re-activation of neuroblast self-renewal factors. Additional functional and biochemical experiments in the future will be required to validate this hypothesis.
MATERIALS AND METHODS
Fly strains
Mutant and transgenic fly strains used include erm1, erm2 (Weng et al., 2010), kluR51 (Kaspar et al., 2008), dpn1 (Younger-Shepherd et al., 1992), brat150 (Betschinger et al., 2006), numb15 (Berdnik et al., 2002), Erm-GAL4(II) and Erm-GAL4(III) (Pfeiffer et al., 2008; Weng et al., 2010), Wor-GAL4 (Lee et al., 2006a), UAS-erm-HA (Weng et al., 2010), UAS-klu-HA (Xiao et al., 2012), UAS-brat-myc (Xiao et al., 2012), UAS-dpn (Wallace et al., 2000), UAS-E(spl)mγ (Ligoxygakis et al., 1998) and UAS-brmDN (Herr et al., 2010). The following fly stocks were obtained from the Bloomington Drosophila Stock Center: Oregon R, bratDG19310, bratk06028, brat11, brm2 and mor1, osa308, Act-FRT-Stop-FRT-GAL4, tub-GAL80, UAS-mCD8-GFP, FRTG13, FRT2A and hs-flp, tub-GAL80ts, Elav-GAL4, Act-FRT-stop-FRT-lacZ(nls), UAS-GFP(nls) and UAS-flp. numbNP2301 was obtained from the Kyoto Stock Center.
The P[acman] BAC CH321-65B19 construct was used to generate erm-flag and erm-rfp transgenic fly lines following previously established protocol (Venken et al., 2006; Bischof et al., 2007; Venken et al., 2009).
Immunofluorescence staining and antibodies
Larval brains were dissected in PBS. Larval brains were fixed in 100 mM PIPES (pH 6.9), 1 mM EGTA, 0.3% Triton X-100, 1 mM MgSO4 containing 4% formaldehyde for 23 minutes and processed for immunofluorescence staining according to a previously published protocol (Weng et al., 2012). Antibodies used in this study include rabbit anti-Erm (1:100; this study), guinea pig anti-Ase (1:1000; this study), rat anti-Dpn (1:2) (Xiao et al., 2012), rat anti-Wor (1:2) (Lee et al., 2006a), rabbit anti-Ase (1:400) (Weng et al., 2010), mouse anti-Pros (MR1A, 1:100) (Lee et al., 2006b), mouse anti-Elav [1:100; 9F8A9, Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Dlg (1:50; 4F3E3E9, DSHB), mouse anti-Osa (1:2) (Treisman et al., 1997), rabbit anti-Brm (1:2000) (Nakayama et al., 2012), chicken anti-GFP (1:2000; cat. no. 1020, Aves Labs), chicken anti-β-gal (1:2000; cat. no. 1040, Aves Labs) and rabbit anti-RFP (1:100; cat. no. 600-401-379, lot 25003, Rockland). Species-specific fluorophore-conjugated secondary antibodies (Jackson ImmunoResearch, 703-545-155, 112-605-167; Life Technologies, A-11034, A-11035, A-11074, A31553, A-31556) were used at 1:500. We used Rhodamine phalloidin (1:100; Invitrogen) to visualize cortical actin. The confocal images were acquired on a Leica SP5 scanning confocal microscope.
Generation of a polyclonal antibody against Erm
The cDNA region encoding the C-terminal 332-611 amino acids of Earmuff (Erm-C) was amplified by PCR and subsequently cloned into EcoRI and SalI sites of pGEX-4T-1, using the In-Fusion HD Cloning Kit (Clontech, cat. no. 639649). The primers used were: 5′-TGGATCCCCGGAATTCCTCACCCGCCACATGCCC-3′ (forward) and 5′-GGCCGCTCGAGTCGACCTAAAACACCTTGGCTATGA-3′ (reverse). The expression of GST-Erm-C was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) and purified using glutathione-Sepharose (GE Healthcare, cat. no. 71-5027-54) and eluted with glutathione. GST-Erm-C was injected into one rabbit and purified by GenScript (Hong Kong).
Clonal analysis
To induce the lineage clone derived from a single Ase+ immature INP or INP: bratDG19310/+, bratDG19310/11, erm1/+, bratDG19310/11 or erm1/2 larvae carrying the UAS-flp, Erm-Gal4(III), Tub-gal80ts and Act-FRT-stop-FRT-lacZ(nls)(III) transgenes were genotyped at hatching, raised at 25°C and dissected at 96 hours after larval hatching. We empirically determined the experimental condition to obtain a small number of clones per brain lobe (supplementary material Fig. S2).
The protocol to examine the expression pattern of Erm-Gal4(II) or Erm-Gal4(III) in brat or erm hypomorphic brains is described in the legend for supplementary material Figs S1 and S3.
GFP-marked mosaic clones derived from single mutant neuroblasts in the various genetic background was induced following a standard protocol (Lee and Luo, 2001).
Overexpression of UAS transgene
Mutant larvae carrying the Wor-Gal4 and Tub-Gal80ts in combination with the UAS transgene were genotyped at hatching, and raised at 31°C for 72 hours after larval hatching. Larvae were dissected and processed for immunofluorescence staining.
Three-dimensional modeling of clones
The model was generated using the Mimics software from Materialize. Confocal images were acquired using a z-step size of 1 μm and the identity of each cell within a clone was determined.
Supplementary Material
Acknowledgments
We thank Drs C. Delidakis, C. Doe, S. Hirose, H. Richardson, G. Rubin, J. Treisman, H. Vaessin and Y. N. Jan for fly stocks and antibody reagents; the Bloomington Drosophila Stock Center, Kyoto Stock Center and the Developmental Studies Hybridoma Bank for fly stocks and antibodies; BestGene Inc. for generating transgenic fly lines; Krista L. Golden for technical assistance throughout the course of this work and proofreading the manuscript; the members of the Lee lab for their intellectual input during the course of this study.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
D.H.J., H. K. and C.-Y.L. designed experiments. D.H.J., H.K. and D.G. performed experiments. K.C., C.T.K. and H.W. generated guinea pig Ase and Erm antibodies. D.H.J., H.K., D.G. and C.-Y.L. analyzed and interpreted data. D.H.J., H.K. and C.-Y.L. wrote the manuscript.
Funding
D.H.J. was supported by a Cellular and Molecular Biology training grant [T32-GM007315]; H.K. was supported by a fellowship from the Japan Society for the Promotion of Science; D.G. would like to express his gratitude to the Marshallplan Foundation for the generous financial support during his stay in the Lee lab; C.-Y.L. is supported by the University of Michigan start-up fund, a Sontag Foundation Distinguished Scientist Award and a National Institutes of Health grant [R01-GM092818]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.106534/-/DC1
References
- Bello B. C., Izergina N., Caussinus E., Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D., Török T., González-Gaitán M., Knoblich J. A. (2002). The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3, 221–231 [DOI] [PubMed] [Google Scholar]
- Berger C., Harzer H., Burkard T. R., Steinmann J., van der Horst S., Laurenson A. S., Novatchkova M., Reichert H., Knoblich J. A. (2012). FACS purification and transcriptome analysis of drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G., Knoblich J. A. (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayer K. J., Segal D. J. (2008). Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50, 111–131 [DOI] [PubMed] [Google Scholar]
- Carrera I., Zavadil J., Treisman J. E. (2008). Two subunits specific to the PBAP chromatin remodeling complex have distinct and redundant functions during drosophila development. Mol. Cell. Biol. 28, 5238–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. C., Wang C., Wang H. (2012). Balancing self-renewal and differentiation by asymmetric division: insights from brain tumor suppressors in Drosophila neural stem cells. Bioessays 34, 301–310 [DOI] [PubMed] [Google Scholar]
- Franco S. J., Müller U. (2013). Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 77, 19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenfler J. M., Kuang C., Lee C. Y. (2012). Cortical aPKC kinase activity distinguishes neural stem cells from progenitor cells by ensuring asymmetric segregation of Numb. Dev. Biol. 365, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A., Mckenzie L., Suryadinata R., Sadowski M., Parsons L. M., Sarcevic B., Richardson H. E. (2010). Geminin and Brahma act antagonistically to regulate EGFR-Ras-MAPK signaling in Drosophila. Dev. Biol. 344, 36–51 [DOI] [PubMed] [Google Scholar]
- Hirata T., Nakazawa M., Muraoka O., Nakayama R., Suda Y., Hibi M. (2006). Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development 133, 3993–4004 [DOI] [PubMed] [Google Scholar]
- Homem C. C., Knoblich J. A. (2012). Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297–4310 [DOI] [PubMed] [Google Scholar]
- Kaspar M., Schneider M., Chia W., Klein T. (2008). Klumpfuss is involved in the determination of sensory organ precursors in Drosophila. Dev. Biol. 324, 177–191 [DOI] [PubMed] [Google Scholar]
- Komori H., Xiao Q., McCartney B. M., Lee C. Y. (2014). Brain tumor specifies intermediate progenitor cell identity by attenuating β-catenin/Armadillo activity. Development 141, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Robinson K. J., Doe C. Q. (2006a). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594–598 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Wilkinson B. D., Siegrist S. E., Wharton R. P., Doe C. Q. (2006b). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P., Yu S. Y., Delidakis C., Baker N. E. (1998). A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125, 2893–2900 [DOI] [PubMed] [Google Scholar]
- Liu C., Sage J. C., Miller M. R., Verhaak R. G., Hippenmeyer S., Vogel H., Foreman O., Bronson R. T., Nishiyama A., Luo L., et al. (2011). Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 146, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J. H., Hansen D. V., Kriegstein A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G. L., Song H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann L., Langenberg K., Krijgsveld J., Kal A. J., Heck A. J., Verrijzer C. P. (2004). Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24, 3077–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Shimojima T., Hirose S. (2012). The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development 139, 4582–4590 [DOI] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., Knoblich J. A. (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Jenett A., Hammonds A. S., Ngo T. T., Misra S., Murphy C., Scully A., Carlson J. W., Wan K. H., Laverty T. R., et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Juán B. P., Baonza A. (2011). The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 352, 70–82 [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Fingerle A. A., Cammareri P., Nebelsiek T., Göktuna S. I., Ziegler P. K., Canli O., Heijmans J., Huels D. J., Moreaux G., et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 [DOI] [PubMed] [Google Scholar]
- Treisman J. E., Luk A., Rubin G. M., Heberlein U. (1997). eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 11, 1949–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., Spokony R., Wan K. H., Koriabine M., de Jong P. J., White K. P., et al. (2009). Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6, 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Liu T. H., Vaessin H. (2000). The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis 26, 77–85 [DOI] [PubMed] [Google Scholar]
- Weng M., Lee C. Y. (2011). Keeping neural progenitor cells on a short leash during Drosophila neurogenesis. Curr. Opin. Neurobiol. 21, 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Golden K. L., Lee C. Y. (2010). dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev. Cell 18, 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Komori H., Lee C. Y. (2012). Identification of neural stem cells in the Drosophila larval brain. Methods Mol. Biol. 879, 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Komori H., Lee C. Y. (2012). klumpfuss distinguishes stem cells from progenitor cells during asymmetric neuroblast division. Development 139, 2670–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Dong Z., Guo S. (2012). Fezf2 regulates multilineage neuronal differentiation through activating basic helix-loop-helix and homeodomain genes in the zebrafish ventral forebrain. J. Neurosci. 32, 10940–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger-Shepherd S., Vaessin H., Bier E., Jan L. Y., Jan Y. N. (1992). deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell 70, 911–922 [DOI] [PubMed] [Google Scholar]
- Zacharioudaki E., Magadi S. S., Delidakis C. (2012). bHLH-O proteins are crucial for Drosophila neuroblast self-renewal and mediate Notch-induced overproliferation. Development 139, 1258–1269 [DOI] [PubMed] [Google Scholar]
- Zhu S., Wildonger J., Barshow S., Younger S., Huang Y., Lee T. (2012). The bHLH repressor Deadpan regulates the self-renewal and specification of Drosophila larval neural stem cells independently of Notch. PLoS ONE 7, e46724 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.