Abstract
Here we report the design and production of an antibody-fluorophore conjugate (AFC) as a non-toxic model of an antibody-drug conjugate (ADC). This AFC is based on the conjugation of dansyl sulfonamide ethyl amine (DSEA)-linker maleimide on interchain cysteines of trastuzumab used as a reference antibody. The resulting AFC was first characterized by routine analytical methods (SEC, SDS-PAGE, CE-SDS, HIC and native MS), resulting in similar chromatograms, electropherograms and mass spectra to those reported for hinge Cys-linked ADCs. IdeS digestion of the AFC was then performed, followed by reduction and analysis by liquid chromatography coupled to mass spectrometry analysis. Dye loading and distribution on light chain and Fd fragments were calculated, as well as the average dye to antibody ratio (DAR) for both monomeric and multimeric species. In addition, by analyzing the Fc fragment in the same run, full glyco-profiling and demonstration of the absence of additional conjugation was easily achieved.
As for naked antibodies and Fc-fusion proteins, IdeS proteolytic digestion may rapidly become a reference analytical method at all stages of ADC discovery, preclinical and clinical development. The method can be routinely used for comparability assays, formulation, process scale-up and transfer, and to define critical quality attributes in a quality-by-design approach.
Keywords: ADC, antibody-drug conjugates, AFC, antibody-fluorophore conjugates, DAR, multimers, IdeS, N-linked glycosylation, mass spectrometry
Introduction
Antibody-drug conjugates (ADCs), or immunoconjugates aiming to combine the potency of cytotoxic drugs with the high specificity of a monoclonal antibody (mAb), are becoming increasingly important as new targeted therapies in oncology.1 Two ADCs, brentuximab vedotin (Adcetris®) and ado-trastuzumab emtansine (Kadcyla®), have been recently approved by the US Food and Drug Administration, and more than 30 are currently being investigated in clinical trials.2-5 To construct ADCs, highly potent cytotoxic agents are chemically attached to mAbs specific to tumor-related antigens with cleavable or non-cleavable linkers. Depending on the conjugation chemistries, different ADC structures have been developed. The drug payloads may be, for example, randomly attached to surface-exposed lysine residues distributed on both light and heavy chains (average of 80 to 95 per IgG), as illustrated by ado-trastuzumab emtansine.6 Alternatively, site-specific coupling to two or more of the eight cysteine residues involved in inter-chain disulfide bridges of chimeric, humanized or human IgG1 after mild reduction may be used, as illustrated in the case of brentuximab vedotin.7 For the same purpose, the glycan moiety can be subjected to mild oxidation and used for site-selective conjugation via a hydrazone linkage.8 As further improvements for the third-generation ADCs, design of optimized antibodies (e.g., engineered to contain free surface-exposed cysteines for a site-specific drug linkage) and drug loading level optimization are investigated to maintain the natural favorable pharmacokinetics (PK) of chimeric, humanized, and human IgGs. Site-specific conjugation to antibodies results in more homogeneous drug loading and avoids ADC subpopulations with altered antigen-binding caused by cross-linking in the CDRs or altered PK caused by cross-linking at Fc domain binding sites of FcRn. Several such attempts have recently been described, including the addition of two cysteines in the antibody variable domains9 or in the constant domains.10-12
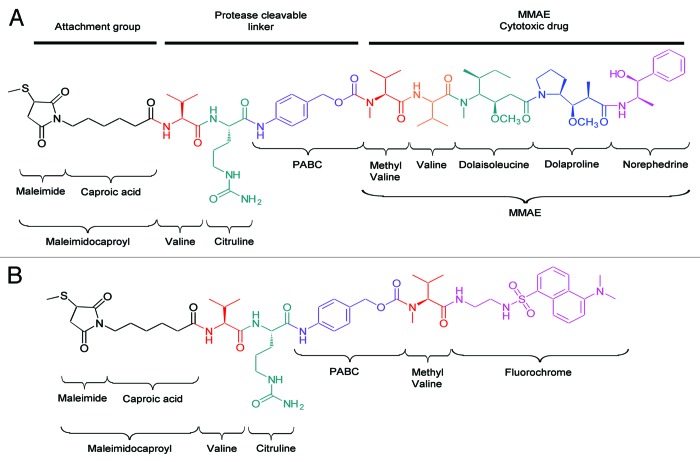
Conjugation of drugs to mAbs increases the structural complexity of the resulting molecule, which triggers the need for improved characterization methods.13 Antibody-fluorophore conjugates (AFCs) using the same linkers and conjugation chemistries as ADCs, but with a non-toxic cargo, are valuable molecular tools for mechanistic studies and PK evaluation as recently illustrated in several reports (e.g., Alexa48814 and Alexa350,15 or biotin16). Here, we report the design and production of an AFC as a non-toxic ADC model to extend the analytical platform for optimization of next-generation ADCs (OptimADCs). Our AFC is based on the conjugation of dansyl sulfonamide ethyl amine (DSEA)-linker maleimide on interchain cysteines of trastuzumab. Trastuzumab is frequently used as a reference, both as a naked antibody and as an ADC.17,18 DSEA-linker maleimide payload (Fig. 1B) was designed and synthesized (Fig. S1) to mimic the chemistry and the linker of brentuximab vedotin and the majority of ADCs in clinical trials (Fig. 1A).
Figure 1. Structures of (A) linker-cytotoxic (mc_MMAE) and (B) linker-fluorescent (mc_DSEA) payloads.
As proof of principle, the trastuzumab-mc-DSEA conjugate was initially analyzed by the current analytical methods used for antibody conjugated to maleimidocaproyl valine-citruline monomethylauristatin E (vc-MMAE) (Fig. 1A)19 to demonstrate the comparability of the resulting profiles,20 including size-exclusion chromatography (SEC) (Fig. 2), sodium dodecyl sulfate – polyacrylamide Gel (SDS-PAGE) (Fig. 3), capillary electrophoresis sodium dodecyl sulfate (CE-SDS) (Fig. 4), hydrophobic interaction chromatography (HIC) (Fig. 5), and high performance liquid chromatography with UV and mass spectrometry detection (HPLC-UV/MS) in both denaturating conditions and native (Fig. 6).21-23
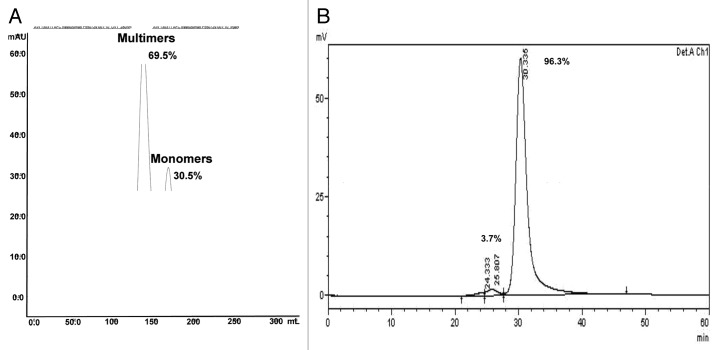
Figure 2. Purification of trastuzumab-mc_DSEA. (A) Preparative SEC. (B) SEC analysis of monomeric AFC.
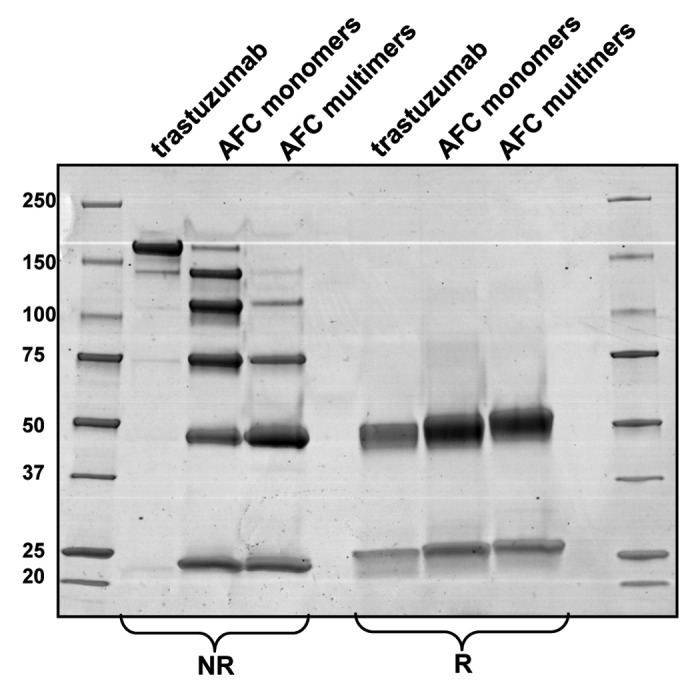
Figure 3. SDS-PAGE of the purified monomeric and multimeric forms of trastuzumab-mc_DSEA in non-reducing (NR) and reducing (R) conditions.
Figure 4. CE-SDS-PAGE of the purified (A) monomeric and (B) multimeric forms of trastuzumab-mc_DSEA in non-reducing (NR) and reducing (R) conditions.
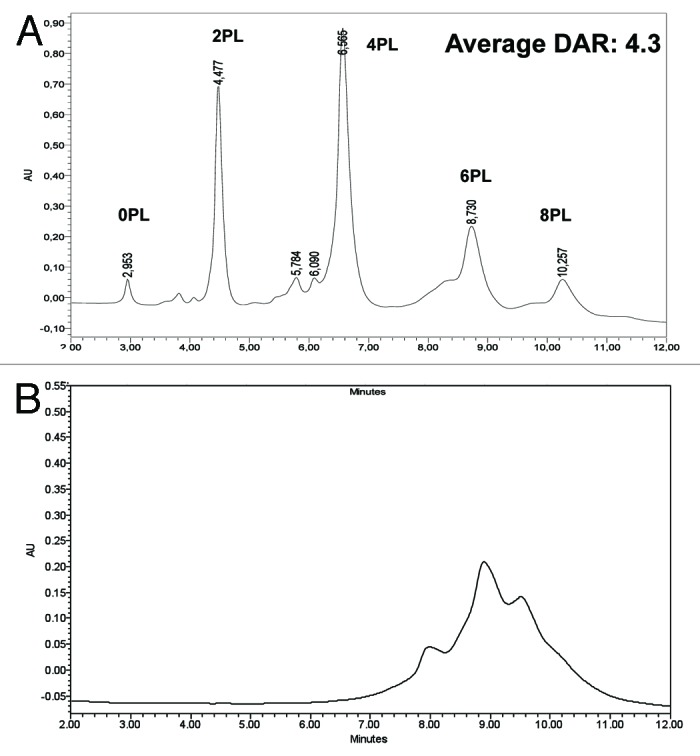
Figure 5. Drug loading distribution and average DAR calculation by HIC. (A) Monomeric trastuzumab-mc_DSEA. (B) Multimeric trastuzumab-mc_DSEA. Average DAR is calculated by quantifying the various loaded forms based on the peak areas (A) of the UV chromatogram at 210 nm.
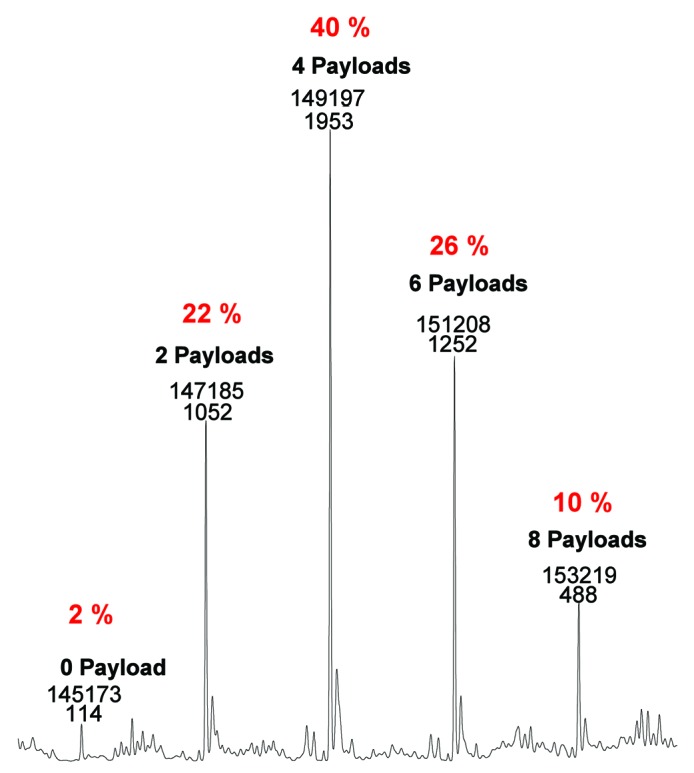
Figure 6. Drug loading distribution and average DAR calculation by native mass spectrometry.
Subsequently, digestion of the AFC by immunoglobulin-degrading enzyme of Streptococcus pyogenes (IdeS), followed by LC-MS was investigated. IdeS specifically cleaves immunoglobulin G under its hinge domain.24-26 In his 2012 published notes, Chevreux et al. described the advantages of IdeS proteolytic digestion prior to LC-MS of recombinant mAbs.27 Under reducing conditions, IdeS digestion of mAbs results in three polypeptide chains of around 25 kDa each. With minimal sample preparation, and within a single shot analysis, the method provides efficient LC and MS resolution that potentially results in relevant information on N-glycan profiling, charge state variants such as C-terminal lysine truncation, pyroglutamylation, oxidation and product cleavages.
The use of IdeS is becoming increasingly popular for the fast characterization of antibody by mass spectrometry,27,28 including correct sequence assessment,29,30 antibody Fab and Fc glyco-profiling,31 biosimilar comparability studies and Fc-fusion protein studies.32 Here, we report and discuss the potential of IdeS for ADC and AFC fast characterization using the trastuzumab-mc-DSEA conjugate and allowing glycoprofilling which may be important to retains the effector function of the naked parent antibody.33
Results
The raw trastuzumab-mc_DSEA analyzed by SEC (Fig. 2A) showed the presence of two main peaks interpreted as multimeric (69.5%) and monomeric species (30.5%). Both populations were separated by a preparative SEC and further characterized by SDS-PAGE (Fig. 3), CE-SDS (Fig. 4), HIC (Fig. 5), native mass spectrometry (Fig. 6), and liquid chromatography coupled to electrospray time of flight mass spectrometry MS (LC-ESI-TOF) after IdeS digestion and reduction. We investigated the structural characterization of both purified populations to establish a potential relationship between aggregation of the AFC and its average dye-to-antibody ratio (DAR). The SEC chromatogram of the purified monomeric products performed at an analytical scale is presented in Figure 2B and shows a main peak of monomers at 30.3 min (96.3%) and dimers (3.7%).
Monomeric and multimeric fractions of the AFC vs. non-conjugated trastuzumab were analyzed by SDS-PAGE under non-reducing and reducing conditions (Fig. 3). Unreduced trastuzumab displays a main band at ~150 kDa that corresponds to the H2L2 form and another lighter band of lower molecular weight likely to correspond to the molecule lacking one light chain, which is commonly described as a result of the production process. The pattern of the non-reduced monomeric AFC fraction is distributed as following: six bands of apparent molecular weight around 150, 125, 100, 75, 50, and 25 kDa, which fit to H2L2 and H2L, H2, HL, H and L, respectively, each carrying payloads. Indeed, because some of the interchain disulfide bridges are disrupted by the conjugation, the structure of the typical heterodimeric mAb (H2L2) is no longer maintained in the presence of SDS.
The profile of the non-reduced multimeric AFC fraction looks different in its distribution because only H2, HL, H, L plus payloads are present. The intensity of each band is also differently distributed with less intense H2 and HL bands with respect to H and L. Altogether, these results highlight a different payload distribution between monomeric and multimeric fractions, with a higher level of conjugation for the multimeric AFC. The SDS-PAGE pattern under reducing conditions confirms these observations. Indeed, the apparent molecular weight of the light and heavy chains increases between unconjugated trastuzumab, monomeric AFC and multimeric AFC, respectively. The heavy chain, which has three cysteines capable of forming interchain disulfides, may carry up to three payloads, resulting in an increasing distribution in our case. On the other hand, the light chain may carry a maximum of one payload because it only has one interchain disulfide-forming cysteine. This does not fit with the slightly elevated molecular weight observed for the light chain of the multimeric fraction compared with the monomeric one, and may be the result of over-payload conjugation. This will be discussed in the following sections.
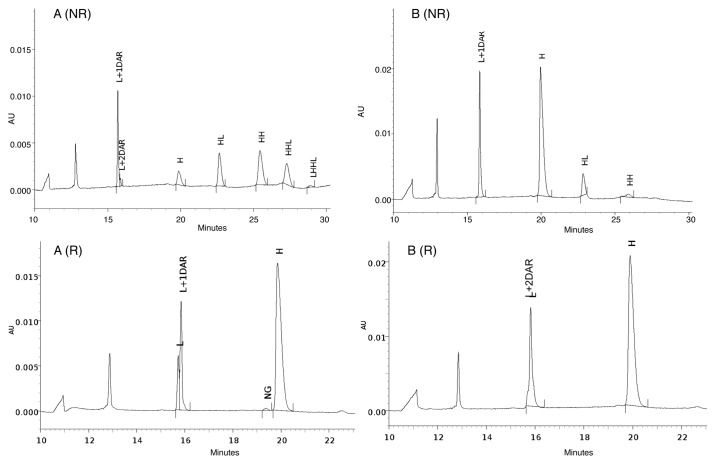
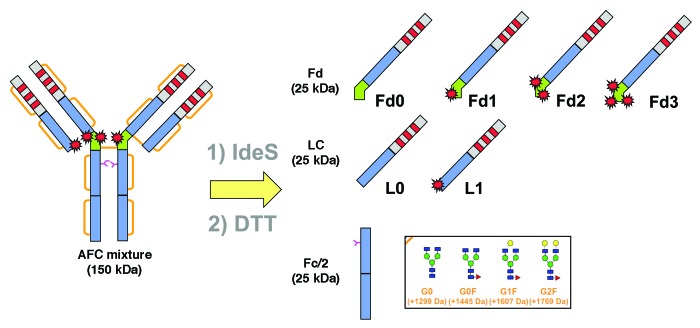
IdeS digestion of an ADC/AFC followed by a reduction step can generate seven fragments as illustrated in Figure 7. Fd and LC exist as naked or conjugated forms carrying up to 3 payloads (depending on the number of conjugated interchain disulfide-forming cysteines) plus Fc/2, which is theoretically free of payload and carrying the glycosylation heterogeneity. Figure 8 shows the UV chromatogram at 280 nm resulting from the elution of trastuzumab that was digested with IdeS then reduced. The three resulting fragments were well-resolved and identified according to their MS profile (Fig. 8B‒D; Table 1). The first eluting peak at 11.3 min corresponds to the Fc/2 fragments reflecting the pattern of trastuzumab being N-glycosylated by biantennary glycans. Mainly agalactosylated fucosylated (G0F) and monogalactosylated fucosylated (G1F) species were detected (at 25 237 and 25 399 Da respectively), whereas only a small amount of afucosylated (G0) and bigalactosylated fucosylated (G2F) forms were found (low intensity signals at 25 091 and 25 560 Da). This is as expected for an antibody produced in a CHO cell line. Eluting at 13.6 min and 17.5 min respectively, we identified LC and Fd fragments with low-intensity Na adducts and loss of water molecule for the Fd fragment.
Figure 7. Generation of seven fragments of near 25 kDa after IdeS digestion and DTT reduction of trastuzumab-mc_DSEA.
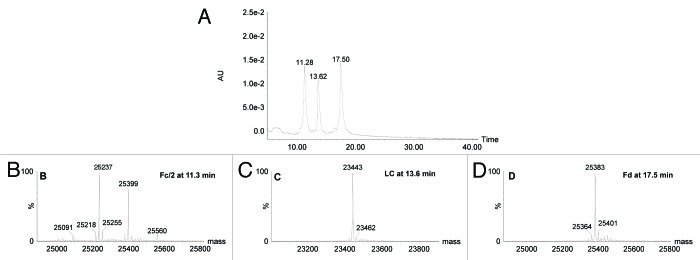
Figure 8. LC-UV-MS of trastuzumab after IdeS digestion and reduction. (A) UV chromatogram at 280 nm. (B‒D) mass spectra of peaks eluting at 11.3, 13.6 and 17.5 min, respectively.
Table 1. Mass assignment of species eluting in the chromatogram presented in Figure 8.
| Time (min) | Assignment | Theoretical masses (Da) | Experimental masses (Da) |
|---|---|---|---|
| 11.3 | Fc/2-G0 | 25 086 | 25 091 |
| Fc/2-G0F(-H2O) | 25 214 | 25 218 | |
| Fc/2-G0F | 25 232 | 25 237 | |
| Fc/2-G0F(+Na) | 25 254 | 25 255 | |
| Fc/2-G1F | 25 394 | 25 399 | |
| Fc/2-G2F | 25 556 | 25 560 | |
| 13.6 | LC | 23 439 | 23 443 |
| LC(+Na) | 23 461 | 23 462 | |
| 17.5 | Fd (-H2O) | 25 366 | 25 364 |
| Fd | 25 384 | 25 383 | |
| Fd(+Na) | 25 406 | 25 401 |
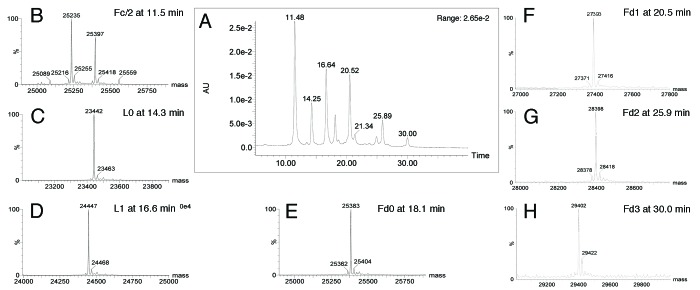
Figure 9A depicts the UV chromatogram at 280 nm resulting from the elution of trastuzumab-mc_DSEA digested with IdeS then reduced. Seven species were resolved and identified by the mass measurement (Fig. 9B‒H; Table 2). As previously described for unconjugated trastuzumab, the Fc/2 fragments elute at 11.5 min with their typical glycoform distribution. Eluting at 14.3 and 16.6 min, within two completely resolved chromatographic peaks, we observed L0 and L1 differing from each other by a mass increment of 1,005 Da. This corresponds to the conjugation of one mc-DSEA payload to the cysteine residue normally responsible for the formation of an inter-chain disulfide with the trastuzumab heavy chain. In a similar manner, we detected the Fd fragment from IdeS digestion coupled with 0, 1, 2, or 3 payloads and eluting at 18.1, 20.5, 25.9, and 30 min. The mass spectra look quite homogeneous, with low-intensity Na adducts (+21 Da) and some loss of a water molecule. The number of additional payloads detected on the Fd fragment fits with the number of possible free thiol residues oxidized during preparation of the AFC (one cysteine linking the LC and two cysteines in the hinge area). In our chromatographic conditions, Fd1 and Fd2 are each split in two UV peaks sharing the same mass, which can be interpreted as positional isomers.
Figure 9. LC-UV-MS of monomeric trastuzumab-mc_DSEA after IdeS digestion and reduction. (A) UV chromatogram at 280 nm. (B‒H) Mass spectra of peaks eluting at 11.5, 14.3, 16.6, 18.1, 20.5, 25.9 and 30 min, respectively.
Table 2. Mass assignment of species eluting in the chromatogram presented in Figure 9.
| Time (min) | Assignment | Theoretical masses (Da) | Experimental masses (Da) |
|---|---|---|---|
| 11.5 | Fc/2-G0 | 25 086 | 25 089 |
| Fc/2-G0F(-H2O) | 25 214 | 25 216 | |
| Fc/2-G0F | 25 232 | 25 235 | |
| Fc/2-G0F(+Na) | 25 254 | 25 255 | |
| Fc/2-G1F | 25 394 | 25 397 | |
| Fc/2-G1F(+Na) | 25 416 | 25 418 | |
| Fc/2-G2F | 25 556 | 25 559 | |
| 14.3 | L0 | 23 439 | 23 442 |
| L0(+Na) | 23 461 | 23 463 | |
| 16.6 | L1 | 24 444 | 24 447 |
| L1(+Na) | 24 466 | 24 468 | |
| 18.1 | Fd0 (-H2O) | 25 366 | 25 362 |
| Fd0 | 25 384 | 25 383 | |
| Fd0(+Na) | 25 406 | 25 404 | |
| 20.5 | Fd1(-H2O) | 26 371 | 26 368 |
| Fd1 | 26 389 | 26 388 | |
| Fd1(+Na) | 26 411 | 26 408 | |
| 25.9 | Fd2(-H2O) | 27 376 | 27 373 |
| Fd2 | 27 394 | 27 392 | |
| Fd2(+Na) | 27 416 | 27 413 | |
| 30.0 | Fd3(-H2O) | 28 379 | 28 378 |
| Fd3 | 28 397 | 28 397 | |
| Fd3(+Na) | 28 419 | 28 418 | |
| Fd3(+2Na) | 28 441 | 28 442 |
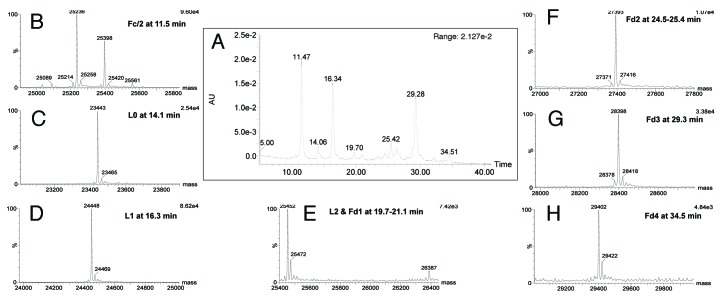
A similar LC-MS experiment was performed on the IdeS-digested, reduced multimeric trastuzumab-mc_DSEA. Chromatogram and mass spectra are presented in Figure 10 (see also Table 3). The resulting profile looks qualitatively and quantitatively different than the previously studied monomeric fraction. Indeed, co-eluting together with the Fd1 fragment between 19.7 and 21.1 min, we detect some light chain carrying two payloads (L2) with a mass of 25 452 Da. An additional species at 34.5 min corresponds to Fd4 (29 402 Da). As predicted by the SDS-PAGE results, the multimeric fraction seems to contain highly conjugated AFC with an unexpected payload distribution.
Figure 10. LC-UV-MS of multimeric trastuzumab-mc_DSEA after IdeS digestion and reduction. (A) UV chromatogram at 280 nm. (B‒H) Nass spectra of peaks eluting at 11.5, 14.1, 16.3, 19.7–21.1, 24.5–25.4, 29.3 and 34.5 min, respectively.
Table 3. Mass assignment of species eluting in the chromatogram presented in Figure 10.
| Time (min) | Assignment | Theoretical masses (Da) | Experimental masses (Da) |
|---|---|---|---|
| 11.5 | Fc/2-G0 | 25 086 | 25 089 |
| Fc/2-G0F(-H2O) | 25 214 | 25 214 | |
| Fc/2-G0F | 25 232 | 25 236 | |
| Fc/2-G0F(+Na) | 25 254 | 25 258 | |
| Fc/2-G1F | 25 394 | 25 398 | |
| Fc/2-G1F(+Na) | 25 416 | 25 420 | |
| Fc/2-G2F | 25 556 | 25 561 | |
| 14.1 | L0 | 23 439 | 23 443 |
| L0(+Na) | 23 461 | 23 465 | |
| 16.3 | L1 | 24 444 | 24 448 |
| L1(+Na) | 24 466 | 24 469 | |
| 19.7–21.1 | L2 | 25 453 | 25 452 |
| L2(+Na) | 25 474 | 25 472 | |
| Fd1 | 26 389 | 26 387 | |
| 24.4–25.4 | Fd2(-H2O) | 27 376 | 27 371 |
| Fd2 | 27 394 | 27 393 | |
| Fd2(+Na) | 27 416 | 27 416 | |
| 29.3 |
Fd3(-H2O) Fd3 Fd3(+Na) |
28 380 28 397 28 419 |
28 378 28 398 28 418 |
| 30.0 | Fd4 | 29 404 | 29 402 |
| Fd4(+Na) | 29 426 | 29 422 |
To confirm this assumption, we further investigated the payload distribution, the results of which are presented in Figure 11. By analysis of the peak areas after integration, we determined the percentage of L0 and the conjugated forms for the light chain, fragments, as well as conjugation of the Fd region (Fd0, Fd1, Fd2, Fd3 and Fd4). The highest loaded fragments were found in higher concentration in the multimeric fraction compared with the monomeric fraction. Correspondingly, the opposite was true for the less conjugated forms. Thus, whereas L0 and L1 represent 32% and 68% of the light chain in the monomeric fraction, respectively, they are 12% and 75% in the multimeric fraction, with 13% of L2 present. Similarly, Fd forms are represented as following in the monomeric fraction: Fd0 (16%), Fd1 (53%), Fd2 (26%), and Fd3 (5%), whereas we calculated Fd1 (3%), Fd2 (16%), Fd3 (72%) and Fd4 (9%) for the multimeric one.
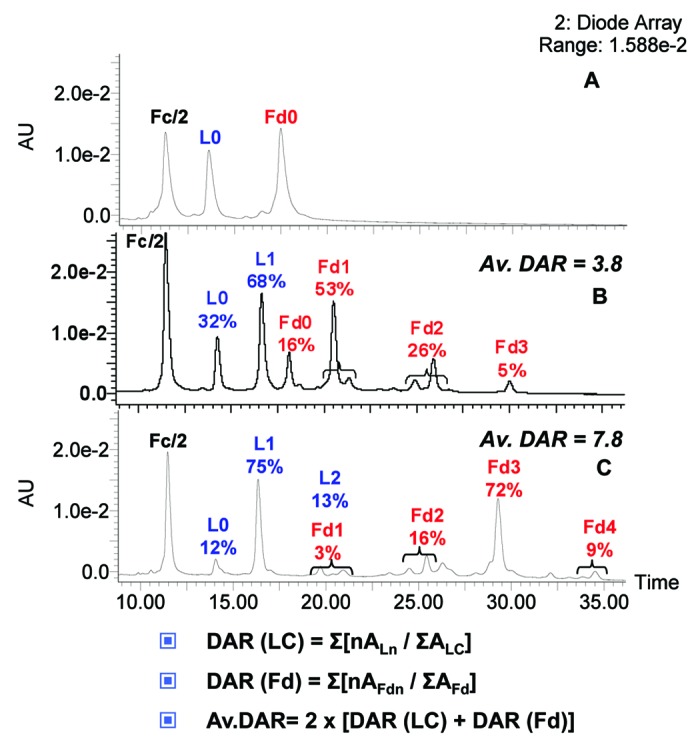
Figure 11. Drug loading distribution and average DAR calculation. (A) Unconjugated trastuzumab. (B) Monomeric. (C) Multimeric trastuzumab-mc_DSEA, respectively. Average DAR is calculated by quantifying the various loaded forms based on the peak areas (A) of the UV chromatogram at 280 nm.
Average DAR was estimated from the latter calculated distributions as described in Figure 9. An average value of 3.8 payloads per antibody was determined for monomeric AFC whereas the average for multimeric AFC was 7.8.
Discussion
Broadening the application to mAbs first described by Chevreux et al.,27 the use of IdeS for characterization of antibody conjugates by LC-MS has been described. The method has all the advantages as for unconjugated mAbs, with the added bonus of access to supplementary structural information specific to AFCs or ADCs. Within a simple and fast sample preparation, the method generates fragments of reasonable size (25 kDa). They are easily ionized and analyzed by ESI-TOF-MS, and in a better manner than for the heavy chain resulting from a single reduction process, for example. In addition, the approach can also monitor variants such as C-terminal lysine truncation, pyroglutamylation, oxidation and degradation products. Whereas LC and Fd fragments bear information related to conjugation and thus to ADC loading, Fc/2 affords accurate profiling of N-glycosylation, which is important because an ADC keeps the same effector functions as the naked antibody, as reported for trastuzumab and ado-trastuzumab emtansine.34 On the other hand, the LC-MS profile of the Fc/2 fragment should be unchanged throughout the ADC production process and could therefore be used as an internal reference, since the various loaded forms of Fd and LC fragments can be quantified relative to the proportion of Fc/2.
As depicted in the chromatographic profiles of our IdeS-reduced AFC, Fd1 and Fd2 split into two peaks. Indeed, the process of conjugation yields a controlled but heterogeneous population in terms of the number of payloads linked and the position of the linkage on the interchain disulfide cysteine residues. Under the reducing conditions of the conjugation reaction, only the Fd fragment carrying three of those cysteines may exhibit such positional isomers. In a remarkable manner, Le et al. developed a mathematical approach to quantify the proportion of each, and validated their systems by crossing calculated values with experimental results for several antibodies conjugated to vcMMAE.20 They showed that whatever the average DAR (ranking from 2.0 to 5.5), the most represented ADC isomers were so-called DAR2f, DAR4ff and DAR6fhh, where f and h are conjugation sites (f refers to the disulfide bridge between light and heavy chain and h to one in the hinge region). From these observations, we may assume that the most represented isomers in our chromatogram correspond to Fd1f and Fd2fh, whereas smaller peaks would be the Fd1h and Fd2hh isomers. These observations are also in agreement with data reported by H. Liu and colleagues on the ranking of the susceptibility of disulfide bonds in human IgG1 antibodies.34
Finally, the method can be applied routinely to the determination of payload distribution and calculation of average DAR, which has not been described until now. In our case, it allowed comparison between two SEC fractions purified from the same production batch targeting an average DAR of 4. We highlighted a very significant difference between both samples because the multimeric fraction consists of highly loaded forms (average DAR of 7.8), whereas the monomeric fraction more closely resembles the targeted distribution (average DAR of 3.8). In our case, there was a correlation between the number of conjugated payloads and the tendency for aggregation. We have also observed this for other mAbs and other payloads (data not shown). The hydrophobicity of the payload might also affect the ratio of resulting aggregates. By LC-MS analysis, we described fragments with more payloads than available interchain disulfide-forming cysteines (Fd4, L2), showing that the conjugation reaction may lead to uncontrolled alkylation. We presume that intrachain disulfide bridges were also reduced and conjugated during the reaction process, showing that the mild reduction conditions are somehow difficult to control. An extensive analysis by LC-MS peptide mapping is in progress to further investigate the position of payloads.
In summary, as for naked antibodies and Fc-fusion proteins, IdeS proteolytic digestion may rapidly become a reference analytical method at all stages of ADC discovery, preclinical and clinical development. The method can be routinely used for comparability assays, formulation, process scale-up and transfer of ADCs, and to define critical quality attributes in a quality-by-design approach.
Material and Methods
Antibody and linker-payload production and purification
The trastuzumab used in this study is the European Medicines Agency-approved version and formulation (21 mg/mL). The linker-fluorophore payload was designed to mimic the linker-drug most frequently used in ADC clinical trials. The synthesis is briefly reported in the supplemental material. It consists of maleimide-caproic acid dansyl sulfonamide ethyl amine (mc_DSEA, see structure Fig. 1) with a valine-citruline linker that mimics the cytotoxic agent and linker conjugated to mAbs through reduced interchain cysteine via the maleimide function.
Mild reduction of trastuzumab and coupling of DSEA-linker were performed as previously described.19 Briefly, trastuzumab was reduced with 2.75 equivalents of TCEP in 10 mM borate pH 8.4 buffer containing 150 mM NaCl and 2 mM EDTA for 2 h at 37 °C. The concentration of free thiols was determined by using the Ellman’s reagent with l-cysteine as standard, typically resulting in around 5 thiols per antibody. To target a DAR of 4, the partially reduced trastuzumab was then alkylated with 2 equivalents of DSEA-linker per thiol in the same buffer for 1 h at room temperature. N-acetyl-cysteine (1.5 equivalents / DSEA-linker) was used to quench any unreacted DSEA-linker. The AFC was purified by size exclusion chromatography on a Superdex 200 pg column (GE Life Sciences) eluted with 25 mM histidine pH 6.5 buffer containing 150 mM NaCl, by using an AKTA Avant biochromatography system (GE Life Sciences). Fractions corresponding to AFC monomers and multimers were collected and concentrated. The protein concentration of the monomeric and multimeric AFC solutions was determined by BCA with bovine immunoglobulins as standard. The labeled antibodies were further analyzed by SDS-PAGE on a 4–15% gradient gel (BioRad) under reducing and non-reducing conditions. The gel was stained with Coomassie blue. For analytical SEC, an Agilent HPLC system (Les Ulis) was used. Samples were analyzed on a Superdex 200 GL column (10 × 300 mm, GE Life Sciences) at room temperature. They were eluted at a flow rate of 0.4 ml/min with 25 mM histidine pH 6.5 buffer containing 150 mM NaCl as mobile phase. The elution was monitored at 280 nm.
CE-SDS characterization
The buffer of 100 µg of ADC sample was exchanged with SDS-MW sample buffer by using the Microcon YM30 centrifuge filter unit to have a final volume at 95 µL. Two µL of internal standard 10kDa were added with 5 µL of 250 mM iodoacetamide for non-reduced sample or 5 µL of 2-mercaptoethanol for reduced sample. All tubes were heated at 70 °C for 10 min and centrifuged at 12000 g for 6 min. Samples were then analyzed on ProteomeLab PA800 (Beckman) equipped with a UV detector. The detection was conducted at 220 nm
HIC characterization
HIC was performed using a liquid chromatography system (Alliance HPLC) coupled to a detector UV (2489 dual absorbance). Analyses were realized on a TSKButyl-NPR, 2.1 x 4.6 mm column (Tosoh bioscience) set at room temperature. The mobile phase A consisted of 1.5 M ammonium sulfate, 25 mM potassium phosphate pH7.0, and the mobile phase B consisted of a mixture 25 mM potassium phosphate and 25% isopropanol at pH7. Separation was obtained with a linear gradient of 10–100% B over 12 min at flow rate of 0.8 mL/min. Analysis were obtained by injecting 20 µg of sample diluted at 1 mL/min with mobile phase A and integrating the UV area at 210 nm for each species.
Native mass spectrometry characterization
Prior to native mass spectrometry experiments, an aliquot of 300 µg monomeric trastuzumab-mc_DSEA was deglycosylated by adding 0.6 µL PNGaseF (New England Biolabs) followed by incubation overnight at 37 °C. Thirty µg of deglycosylated sample were injected on a PolyHYDROXYETHYL A™ column (PolyLC), 150 × 1 mm, 5 µm, 300 A°. Elution from the column was performed within a 6 min isocratic step at 0.1 mL/min with 200 mM ammonium acetate solution buffered at pH7. During sample elution, chromatography was coupled online with an electrospray time-of-flight mass spectrometer (LCT PremierTM, Waters) operating in V positive ion mode with a capillary voltage of 3000 V and a sample cone voltage of 40 V. Acquisitions were performed on the mass range m/z 1000–12000 with 1 s scan time. Native desalting conditions maintain the intact bivalent structure of the AFC. Well-resolved peaks were sufficient for confirmation of the identity of the AFC (0, 2, 4, 6, 8 payloads) and to determine the relative distribution of the drug-loaded species. The average DAR deduced from those measurements is 4.4.
IdeS digestion
Twenty UI of IdeS enzyme were added to 20 µg of AFC solution following the instructions of the enzyme kit (FabRICATOR Kit, Genovis). The mixture was then incubated at 37 °C for 30 min. Before reduction, the digested sample is diluted twice with a buffer containing 6 M guanidine-HCl, 2 mM EDTA, TRIS-HCl 0.1 M. DTT is added to reach a final concentration of 10 mM. After 45 min incubation at 56 °C the reaction is quenched with 1 µL acetic acid.
LC-MS analysis
Reverse-phase high performance liquid chromatography (RP-HPLC) was performed using an ultrahigh-performance liquid chromatography system (Acquity UPLC, Waters) coupled to an electrospray time-of-flight mass spectrometer (LCT PremierTM, Waters). A volume equivalent to 8 µg of sample preparation was injected on a PLRP-S 1000A°, 8µm, 150 × 2.1 mm column (Agilent) set at 80 °C. The gradient was generated at a flow rate of 0.25 mL/min using 0.05% trifluoroacetic acid (TFA) for mobile phase A and acetonitrile containing 0.05% TFA for mobile phase B. B was raised from 5% to 30% in 8 min and to 50% in an additional 40 min followed by a 2 min washing step at 95% B and a 10 min reequilibration period. Elution was monitored spectrophotometrically at 280 nm. The LCT Premier was operated in W positive ion mode with a capillary voltage of 3000V and a sample cone voltage of 120 V. Acquisitions were performed on the mass range m/z 1000–4000 with a 1 s scan time. Calibration was performed using the singly charged ions produced by a 2 mg/L solution of cesium iodide in 2-propanol/water (1/1). Data analysis was performed with MassLynx 4.0 (Waters).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Laura Morel-Chevillet, Anne Viaud, and Anne Humbert for their technical support, Thierry Champion for structure modeling, Amandine Boeuf and Christine Klinguer-Hamour for helpful discussions, and Claire Catry for her assistance.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/mabs/article/26773
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/26773
References
- 1.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–52. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 2.Beck A, Senter P, Chari R. World Antibody Drug Conjugate Summit Europe: February 21-23, 2011; Frankfurt, Germany. MAbs. 2011;3:331–7. doi: 10.4161/mabs.3.4.16612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck A, Lambert J, Sun M, Lin K. Fourth World Antibody-Drug Conjugate Summit: February 29-March 1, 2012, Frankfurt, Germany. MAbs. 2012;4:637–47. doi: 10.4161/mabs.21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullard A. Maturing antibody-drug conjugate pipeline hits 30. Nat Rev Drug Discov. 2013;12:329–32. doi: 10.1038/nrd4009. [DOI] [PubMed] [Google Scholar]
- 5.Beck A, Carter PJ, Gerber HP, Lugovskoy AA, Wurch T, Junutula JR, et al. 8 (th) Annual European Antibody Congress 2012: November 27-28, 2012, Geneva, Switzerland. MAbs. 2013; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck A, Haeuw JF, Wurch T, Goetsch L, Bailly C, Corvaïa N. The next generation of antibody-drug conjugates comes of age. Discov Med. 2010;10:329–39. [PubMed] [Google Scholar]
- 7.Alley SC, Anderson KE. Analytical and bioanalytical technologies for characterizing antibody-drug conjugates. Curr Opin Chem Biol. 2013;17:406–11. doi: 10.1016/j.cbpa.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Beck A, Wagner-Rousset E, Bussat MC, Lokteff M, Klinguer-Hamour C, Haeuw JF, Goetsch L, Wurch T, Van Dorsselaer A, Corvaïa N. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol. 2008;9:482–501. doi: 10.2174/138920108786786411. [DOI] [PubMed] [Google Scholar]
- 9.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26:925–32. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 10.Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30:184–9. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 11.Jeffrey SC, Burke PJ, Lyon RP, Meyer DW, Sussman D, Anderson M, Hunter JH, Leiske CI, Miyamoto JB, Nicholas ND, et al. A potent anti-CD70 antibody-drug conjugate combining a dimeric pyrrolobenzodiazepine drug with site-specific conjugation technology. Bioconjug Chem. 2013;24:1256–63. doi: 10.1021/bc400217g. [DOI] [PubMed] [Google Scholar]
- 12.Kung Sutherland MS, Walter RB, Jeffrey SC, Burke PJ, Yu C, Kostner H, Stone I, Ryan MC, Sussman D, Lyon RP, et al. SGN-CD33A: a novel CD33-targeting antibody-drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood. 2013;122:1455–63. doi: 10.1182/blood-2013-03-491506. [DOI] [PubMed] [Google Scholar]
- 13.Wakankar A, Chen Y, Gokarn Y, Jacobson FS. Analytical methods for physicochemical characterization of antibody drug conjugates. MAbs. 2011;3:161–72. doi: 10.4161/mabs.3.2.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 2012;30:184–9. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 15.Strop P, Liu SH, Dorywalska M, Delaria K, Dushin RG, Tran TT, Ho WH, Farias S, Casas MG, Abdiche Y, et al. Location matters: site of conjugation modulates stability and pharmacokinetics of antibody drug conjugates. Chem Biol. 2013;20:161–7. doi: 10.1016/j.chembiol.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Acchione M, Kwon H, Jochheim CM, Atkins WM. Impact of linker and conjugation chemistry on antigen binding, Fc receptor binding and thermal stability of model antibody-drug conjugates. MAbs. 2012;4:362–72. doi: 10.4161/mabs.19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, Kazane SA, Halder R, Forsyth JS, Santidrian AF, Stafin K, et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci U S A. 2012;109:16101–6. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gianolio DA, Rouleau C, Bauta WE, Lovett D, Cantrell WR, Jr., Recio A, 3rd, Wolstenholme-Hogg P, Busch M, Pan P, Stefano JE, et al. Targeting HER2-positive cancer with dolastatin 15 derivatives conjugated to trastuzumab, novel antibody-drug conjugates. Cancer Chemother Pharmacol. 2012;70:439–49. doi: 10.1007/s00280-012-1925-8. [DOI] [PubMed] [Google Scholar]
- 19.Sun MM, Beam KS, Cerveny CG, Hamblett KJ, Blackmore RS, Torgov MY, Handley FG, Ihle NC, Senter PD, Alley SC. Reduction-alkylation strategies for the modification of specific monoclonal antibody disulfides. Bioconjug Chem. 2005;16:1282–90. doi: 10.1021/bc050201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le LN, Moore JM, Ouyang J, Chen X, Nguyen MD, Galush WJ. Profiling antibody drug conjugate positional isomers: a system-of-equations approach. Anal Chem. 2012;84:7479–86. doi: 10.1021/ac301568f. [DOI] [PubMed] [Google Scholar]
- 21.Valliere-Douglass JF, McFee WA, Salas-Solano O. Native intact mass determination of antibodies conjugated with monomethyl Auristatin E and F at interchain cysteine residues. Anal Chem. 2012;84:2843–9. doi: 10.1021/ac203346c. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Yin S, Wu Y, Ouyang J. Development of a native nanoelectrospray mass spectrometry method for determination of the drug-to-antibody ratio of antibody-drug conjugates. Anal Chem. 2013;85:1699–704. doi: 10.1021/ac302959p. [DOI] [PubMed] [Google Scholar]
- 23.Rosati S, van den Bremer ET, Schuurman J, Parren PW, Kamerling JP, Heck AJ. In-depth qualitative and quantitative analysis of composite glycosylation profiles and other micro-heterogeneity on intact monoclonal antibodies by high-resolution native mass spectrometry using a modified Orbitrap. MAbs. 2013;5:915–22. doi: 10.4161/mabs.26282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Pawel-Rammingen U, Johansson BP, Björck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 2002;21:1607–15. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wenig K, Chatwell L, von Pawel-Rammingen U, Björck L, Huber R, Sondermann P. Structure of the streptococcal endopeptidase IdeS, a cysteine proteinase with strict specificity for IgG. Proc Natl Acad Sci U S A. 2004;101:17371–6. doi: 10.1073/pnas.0407965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincents B, von Pawel-Rammingen U, Björck L, Abrahamson M. Enzymatic characterization of the streptococcal endopeptidase, IdeS, reveals that it is a cysteine protease with strict specificity for IgG cleavage due to exosite binding. Biochemistry. 2004;43:15540–9. doi: 10.1021/bi048284d. [DOI] [PubMed] [Google Scholar]
- 27.Chevreux G, Tilly N, Bihoreau N. Fast analysis of recombinant monoclonal antibodies using IdeS proteolytic digestion and electrospray mass spectrometry. Anal Biochem. 2011;415:212–4. doi: 10.1016/j.ab.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 28.Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S. Characterization of therapeutic antibodies and related products. Anal Chem. 2013;85:715–36. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- 29.Beck A, Diemer H, Ayoub D, Debaene F, Wagner-Rousset E, Carapito C, et al. Analytical characterization of biosimilar antibodies and Fc-fusion proteins. TrAC Trends in Analytical Chemistry. 2013;48:81–95. doi: 10.1016/j.trac.2013.02.014. [DOI] [Google Scholar]
- 30.Ayoub D, Jabs W, Resemann A, Evers W, Evans C, Main L, Baessmann C, Wagner-Rousset E, Suckau D, Beck A. Correct primary structure assessment and extensive glyco-profiling of cetuximab by a combination of intact, middle-up, middle-down and bottom-up ESI and MALDI mass spectrometry techniques. MAbs. 2013;5:699–710. doi: 10.4161/mabs.25423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janin-Bussat MC, Tonini L, Huillet C, Colas O, Klinguer-Hamour C, Corvaïa N, et al. Cetuximab Fab and Fc N-Glycan Fast Characterization Using IdeS Digestion and Liquid Chromatography Coupled to Electrospray Ionization Mass Spectrometry. In: Beck A, editor. Glycosylation Engineering of Biopharmaceuticals. Springer, 2013. [DOI] [PubMed] [Google Scholar]
- 32.Lynaugh H, Li H, Gong B. Rapid Fc glycosylation analysis of Fc fusions with IdeS and liquid chromatography mass spectrometry. MAbs. 2013;5:641–5. doi: 10.4161/mabs.25302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junttila TT, Li G, Parsons K, Phillips GL, Sliwkowski MX. Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib insensitive breast cancer. Breast Cancer Res Treat. 2011;128:347–56. doi: 10.1007/s10549-010-1090-x. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Chumsae C, Gaza-Bulseco G, Hurkmans K, Radziejewski CH. Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem. 2010;82:5219–26. doi: 10.1021/ac100575n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.