Summary
The molecular mechanisms regulating olfactory receptor (OR) expression in the mammalian nose are not yet understood. Here, we identify the transient expression of histone demethylase LSD1, and the OR-dependent expression of Adenylyl Cyclase 3 (Adcy3) as requirements for initiation and stabilization of OR expression. As a transcriptional co-activator, LSD1 is necessary for de-silencing and initiating OR transcription, but as a transcriptional co-repressor, it is incompatible with maintenance of OR expression and its downregulation is imperative for stable OR choice. Adcy3, a sensor of OR expression and a transmitter of an OR-elicited feedback, mediates the downregulation of LSD1 and promotes the differentiation of olfactory sensory neurons (OSNs). This novel, three-node signaling cascade locks the epigenetic state of the chosen OR, stabilizes its singular expression, and prevents the transcriptional activation of additional OR alleles for the life of the neuron.
Introduction
Olfactory receptors (ORs) are G protein-coupled receptors that detect odors and regulate the projection of olfactory sensory neurons (OSNs) to the brain (Buck and Axel, 1991). In the mouse, ORs are encoded by ~1400 genes (Young et al., 2002) organized in gene clusters found on most chromosomes (Sullivan et al., 1996; Zhang and Firestein, 2002). OR genes have promoter elements that share similar regulatory sequences (Clowney et al., 2011; Michaloski et al., 2006) and are expressed in the main olfactory epithelium (MOE) in a monogenic and monoallelic fashion (Chess et al., 1994). An unusual, MOE-specific epigenetic signature of OR loci, characterized by enrichment for H3K9me3 and H4K20me3, likely governs what is a seemingly stochastic OR expression pattern (Magklara et al., 2011). This epigenetic silencing is reinforced by the aggregation of silenced OR genes in a few heterochromatic foci that preserve the expression of only one OR allele in each OSN (Clowney et al., 2012). The active OR allele in each OSN escapes from the OR aggregates and relocates to a euchromatic territory where it frequently interacts with a distant OR enhancer, the H enhancer (Clowney et al., 2012; Lomvardas et al., 2006). The singularity of OR expression is essential for olfactory perception because ORs are localized in the OSN dendrites and axons (Barnea et al., 2004) and participate in both odorant detection and axon targeting (Wang et al., 1998). Neurons expressing the same OR converge their axons in distinct glomeruli of the olfactory bulb (Mombaerts et al., 1996), by a process that relies on the identity of the OR protein and its basal activity levels (Mori and Sakano, 2011). Thus, maintaining the stable and singular expression of the same OR throughout the life of the neuron is necessary for the integrity of the topographic map in the olfactory bulb, such that coherent OR expression may be required for proper odor decoding in the brain.
Although the molecular mechanisms that stabilize OR expression are not known, it is established that the expression of transgenic ORs elicits a negative feedback that prevents the expression of endogenous OR genes (Lewcock and Reed, 2004; Nguyen et al., 2007; Serizawa et al., 2003). Moreover, lineage tracing experiments monitoring the expression of OR alleles from their endogenous loci showed that OR expression elicits a positive feedback signal that stabilizes its own choice and prevents OR gene switching in most OSNs (Shykind et al., 2004). Together, these observations raise questions regarding possible mechanisms that could stabilize the transcription of one OR allele, while simultaneously preventing the expression of all the other OR genes. A simple model that could account for the existence of an OR-elicited feedback signal emerged from the discovery that epigenetic silencing of ORs occurs prior to detectable OR transcription, and that OR choice coincides with a switch from the repressive histone H3K9 methylation to the activating H3K4 methylation (Magklara et al., 2011). Based on these observations, singular OR expression could become permanent if the choice of an intact OR allele suppresses H3K9 and H3K4 demethylases, so that one active OR allele and ~1000 silent OR genes preserve their distinct epigenetic states for the life of the neuron.
Demethylation of H3K9me3 and H3K4me3 are both stepwise processes that require removal of one methyl-group first, creating H3K9me2 and H3K4me2 respectively, which are then further demethylated by different enzymes. LSD1 (Lysine Specific Demethylase 1, Kdm1a), an amine oxidase, is the only protein with the enzymatic ability to catalyze lysine demethylation reactions for both intermediates, H3K9me2 and H3K4me2, and therefore act as transcriptional coactivator or corepressor, respectively (Metzger et al., 2005; Shi et al., 2004). The exact mode of action of LSD1 is influenced by the context of the transcription factor that recruits it to a specific locus and the nature of the local histone modifications. For example, LSD1 recruitment by androgen receptor to specific loci results in H3K9me2 demethylation and transcriptional activation, whereas LSD1 demethylates H3K4me2 and represses transcription as a component of the CoREST complex (Metzger et al., 2005; Shi et al., 2005; Wang et al., 2007; Wissmann et al., 2007).
Here, we show that LSD1, which is transiently expressed during OSN differentiation, is involved in both OR gene activation and post-choice gene switching, and, thus, plays a dual role in OR regulation. Genetic ablation of LSD1 activity prior to OR choice results in widespread loss of OR expression and failure of the OSNs to mature and to project axons to the brain. Deletion of LSD1 immediately after OR activation has no detectable consequences in OR expression and OSN targeting, suggesting LSD1 activity is needed only during the initial de-repression of the selected OR. OR expression induces the subsequent expression of Adenylyl Cyclase 3 (Adcy3), which promotes OSN maturation and LSD1 downregulation. Lineage tracing experiments reveal increased OR gene switching in Adcy3 KO mice, suggesting a requirement for timely LSD1 downregulation for the stabilization of OR expression. Ectopic expression of transgenic LSD1 in mature OSNs also perturbs the stability of OR choice, suggesting that Adcy3 stabilizes OR transcription by downregulating LSD1. Thus, our data connect OR choice with the terminal differentiation of olfactory neurons and uncover the molecular underpinnings of a feedback loop that preserves the epigenetic state of active and silent ORs for the life of the neuron.
Results
Dynamic LSD1 expression is required for initiation but not maintenance of OR expression
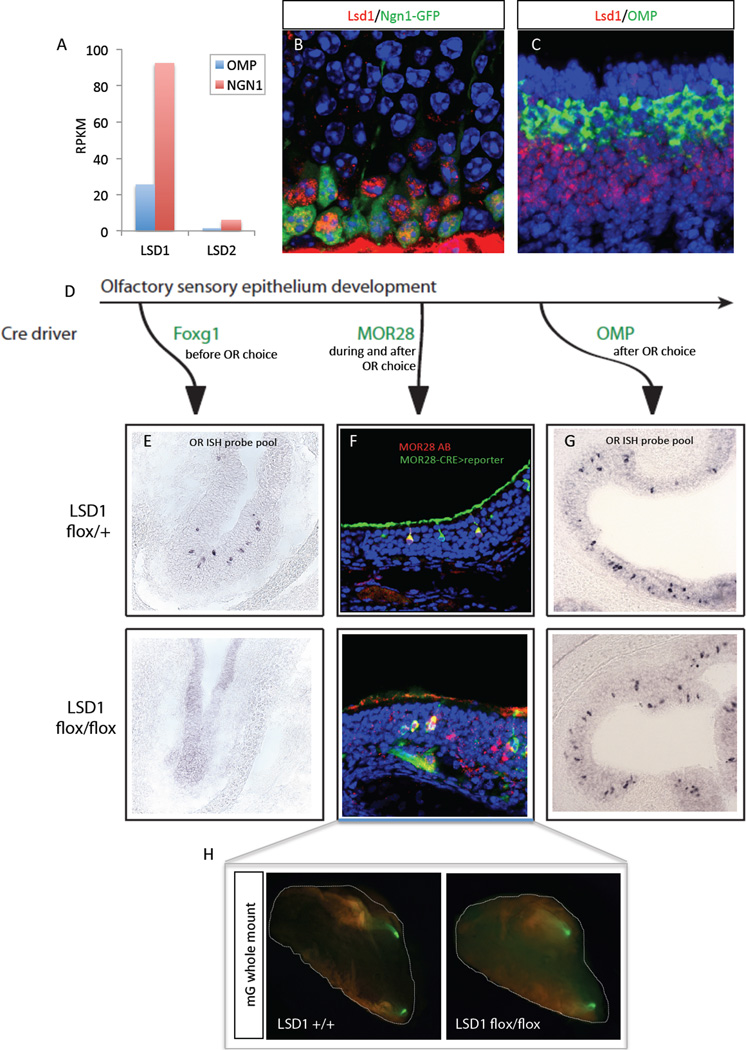
We hypothesized that initiation of OR transcription requires H3K9 demethylation, whereas stabilization of singular OR transcription requires suppression of H3K9 and H3K4 demethylases involved in OR regulation. Because LSD1 has both H3K9 and H3K4 demethylase activities, we asked whether its expression pattern is compatible with such a role in OR regulation. RNA-seq analysis (Magklara et al., 2011) from mature OSNs (mOSNs, Olfactory Marker Protein (OMP)-positive) and progenitor/immature neuronal populations (Neurogenin1 (Ngn1)-positive) shows that LSD1 is expressed in the Neurogenin-1 positive cells but reduced by 3.6 fold during differentiation to the OMP positive stage (Figure 1A). Immunofluorescence (IF) reveals high levels of LSD1 protein in the nuclei of Neurogenin-1 positive cells and their immediate progeny, and significant reduction of LSD1 in more apical, mOSN population of the adult MOE (Figure 1B). Two-color RNA FISH experiments demonstrate the mutually exclusive expression patterns of LSD1 and OMP, verifying the transcriptional downregulation of LSD1 in OR-expressing mOSNs (Figure 1C). The dynamic pattern of LSD1 expression, together with recent microarray analyses showing that injury-induced neurogenesis in the MOE coincides with LSD1 upregulation (Krolewski et al., 2013), support a role for this protein in OR regulation.
Figure 1.
Transient LSD1 expression is required for OR expression
(A) mRNA-seq reads per million mapped per thousand basepairs of exon model (RPKM) for LSD1 and LSD2 in the mature versus immature/globose basal cells (Ngn1+).
(B) LSD1 immunofluorescence (IF, red) in the Ngn1-GFP+ MOE at PND30.
(C) LSD1 and OMP 2-color RNA in situ hybridization (ISH) at PND5. DAPI nuclear stain is shown in blue.
(D) Removal of LSD1 over developmental time with 3 different MOE-specific Cre recombinase mouse lines.
(E) OR ISH probe pool for 8 Class II OR genes in Foxg1-cre; LSD1flox/+ and Foxg1;cre;flox/flox (Class I OR ISH is shown in Figure S1).
(F) MOR28-IRES-Cre mediated Cre reporter (green) in MOE with MOR28 immunofluorescence (red); coexpressing cells are stably expressing MOR28 in the absence of LSD1
(G) Class II OR ISH in OMP-IRES-Cre; LSD1 flox/+ and flox/flox MOE at PND1.
(H) Olfactory bulbs of MOR28-IRES-Cre; LSD1+/+ and MOR28-IRES-Cre; LSD1flox/flox animals at PND30 with a 2-color membrane-bound Cre-reporter: mT before Cre; mG after Cre (mT/mG; Muzumdar et al. 2007). See also Figure S1.
To functionally test the role of LSD1 in initiation and maintenance of OR gene expression, we used a conditional LSD1 KO (Wang et al., 2007) which we deleted at three distinct developmental time points: prior, during, and after OR gene activation, using Foxg1-Cre (Hebert and McConnell, 2000), MOR28-IRES-Cre (Shykind et al., 2004) and OMP-IRES-Cre (Eggan et al., 2004), respectively (Figure 1D). LSD1 deletion before OR expression results in widespread loss of OR expression, based on both ISH with a pool of OR RNA probes and IF with OR antibodies as well as a general targeting deficit of the OSN axons (Figure 1E and supplemental Figure S1A, B). This analysis was performed in E18.5 MOE sections due to perinatal lethality. In contrast to the early LSD1 KO, IF for MOR28 shows that LSD1 deletion immediately after MOR28 activation has no measurable effects on OR expression, or OSN targeting (Figure 1F-H and S1C). Similarly, RNA ISH and IF as above show that LSD1 deletion in mOSNs has no detectable effects on OR expression (Figure 1G and S1D). These data suggest that LSD1 activity is necessary for OR de-silencing and initiation of OR transcription but dispensable for OSN function following OR choice, at least within the kinetic restrictions imposed by available genetic strategies.
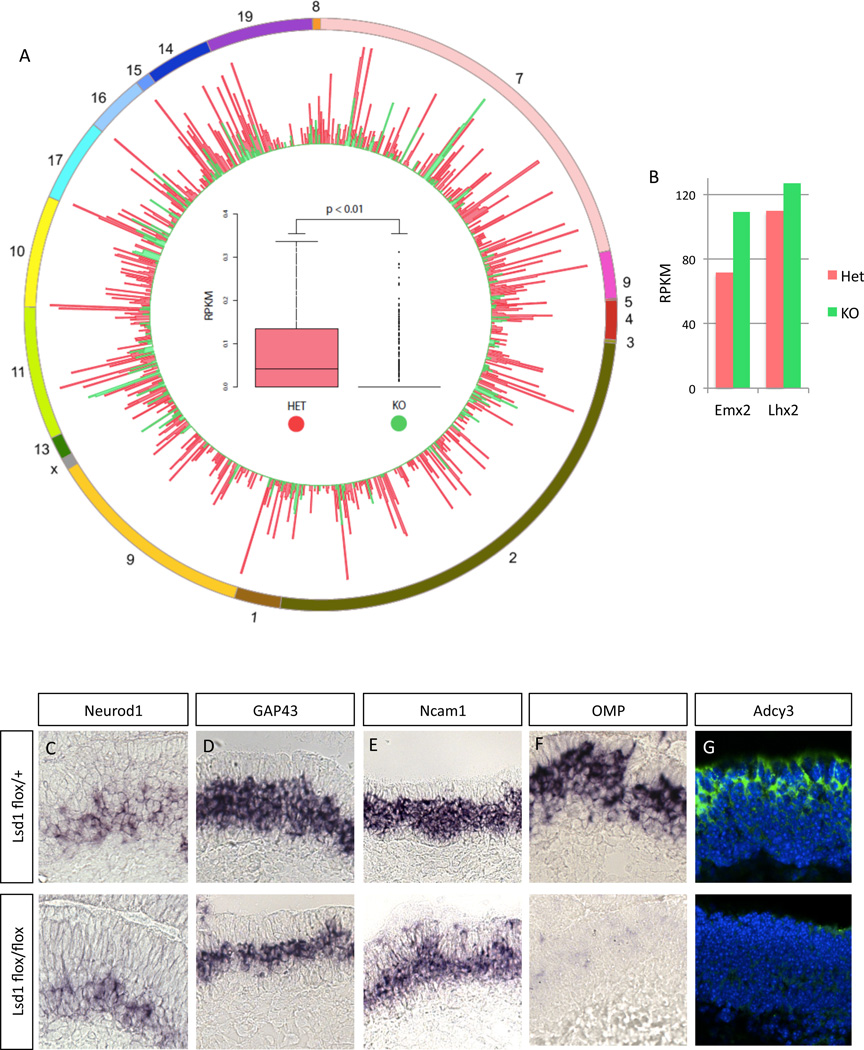
To determine the extent of the transcriptional effects on OR expression we performed RNA-seq analysis using cDNA libraries prepared from the MOE of control and LSD1 KO mice at E18.5. This approach also shows significant reduction of OR transcription, both regarding the total number of OR mRNA reads and the number of ORs that can be detected in the LSD1 KO mice (Figure 2A). In control mice we detect 662 Ors and in LSD1 KO mice we only detect 212 ORs with ~4-fold fewer reads in those ORs that are expressed in LSD1 KO MOEs. This analysis also revealed that transcription factors known or suspected to activate OR transcription, such as Emx2 and Lhx2 (Hirota and Mombaerts, 2004; McIntyre et al., 2008) are still expressed in the LSD1 KO (Figure 2B), supporting a direct role of LSD1 in OR regulation. Developmental markers of progenitor cells, such as Neurod1 are not affected by LSD1 deletion (Figure 2C). Importantly, developmental markers that are post-mitotic and synchronous to OR expression such as GAP43 and NCAM1, or Stmn1, Dpysl5, Marcksl1, and Ablim1 (Iwema and Schwob, 2003; Krolewski et al., 2013; Nickell et al., 2012), are also, only moderately affected by LSD1 deletion, based on our RNAseq and ISH experiments (Figure 2C and S2, respectively). This suggests that the loss of LSD1 activity and not some downstream developmental deficit is the cause of OR downregulation in the LSD1 KO MOE. In contrast, mOSN markers are markedly downregulated in the LSD1 KO MOEs (Figure 2F,G). The loss of the mOSN layer in the LSD1 KO likely explains the thinner expression pattern of GAP43 and NCAM1 and the ~1-fold reduction observed by RNAseq (Figure 2C and S2), since at this developmental stage there is some overlap of these immature markers with mOSN markers.
Figure 2.
Early deletion of LSD1 with Foxg1-Cre causes massive reduction in OR gene expression and developmental arrest at a differentiation stage synchronous to the onset of OR transcription.
(A) mRNA-seq RPKM for each Refseq OR in mouse genome from E18.5 MOE sample. Each spoke of a given color is the value for that OR in MOE of that genotype (red: Foxg1-Cre; LSD1flox/+; green: Foxg1-Cre; Lsd1flox/flox). External doughnut represents relative chromosomal location of each OR gene. Summary boxplot is shown within Circos plot; student’s paired t-test used for significance testing.
(B) RPKM values of the 2 known transcriptional activators of OR genes in the LSD1 heterozygote and knockout.
(C-F) Chromogenic ISH for developmental markers in LSD1 heterozygote (top panels) and knockout (bottom panels), respectively: Neurod1, GAP43, NCAM1, OMP.
(G) IF for Adcy3 at same embryonic stage, DAPI nuclear stain is shown in blue. See also Figure S2.
ORs induce Adcy3 expression
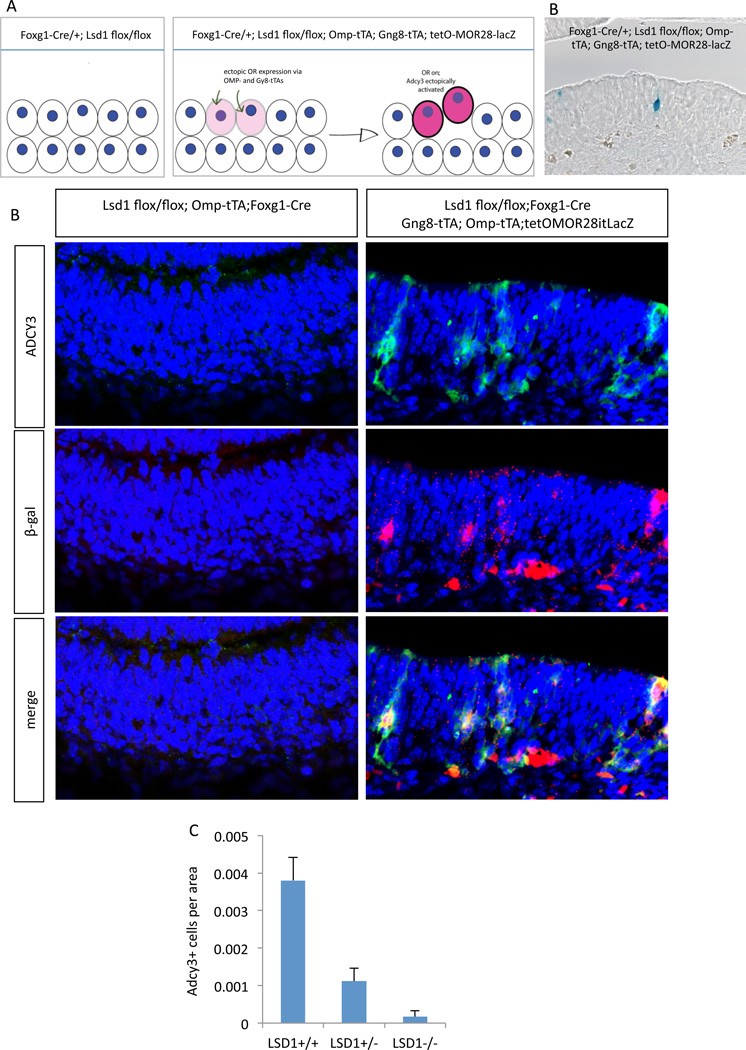
This developmental arrest invites the hypothesis that OR expression might be a prerequisite for OSN maturation. Consistent with this, an “empty” OSN that does not express OR protein, like ORNs generated in Drosophila (Hallem and Carlson, 2006), has yet to be described in the mouse, where neurons with detectable pseudogene OR expression are immature and OMP-negative (Shykind et al., 2004). Thus, we sought to rescue the LSD1 KO phenotype by ectopic OR expression (Figure 3A). We crossed a tetO-MOR28-IRES-LacZ transgene (Clowney et al., 2012) to two tTA drivers: Gγ8-tTa (Nguyen et al., 2007), which is not affected by the LSD1 deletion since its expression initiates in immature OSNs (Ryba and Tirindelli, 1995) and OMP-IRES-tTA (Yu et al., 2004), which might preserve the expression of transgenic MOR28 in mOSNs. These three alleles were put into the Foxg1-Cre;LSD1 KO background (Figure 3B). Embryos were collected at E18.5 and subjected to whole mount X-gal staining (Figure S3). Many LacZ positive neurons are detected in the MOE of these mice and the X-gal stained cells have dendrites that reach the lumen of the olfactory epithelium (Figure 3B and supplemental Figure S3). Strikingly, IF shows that ectopic expression of MOR28 restores Adcy3 immunoreactivity in the LSD1 KO mice (Figure 3B), showing that OR expression controls Adcy3 expression.
Figure 3.
Ectopic expression of transgenic MOR28 in the LSD1 KO MOE can rescue the loss of Adcy3 expression.
(A) Model summary of findings from misexpression study. Using two tTA drivers, one active in the immature neuron (Gγ8-tTa), and one active in the mature neuron (OMPitTA), it is possible to express high levels MOR28 in the LSD1 KO MOE, in a sporadic fashion. We find that OR expression is followed by the onset of Adcy3 protein expression.
(B) Xgal staining in sections of LSD1 KO MOE shows infrequent transgenic MOR28 expression under the control of two tTA drivers. Whole-mount image is shown in Figure S3.
(C) Foxg1-Cre; tetO-MOR28-lacZ MOE at E18.5 with either Lsd1 flox/+; OMPitTA (left panels) or Lsd1 flox/flox; Gγ8-tTa; OMPitTA (right panels). Adcy3 IF (green); Beta-galactosidase IF (red); and merge.
(D) LSD1 dosage positively correlates with Adcy3 immunoreactivity. Adcy3+ cells in E18.5 MOE were quantified per unit area in ImageJ. Y axis units are Adcy3+ cells per micron of MOE area considered. Error bars show standard error of 2 quantified regions of MOE from one experiment.
The fact that Adcy3 constitutes a faithful marker for OR expression, in addition to being a marker of OSN maturation, allowed us to also test whether the levels of LSD1 affect the kinetics of OR choice and OSN maturation. We detect a significant reduction in the number of Adcy3-positive cells in heterozygote LSD1 KO mice compared to wild type littermates (Figure 3C). This suggests that the levels of LSD1 are not saturating during OR choice, which may result in a slow and inefficient process of OR activation contributing to the singularity of OR choice.
Adcy3 promotes OR choice stabilization and OSN differentiation via LSD1 downregulation
The intriguing observation that Adcy3 expression is mutually exclusive with LSD1 expression and depends upon OR expression prompted us to test whether this protein plays a role in the downregulation of LSD1 and the stabilization of OR choice. Adcy3 is the main adenylyl cyclase in OSNs and previous reports have shown that Adcy3 KO OSNs have severe targeting defects (Chesler et al., 2007; Col et al., 2007; Zou et al., 2007). A role of Adcy3 in stabilization of OR expression could account for these targeting deficits, together with activity dependent processes that regulate axon guidance. Since gene switching requires the repression of the previously chosen OR and the de-silencing of a new OR allele, both of which could be accomplished in part by the dual enzymatic activities of LSD1, we examined whether Adcy3 deletion affects LSD1 expression.
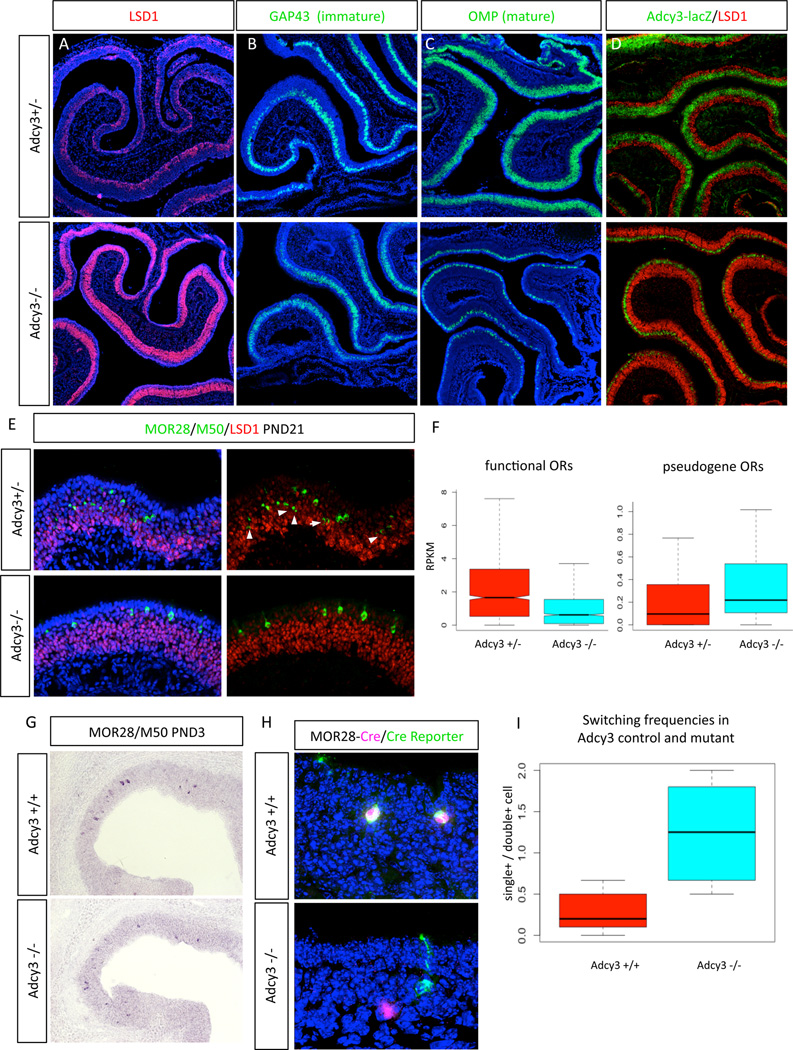
In agreement with an instructive role for Adcy3 in LSD1 gene regulation, we find that Adcy3 deletion causes a dramatic extension of LSD1 immunoreactivity towards the mOSN layers at PND21 (Post-natal day 21) (Figure 4A and S4A). Similarly, GAP43 expression is also apically expanded, showing that Adcy3 deletion delays the terminal differentiation of these neurons (Figure 4B, S4B). RNA ISH for OMP shows that the mOSN layer is reduced and restricted only to the most apical OSN layer, residing below the sustentacular cells (Figure 4C). Similarly, IF for β-galactosidase, which is expressed instead of Adcy3 in this KO strain, shows that transcription of the Adcy3 locus is also restricted to the most apical OSN layer, providing further evidence for a stabilizing positive-feedback loop, whereby stable Adcy3 expression is contingent on OR and Adcy3 proteins (Figure 4D, S4C). Notably, the Adcy3 KO MOEs have a thin mOSN layer at this age, suggesting that eventually a small proportion of neurons settle to a stable and robust choice of a single OR, as previously reported (Zou et al., 2007), possibly via low level, paralogous adenylyl cyclase activity. This is also evident by the OR expression pattern, since at PND21 OR IF shows robust OR expression in the thin LSD1-negative apical layer of the Adcy3 KO MOE (Figure 4E).
Figure 4.
Adcy3 removal triggers upregulation of LSD1 protein levels and increase in OR gene switching.
(A) PND21 sections with IF for LSD1 in Adcy3+/− (top) and −/− (bottom), respectively. See Figure S4 for quantification.
(B,C) Fluorescent RNA ISH for immature (GAP43) and mature (OMP) neurons in Adcy3+/− (top) and −/− (bottom), respectively
(D) Beta-gal (green) and LSD1 (red) IF in Adcy3+/− (top) and −/− (bottom), respectively. A lacZ reporter is knocked into Adcy3 locus. See supplemental experimental procedures for details.
(E) PND21 sections from Adcy3+/− (top) and −/− (bottom), stained with OR (green, MOR28 and M50) and LSD1 (red) antibodies.
(F) RNA-seq RPKM values for all expressed Refseq ORs (n=1072) and OR pseudogenes (n=48) in corresponding genotype from PND21 MOE. RPKMs of unexpressed intact and pseudogene ORs were excluded. See also Fig S4.
(G) PND3 RNA ISH for M50 and MOR28 as in (E). IF for ORs and LSD1 shown in supplemental Figure 4 (H) PND2 MOE from MOR28-IRES-Cre; Cre-reporter mice in Adcy3 wildtype or knockout background. Cre IF (magenta) and mT/mG reporter (green).
(I) Quantification of experiment in (H). Single (CRE or GFP positive) and double positive cells from 10 sections of PND2 wild type and Adcy3 KO mice were counted and plotted as ratio of single to double positive. SE represents variation across sections, single animal, 10 sections quantified. P value=0.007, calculated with Student's unpaired T-test.
Consistent with a delay in the stabilization of OR expression, RNA-seq analysis of wild type and Adcy3 KO MOEs at PND21 shows that overall OR mRNA levels are moderately reduced in a statistical significant manner (Figure 4F, left panel). Because Adcy3 is expressed after OR choice and in an OR-dependent manner, these effects likely do not reflect a developmental delay in the initiation of OR expression but, rather, post-OR choice instability. Indeed, our RNAseq analysis shows that overall expression of pseudogene ORs is not decreased but rather slightly increased, both at absolute and relative levels (Figure 4F, right panel, and supplemental Figure S4D, respectively), supporting the notion that LSD1+ OSNs continue to search ORs, even after the choice of an intact OR. Since OR pseudogenes can be chosen at the same frequency as intact ORs (Shykind et al., 2004), but their expression is less stable, a general deficit in stabilization of intact OR expression would favor the representation of pseudogenized ORs.
To directly test the role of Adcy3 in the stability of OR expression, we performed lineage-tracing experiments (Shykind et al., 2004) in control and Adcy3 KO mice by crossing MOR28-IRES-Cre mice to a Cre inducible GFP reporter (Muzumdar et al., 2007). If intact ORs switch in the absence of Adcy3, then a fraction of GFP positive neurons should stop expressing the MOR28-IRES-Cre allele that recombined and activated the reporter, generating GFP+/Cre- neurons. Moreover, if switching is rapid then a fraction of Cre-expressing neurons should be GFP negative, because Cre-mediated recombination takes 6–24 hours (Hayashi and McMahon, 2002, Nakamura et al., 2006). We performed this analysis at PND2 because the Adcy3 KO mice fail to thrive and mice with all four alleles did not survive beyond this age. At this age, the majority of the OSNs are immature, yet we detect OR expressing OSNs in Adcy3 KO and an apical expansion of LSD1 expression, which is less pronounced than in the older mice (Figure 4G and S4E, F). Lineage tracing, however, shows that Adcy3 KO mice have a ~2 fold increase (Student’s T-test; P=0.007) of single positive (Cre+ or GFP+) over the number of double positive neurons (Figure 4H,I), supporting the frequent switching phenotype suggested by the increase of pseudogene expression.
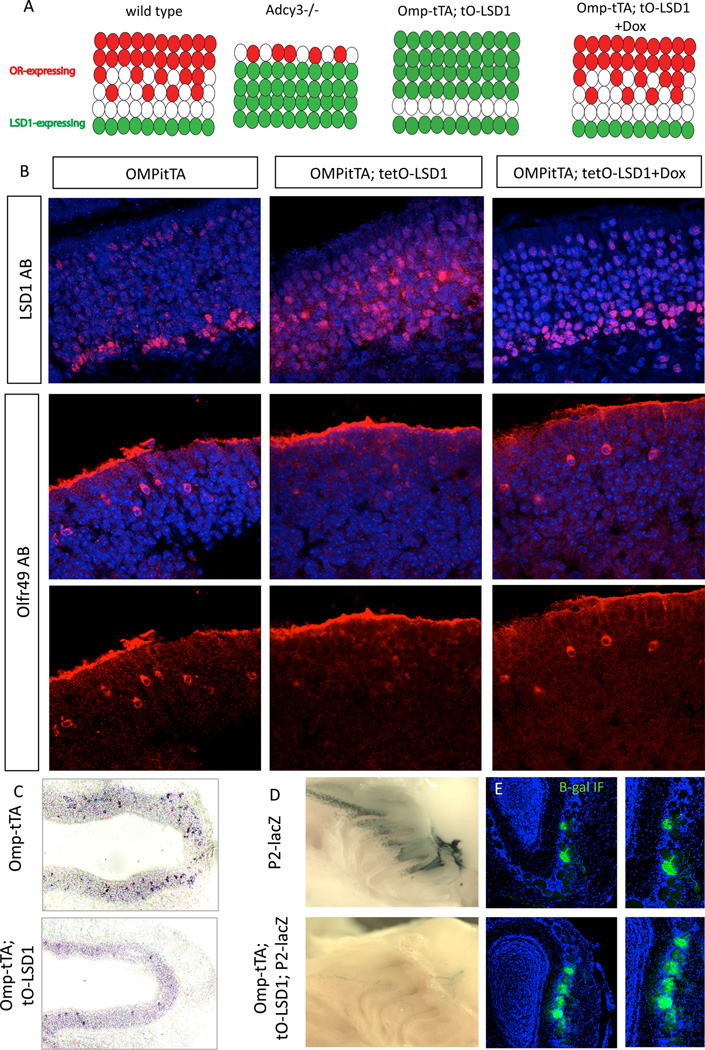
The rapid OR switching phenotype observed in Adcy3 KO pups suggests that the ectopic LSD1 expression in the Adcy3 KO is the cause of post-choice OR downregulation, and that sustained LSD1 expression is incompatible with OR transcription. To directly test the post-choice effects of LSD1 expression, we generated transgenic tetO-LSD1, which we crossed to OMPitTA mice to drive expression of LSD1 in mOSNs (Figure 5A,B). OSNs of these mice retain high levels of LSD1 even after OR choice, causing significant downregulation of OR expression by IF (Figure 5B and quantification in supplemental Figure S5A) and RNA ISH (Figure 5C). Feeding these mice doxycycline for 3 weeks shuts off tTA-driven LSD1 expression in mOSNs and restores robust OR expression (Figure 5B and S5A).
Figure 5.
Ectopic expression of transgenic LSD1 in the mature neuron layer causes reversible destabilization of OR expression.
(A) Model summarizing results in adult MOE regarding the expression pattern of ORs and LSD1 under different genetic manipulations:. OR-expressing OSNs are prevalent in LSD1-negative layer regardless of genotype. Weakly OR-expressing OSNs are present in OMPitTA;tetO-LSD1 mice before dox but robust expression returns following dox and the reduction of LSD1 misexpression.
(B) Adult OMP-tTA; tetO-LSD1 mice were raised until 3 weeks and either placed on doxycycline for 3 weeks to shut off tTA activation, or maintained on dox-free food. Control littermate mice (OMPitTA only) were also placed on doxycycline for 3 weeks. 6 week old MOE were harvested and IF was performed for LSD1 (red top panel) or Olfr49 (C6) (red two bottom panels with or without DAPI). See also Figure S5.
(C) Misexpressing LSD1 in the MOE with OMPitTA reduced OR expression in the MOE. Chromogenic ISH OR pool (15 OR probes total) in OMPitTA (left) and OMPitTA; tetO-LSD1(right).
(D) P2-lacZ (top left) and P2-lacZ;OMPitTA; tetO-LSD1 (bottom left) MOE and bulb following whole mount X-gal staining in PND25 mice. Olfactory bulb sections (right) from the same genotype are shown with bet-agalactosidase IF (green). Despite the low levels of beta-gal at the cell bodies due to switching, the protein appears stable at the axons (Clowney et al, 2013), which allows the visualization of additional glomeruli.
We also brought the P2-ires-taulacZ reporter (Mombaerts et al. 1996) into the LSD1 overexpressing background. Consistent with the aforementioned decrease in OR expression, there was a dramatic reduction of this OR reporter gene (Figure 5D). Moreover, olfactory bulb targeting becomes perturbed in the LSD1 overexpressing MOE, with regional targeting, by and large, unaffected but the total number of targeted glomeruli increasing from 1–2 in the control to roughly a dozen in the OMPitTA; tetO-LSD1 (Figure 5E and data not shown), further supporting that LSD1-overexpressing OSNs are unable to settle on a single OR and switch frequently to other OR alleles from the same zone.
LSD1 triggers guanine oxidation of the active OR DNA
The likely transient interaction of LSD1 with a chosen OR makes technically impossible to detect the binding of LSD1 on an active OR by ChIP. This technical hurdle would be bypassed if we could detect a molecular “apparition” for the presence of LSD1 at the active OR allele. Such a mark could be generated by hydrogen peroxide, which, in addition to formaldehyde, is a localized chemical byproduct of LSD1-mediated lysine demethylation (Anand and Marmorstein, 2007). Importantly, other histone demethylases do not involve the generation of reactive oxygen species like FAD-dependent LSD1 (Hou and Yu, 2010). Hydrogen peroxide tends to selectively oxidize guanosine to 8-oxoguanosine (8-oxodG), thus, we reasoned that the extensive demethylation of an OR locus would generate enough hydrogen peroxide to locally modify guanosines of the chosen allele, as has been implied by the recruitment of OGG1, a DNA repair protein that binds to 8-oxodG at LSD1 regulated promoters (Perillo et al., 2008).
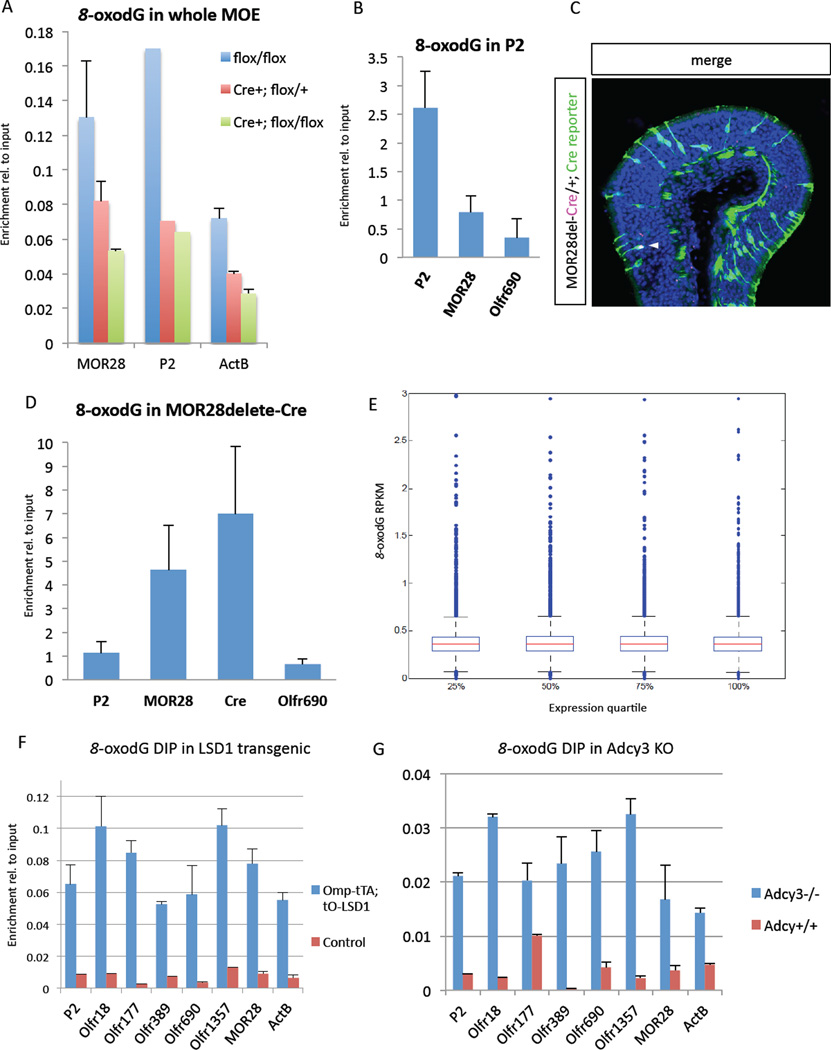
To detect 8-oxodG on a genomic locus we developed a DNA immunoprecipitation (DIP) assay with an antibody specific for this modified base. We performed preliminary titration experiments with a synthetic DNA template derived from the Cre sequence. This analysis showed that a commercially available antibody could immunoprecipitate this PCR-synthesized Cre DNA with a ~10 fold higher efficiency than its unmodified counterpart (Figure S6A; see experimental procedures). Thus, a DIP-based strategy is sensitive and specific enough for the detection of this modified base on an active OR allele. DIP-qPCR analysis with DNA prepared from E18.5 MOEs from wild type, heterozygote and homozygote LSD1 KO mice shows LSD1-dependent 8-oxodG enrichment on two OR loci tested, P2 and MOR28 (Figure 6A). The enrichment levels for 8-oxodG are low in this experiment, and comparable with the enrichment of a control locus, likely because we performed DIP in whole MOE populations in which the two OR alleles are expressed in very low fraction of cells.
Figure 6.
LSD1 generates stable 8-oxodG at active OR genes.
(A) 8-oxodG DIP was performed on sonicated genomic DNA (gDNA) from LSD1 wildtype, heterozygote, and knockout MOE at E18.5.
(B) 8oxodG-DIP-qPCRs from gDNA of P2-GFP sorted cells from PND30 mice.
(C) PND30 MOE of MOR28-del-Cre; Cre-reporter mouse, with Cre IF (magenta) and mT/mG Cre-reporter (Green are cells that have expressed Cre to levels sufficient to recombine reporter locus). DAPI nuclear stain is blue.
(D) 8oxodG-DIP-qPCRs from Cre-reporter-positive neuron gDNA at PND30, as shown in (C).
(E) DIP-seq analysis of an E18.5 wild-type 8-oxodG library. Expression quartiles from the RPKM values generated from Fig. 1 mRNA-seq. y-axis is 8-oxodG RPKM. Boxplots show mean 8-oxodG RPKM for each expression quartile demarcated by horizontal red bar.
(F, G) 8oxodG-DIP-qPCRs from gDNA from whole MOE of LSD1 overexpressing mice and Adcy3 knockout mice, respectively. Error bars are standard error from 2 PCR replicates from one representative experiment. See also Figure S6 for control experiments.
To test whether 8-oxodG enrichment stems from transcriptionally active OR alleles, we FAC-sorted GFP+ neurons expressing olfactory receptor P2 from P2-IRES-GFP knock-in mice. Using DIP-qPCR analysis we quantified the relative enrichment of 8-oxodG on the active and inactive OR alleles. We detect a ~3 fold higher enrichment of 8-oxodG on the P2 allele, compared to the enrichment of this base on the inactive OR genes tested (Figure 6B). Since the majority of the sorted P2 neurons are mature, their transcription was initiated days or weeks before, and thus they have long ago downregulated LSD1. The enrichment levels we obtained imply that 8-oxodG is stable on OR DNA, due to a probably inefficient DNA repair process stemming from the low expression levels of OGG1 and NEIL1 (Klungland and Bjelland, 2007) as shown by our RNA-seq analysis (data not shown). To test this we used a Cre-OR knock-in line whereby Cre replaces the coding sequence of MOR28. MOR28-delete-Cre expressing OSNs treat this allele like a pseudogene OR and switch from it in order to express a functional OR, often the functional MOR28 allele (Shykind et al., 2004). We crossed this delete-Cre line to the membrane-GFP (mT/mG) Cre reporter (Muzumdar et al., 2007) and isolated the GFP-positive neurons with FACS. Although transcription from the deleted MOR28 allele has ceased in most GFP-positive neurons (Figure 6C), we detect significant enrichment for 8-oxodG on the Cre locus in the GFP-positive cells (Figure 6D). Interestingly, we also detect enrichment for this modified base on the wild type MOR28 allele, which is explained by the high frequency by which these OSNs switch to this allele.
To test the possibility that 8-oxodG is a reflection of unprotected DNA due to transcription and not a direct consequence of LSD1-dependent demethylation we performed Illumina sequencing in DIP from the whole MOE. DIP-seq analysis shows that the enrichment for 8-oxodG does not correlate with levels of transcription. We sorted the mouse genes into 4 quartiles of transcription levels and we plotted 8-oxodG levels for each quartile. The mean 8-oxodG RPKMs are essentially identical between the 4 expression quartiles, suggesting that the enrichment of this base on the chosen OR allele is not a byproduct of the unusual transcription rates of OR alleles but rather it is indicative of LSD1’s proximity to that OR locus (Figure 6E).
Finally, we measured the enrichment levels of 8-oxodG in Adcy3 KO and OMPitTA; tetO-LSD1 MOE, which have abnormally high levels of LSD1 protein. We find significant increases of 8-oxodG levels in both the Adcy3 KO and the LSD1 overexpressing mice (Figure 6F,G). Notably, in wild type adults, the baseline 8-oxodG levels are lower than in embryos, which is probably explained by the significantly higher proportion of LSD1-expressing cells in embryonic than adult MOEs. Interestingly, under these overexpression conditions there is an expected loss of specificity at the LSD1-dependent DNA oxidation. Moreover, these results also show that in the OMPitTA; tetO-LSD1 MOE, only a small fraction of OR genes becomes ectopically demethylated. A 10–20 fold increase of 8-oxodG levels in DIPs from mixed MOE populations suggests that each OR allele is H3K9-free in 1–2% of the cells instead of 0.1% that is calculated in wild type MOEs. In agreement with the notion that ORs remain epigenetically silenced in >95% of the cells, and that in each OSN the vast majority of OR genes remain heterochromatinized, ChIP-qPCR for H3K9me3 shows similar enrichment levels between wild type and LSD1 overexpressing MOEs (Figure S6C).
Discussion
Our understanding of OR regulation changed by the realization that OR expression elicits a feedback signal that prevents the expression of additional ORs and/or stabilizes the expression of the chosen one (Lewcock and Reed, 2004; Nguyen et al., 2007; Serizawa et al., 2003; Shykind et al., 2004). However, neither the mechanism of OR gene activation, nor the pathway that stabilizes this activation, were previously understood. We recently showed that ORs undergo heterochromatic silencing in the MOE, at a stage prior to OR expression and that OR choice coincides with an epigenetic switch from H3K9me3 to H3K4me3 at the chosen allele (Magklara et al., 2011). The data presented here demonstrate that the epigenetic signature of active and silent ORs affords the deployment of a feedback mechanism that prevents the activation of additional ORs, while at the same time stabilizes the expression of the chosen allele. This epigenetic switch, combined with the dual function of LSD1 as H3K9 and H3K4 demethylase, not only renders the chosen OR immune to the feedback signal, but makes the subsequent downregulation of LSD1 imperative for the stabilization of OR choice.
These observations pose a significant question: why LSD1 activates OR transcription before an OR is chosen but represses OR transcriptions after OR choice? Before OR choice, ORs are marked only by H3K9 methylation, thus LSD1 can only demethylate H3K9 in an OR locus and activate transcription. However, after OR choice, the chosen OR switches from H3K9 to H3K4 methylation (Magklara et al., 2011). Therefore, at this stage, if LSD1 is still present and recruited to the chosen OR, it can only demethylate H3K4, resulting in the repression of this OR. Thus, the same molecule before OR choice is by default an activator but after choice is a repressor for an already activated OR and a potential activator for the remaining silenced OR genes. Therefore, local epigenetic context determines the exact action of LSD1 and makes the chosen OR susceptible to LSD1-mediated repression. Alternatively, if the OSN cannot support OR transcription from two different loci simultaneously, it is possible that the downregulation of the chosen OR is not a direct consequence of H3K4 demethylation by LSD1, but indirect effect of the fact that an additional OR has been activated. Finally, as in every genetic manipulation, indirect effects from either the deletion or the overexpression of LSD1 could contribute to the observed phenotypes.
It is worth emphasizing, that the genetic manipulations presented here affected only the stability of OR choice and not the singularity of expression, unlike when we disrupted nuclear OR aggregation by ectopic LBR expression (Clowney et al., 2012). The fact that ORs aggregate in large nuclear foci makes the majority of ORs inaccessible to LSD1, explaining why most of ORs remain heterochromatic after LSD1 overexpression. Thus, the mechanism that affords the selection of only one out of thousands of OR alleles is different than the mechanism that makes this selection permanent.
Feedback signal vs feed-forward loop
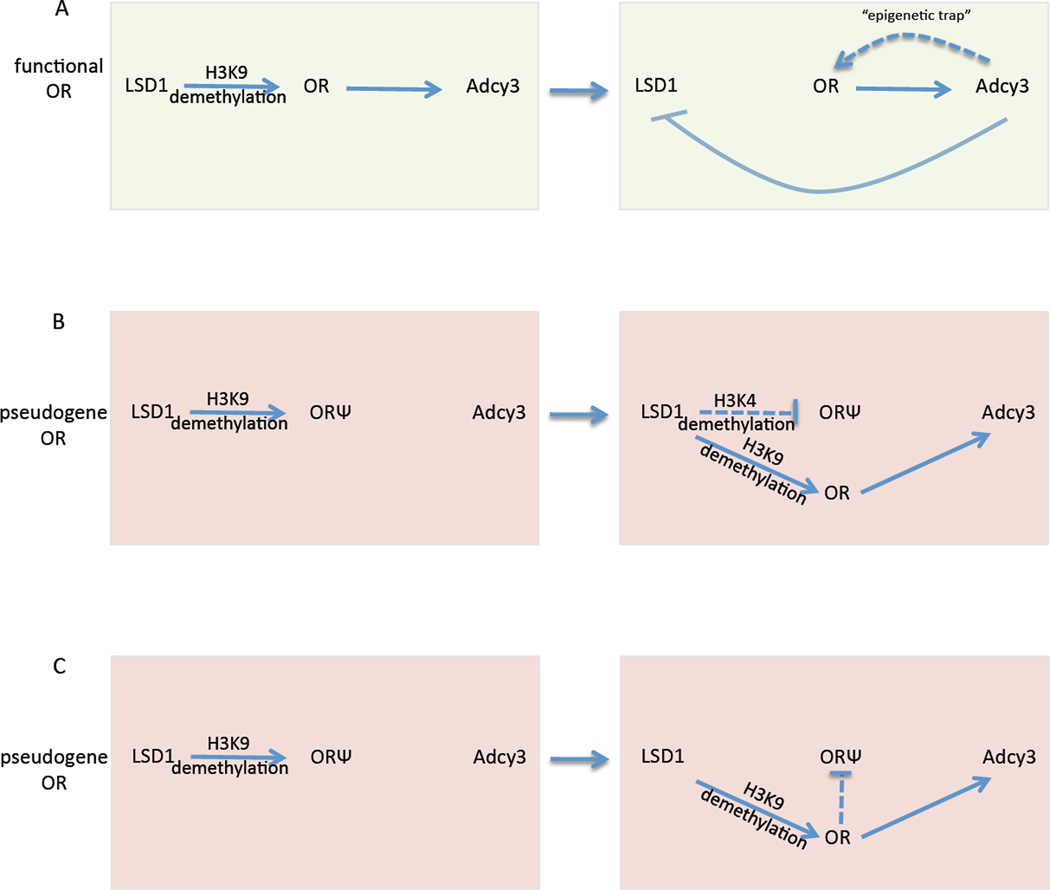
Our data, together with the established requirement of intact OR protein for the generation of the feedback signal, lead to the following regulatory model: LSD1, in complex with an as-yet unidentified H3K9me3 demethylase, de-silence a previously heterochromatinized OR allele, allowing H3K4 trimethylation and transcriptional activation. If this allele encodes an intact OR, then it will induce Adcy3 expression, LSD1 downregulation and OSN maturation, generating an “epigenetic” trap that will preserve OR expression, cellular identity and targeting specificity, as long as the underlying transcription factor milieu remains unaltered (Figure 7A). In contrast, if the initially chosen allele is a pseudogene and does not produce OR protein, then this particular choice cannot induce Adcy3 expression, and LSD1 will not be turned off. With LSD1 activity still present, an additional OR can become de-silenced via H3K9 demethylation, but also the previously chosen OR allele can become demethylated at H3K4 and turned off. Thus, failure to terminate LSD1 activity results in OR gene switching and this process will continue until an intact OR is expressed (Figure 7B).
Figure 7.
A three node signaling cascade combined with a feedback signal that generates epigenetic memory (A) Stabilization of OR expression is achieved by an Adcy3-dependent "trap" such that the functional chosen OR cannot be turned off once LSD1 is downregulated by its induction of Adcy3. This trap is caused by removing LSD1 from the signaling circuit which allows stable transcription to ensue (represented by dashed line that reflects the indirect OR stabilization by Adcy3). (B) Pseudogene ORs (ORΨ) are unable to activate Adcy3 and thus OSNs that have chosen these ORs maintain the ability to re-choose and use LSD1 to transcriptionally silence the ORΨ. (C) Alternatively, LSD1 may not silence directly the previously chosen OR, but causes its repression by activating and additional OR allele.
Different versions of feed-forward developmental loops in transcription factor regulation were recently described in the differentiation of Drosophila photoreceptor neurons (Johnston et al., 2011) and in various examples of mammalian differentiation (Neph et al., 2012). In these systems, as in a plethora of cases where a network of interactions has been mapped (Alon, 2007), establishment of cellular identity did not require downregulation of the activator that initiates a specific differentiation program, as is the case with LSD1 here. A major difference in OR regulation is the existence of an extraordinary number of similar promoters and a strict requirement for singularity in OR expression, which likely makes a feed-forward circuit ineffective. Instead the three node signaling cascade described here, which locks the epigenetic states of the chosen allele and of the silent ORs can assure both singularity and robustness. Using LSD1, which has co-activator and co-repressor activites, as an initiator of this cascade, provides the additional advantage of auto-correction, through the post-choice repression of pseudogenes.
Adcy3 has multiple functions in the MOE
The finding that Adcy3, which requires the LSD1-dependent OR gene activation to be expressed, induces rapid LSD1 downregulation makes this protein both a sensor for a productive OR choice and a transmitter of the feedback signal that stabilizes its own expression and the expression of the chosen OR, while promoting OSN differentiation. It is intriguing that Adcy3 induces LSD1 downregulation, because previous studies failed to implicate OR activity in the feedback signal. Deletion of various components of OR signaling, such as Golf(α) and Cnga2, do not affect OR expression (Belluscio et al., 1998; Brunet et al., 1996), unlike in Drosophila photoreceptor neurons (Vasiliauskas et al., 2011). Furthermore, mutation of the DRY motif, which prevents OR interaction with G(α) proteins, has no impact on the singularity of OR expression (Imai et al., 2006). Although the effects of the DRY mutation in the stability of OR expression were not addressed, it is possible that there are additional, G(α)-independent mechanisms by which an OR activates Adcy3 signaling, or that LSD1 dowregulation requires only OR-dependent Adcy3 expression and not OR-dependent Adcy3 activation. In either scenario, only low levels of cAMP, generated by basal Adcy3 activity might be sufficient to induce LSD1 downregulation and OSN maturation, since in the Adcy3 KO mice, some OSNs eventually mature and turn off LSD1. Low expression of other adenylyl cyclases may eventually generate enough cAMP to elicit a feedback signal in the Adcy3 KO OSNs. In any case, Adcy3 occupies a critical checkpoint role in the development of the peripheral olfactory system: it regulates the stability of OR choice, the targeting of OSN axons, and the longevity of olfactory neurons (Santoro and Dulac, 2012).
A transcriptional regulator with mutagenic potential
The detection of 8-oxodG on the chosen OR allele is suggestive of extensive LSD1-mediated demethylation activity in close proximity to the chosen OR allele. Since ORs are embedded in continuous blocks of methylated H3K9 (Magklara et al., 2011), demethylation of this lysine residue during OR activation is a plausible explanation for the local production of hydrogen peroxide and the accumulation of 8-oxodG at the chosen OR. Guanosine oxidation, by default, would not have consequences in the genomic stability of OSNs, since they are post-mitotic and relatively short-lived. However, were an LSD1 mediated demethylation responsible for OR activation during spermatogenesis (Fukuda et al., 2004), this could provide a mechanistic explanation for the high AT-rich content of OR genes and the extreme intra- and inter-species polymorphisms observed in this gene family, since 8-oxodG frequently pairs with adenosine rather than cytosine during DNA replication (Grollman and Moriya, 1993). Thus, LSD1 mediated derepression in the germ line could explain both the drift towards high AT-content (Glusman et al., 2001) and the evolutionary plasticity of olfactory receptor genes(Clowney et al., 2011; Niimura and Nei, 2007). Moreover, the observation that deletion of Adcy3 results in substantial increase of DNA oxidation in the OSN nuclei invites speculation regarding the role of neuronal activity pathways in protecting CNS neurons from DNA oxidation and its deleterious long-term effects.
In summary, ORs provide an unusual example in biology, whereby a transmembrane receptor protein specialized in odorant detection functions also as a molecular organizer of the sensory neuron. The finding that Adcy3 expression and OSN differentiation depend upon OR expression suggests that there are no temporal restrictions or developmental windows for OR choice; an immature OSN will remain as such until it chooses a functional OR, allowing a slow, inefficient and stochastic process for the choice of only one out of thousands of available alleles. The pleiotropic function of ORs in odor detection, OSN maturation, axonal wiring, and OSN longevity makes the peripheral olfactory system “self organizing” and centered solely around the identity of the OR, which may have facilitated the rapid expansion of this gene family during tetrapod evolution (Niimura and Nei, 2007). To accommodate adaptation in novel and variable ecological niches, olfaction has remained extremely plastic, both at the level of the genomic integrity of the chemoreceptors and at the transmission and interpretation of odorant information in piriform cortex (Choi et al., 2011). For a sensory system that lacks “labeled lines” and where polymorphisms appear constantly, ascribing such a central developmental role to the receptor protein itself, prevents pseudogene ORs from compromising the sensitivity and discriminatory power of the olfactory system (Shykind et al., 2004). The initial screening for OR quality is further fortified by a secondary, activity dependent screen that gradually eliminates OSNs that are seldomly used, affording individualized adaptation to an extremely plastic system (Santoro and Dulac, 2012).
Experimental Procedures
Mice and strains used
All mice were housed in standard conditions with a 12-hour light/dark cycle and access to food and water ad libitum and in accordance with the University of California IACUC guidelines. All strains were maintained on a mixed genetic background. Detailed information on the various mouse strains used is provided in Supplemental Experimental Procedures.
In situ hybridization and Immunofluorescence
IF and ISH was performed as previously described (Clowney et al. 2012). Information on the riboprobes and antibodies used can be found in the Supplemental Experimental Procedures. Confocal images were collected with the Zeiss LSM 700 and brightfield images were collected on the Zeiss Axioskop Plus. All image processing was carried out with ImageJ (NIH).
Chromatin immunoprecipitation
ChIP was performed as described in Magklara et al. (2011).
DNA preparation and immunoprecipitation
Genomic DNA was purified either following FACS or total MOE dissection using DNeasy genomic DNA isolation kit (Qiagen). Purified genomic DNA was sonicated in PBS with 0.5% Tween-20 to a peak around 400-bp fragments using the Bioruptor (Diagenode). For sorted cells, fragmentation of DNA was assumed to be complete following 15 to 30 minutes of sonication using medium to high power output with samples in ice water. 8-oxodG monoclonal antibody (Trevingen) was incubated with DNA rotating overnight at 4°C. Immunoprecipitation and washes were carried out in PBS-Tween 0.05% and DNA elution buffer consisted of 0.1M NaAOc and 1% SDS in TE pH 8.
DNA deep sequencing
Oligonucleotide reads were generated for Lsd1 and Adcy3 mutant and control mRNA libraries as well as 8-oxodG libraries using the Genome Analyzer IIx or HiSeq2000 (Illumina). Sequencing libraries were prepared with standard methods (Magklara et al. 2011) but in the case of the mRNA, the ScriptSeq kit (Epicentre) was used. Detailed information can be found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
Excellent technical support was provided by Zoe Evans. We would like to thank Dr. Geoff Rosenfeld for the LSD1 conditional KO mice, Dr. Nicholas Ryba for the Gγ8 tTA transgenic mice. We would also like to thank Drs. Shah and Ngai, as well as members of the Lomvardas lab, for critical reading of the manuscript. This project was funded by the Roadmap for Epigenomics grant #5R01DA030320-02, and a EUREKA grant #5R01MH091661-02, as well as the Mcknight Foundation.
References
- Alon U. Network motifs: theory and experimental approaches. Nature reviews. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. The Journal of biological chemistry. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- Barnea G, O'Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science (New York, NY. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Zou DJ, Le Pichon CE, Peterlin ZA, Matthews GA, Pei X, Miller MC, Firestein S. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Choi GB, Stettler DD, Kallman BR, Bhaskar ST, Fleischmann A, Axel R. Driving opposing behaviors with ensembles of piriform neurons. Cell. 2011;146:1004–1015. doi: 10.1016/j.cell.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome research. 2011;21:1249–1259. doi: 10.1101/gr.120162.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col JA, Matsuo T, Storm DR, Rodriguez I. Adenylyl cyclase-dependent axonal targeting in the olfactory system. Development (Cambridge, England) 2007;134:2481–2489. doi: 10.1242/dev.006346. [DOI] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. Journal of cell science. 2004;117:5835–5845. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome research. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Developmental biology. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Yu H. Structural insights into histone lysine demethylation. Curr Opin Struct Biol. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science (New York, NY. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Schwob JE. Odorant receptor expression as a function of neuronal maturity in the adult rodent olfactory system. The Journal of comparative neurology. 2003;459:209–222. doi: 10.1002/cne.10583. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Otake Y, Sood P, Vogt N, Behnia R, Vasiliauskas D, McDonald E, Xie B, Koenig S, Wolf R, et al. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Bjelland S. Oxidative damage to purines in DNA: role of mammalian Ogg1. DNA Repair (Amst) 2007;6:481–488. doi: 10.1016/j.dnarep.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Krolewski RC, Packard A, Schwob JE. Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. The Journal of comparative neurology. 2013;521:833–859. doi: 10.1002/cne.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. Emx2 stimulates odorant receptor gene expression. Chemical senses. 2008;33:825–837. doi: 10.1093/chemse/bjn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Michaloski JS, Galante PA, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome research. 2006;16:1091–1098. doi: 10.1101/gr.5185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annual review of neuroscience. 2011;34:467–499. doi: 10.1146/annurev-neuro-112210-112917. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell MD, Breheny P, Stromberg AJ, McClintock TS. Genomics of mature and immature olfactory sensory neurons. The Journal of comparative neurology. 2012;520:2608–2629. doi: 10.1002/cne.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science (New York, NY. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A novel GTP-binding protein gamma-subunit, G gamma 8, is expressed during neurogenesis in the olfactory and vomeronasal neuroepithelia. The Journal of biological chemistry. 1995;270:6757–6767. doi: 10.1074/jbc.270.12.6757. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife. 2012;1:e00070. doi: 10.7554/eLife.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science (New York, NY. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Molecular cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskas D, Mazzoni EO, Sprecher SG, Brodetskiy K, Johnston RJ, Jr, Lidder P, Vogt N, Celik A, Desplan C. Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature. 2011;479:108–112. doi: 10.1038/nature10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Human molecular genetics. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Yu CR, Power J, Barnea G, O'Donnell S, Brown HE, Osborne J, Axel R, Gogos JA. Spontaneous neural activity is required for the establishment and maintenance of the olfactory sensory map. Neuron. 2004;42:553–566. doi: 10.1016/s0896-6273(04)00224-7. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nature neuroscience. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Chesler AT, Le Pichon CE, Kuznetsov A, Pei X, Hwang EL, Firestein S. Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. J Neurosci. 2007;27:6675–6683. doi: 10.1523/JNEUROSCI.0699-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.