Abstract
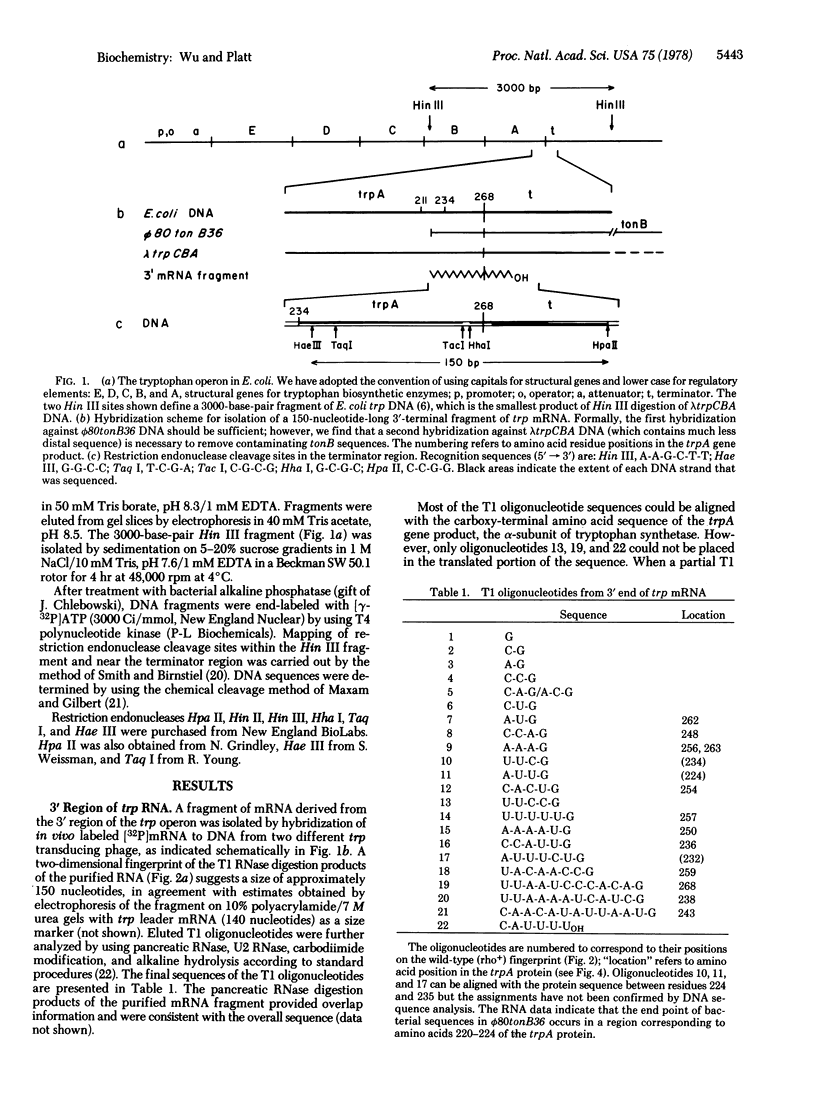
We have determined the RNA and DNA sequences in the region specifying termination of transcription at the end of the tryptophan (trp) operon of Escherichia coli. A 3′-terminal mRNA fragment of about 150 nucleotides yielded oligonucleotide products that could be assigned to the end of trpA (the last structural gene in the operon) by correlation with the amino acid sequence of the protein product. Analysis of the DNA corresponding to this region served to align the few noncoding RNA oligonucleotide sequences and demonstrated that termination of trp transcription occurs in vivo at a site 36 nucleotides after trpA, with greater than 95% efficiency. In two different strains partially defective in the transcription termination factor rho, the purified transcript is much longer and more complex, suggesting that a significant amount of read-through occurs in these strains. This is consistent with evidence [Guarente, L. P., Mitchell, D. H. & Beckwith, J. (1977) J. Mol. Biol. 112, 423-436] that efficient termination in vivo at the end of the trp operon is a rho-dependent event. The trp terminator (trp t) shares several features with other known sites of transcription termination, including (i) a 3′-terminal RNA sequence of several uridine residues, C-A-U-U-U-UOH, (ii) a G·C-rich region in the DNA immediately preceding the site of termination, followed by an A·T-rich region, and (iii) a region of dyad symmetry in the DNA which, in the transcript, is capable of forming a stable hairpin containing seven G·C base pairs and one A·U base pair in its stem.
Keywords: DNA sequence, dyad symmetry, RNA hairpin, rho factor
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bertrand K., Korn L. J., Lee F., Yanofsky C. The attenuator of the tryptophan operon of Escherichia coli. Heterogeneous 3'-OH termini in vivo and deletion mapping of functions. J Mol Biol. 1977 Nov 25;117(1):227–247. doi: 10.1016/0022-2836(77)90032-8. [DOI] [PubMed] [Google Scholar]
- Bertrand K., Korn L., Lee F., Platt T., Squires C. L., Squires C., Yanofsky C. New features of the regulation of the tryptophan operon. Science. 1975 Jul 4;189(4196):22–26. doi: 10.1126/science.1094538. [DOI] [PubMed] [Google Scholar]
- Bertrand K., Yanofsky C. Regulation of transcription termination in the leader region of the tryptophan operon of Escherichia coli involves tryptophan or its metabolic product. J Mol Biol. 1976 May 15;103(2):339–349. doi: 10.1016/0022-2836(76)90316-8. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Guarente L. P., Beckwith J. Mutant RNA polymerase of Escherichia coli terminates transcription in strains making defective rho factor. Proc Natl Acad Sci U S A. 1978 Jan;75(1):294–297. doi: 10.1073/pnas.75.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. P., Mitchell D. H., Beckwith J. Transcription termination at the end of the tryptophan operon of Escherichia coli. J Mol Biol. 1977 May 25;112(3):423–436. doi: 10.1016/s0022-2836(77)80190-3. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Carlton B. C., Yanofsky C. The amino acid sequence of the A protein (alpha subunit) of the tryptophan synthetase of Escherichia coli. I. Tryptic peptides. J Biol Chem. 1967 Nov 25;242(22):5397–5412. [PubMed] [Google Scholar]
- Hardman J. D., Berger H., Goodman M. Tryptophan operon read-through. Isolation and characterization of an abnormally long tryptophan synthetase alpha subunit from a frame-shift mutant of Escherichia coli. J Biol Chem. 1975 Jun 25;250(12):4634–4642. [PubMed] [Google Scholar]
- Hopkins A. S., Murray N. E., Brammar W. J. Characterization of lambdatrp-transducing bacteriophages made in vitro. J Mol Biol. 1976 Nov 15;107(4):549–569. doi: 10.1016/s0022-2836(76)80082-4. [DOI] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Bertrand K., Bennett G., Yanofsky C. Comparison of the nucleotide sequences of the initial transcribed regions of the tryptophan operons of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1978 May 15;121(2):193–217. doi: 10.1016/s0022-2836(78)80005-9. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. H., Reznikoff W. S., Beckwith J. Genetic fusions that help define a transcription termination region in Escherichia coli. J Mol Biol. 1976 Mar 15;101(4):441–457. doi: 10.1016/0022-2836(76)90239-4. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Morse A. N. Dual-control of the tryptophan operon is mediated by both tryptophanyl-tRNA synthetase and the repressor. J Mol Biol. 1976 May 15;103(2):209–226. doi: 10.1016/0022-2836(76)90310-7. [DOI] [PubMed] [Google Scholar]
- Platt T., Yanofsky C. An intercistronic region and ribosome-binding site in bacterial messenger RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2399–2403. doi: 10.1073/pnas.72.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels P. H., van Rotterdam J. In vitro synthesis of enzymes of the tryptophan operon of Escherichia coli. Evidence for positive control of transcription. Mol Gen Genet. 1975;136(3):215–226. doi: 10.1007/BF00334016. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Rose J. K., Yanofsky C. Transcription of the operator proximal and distal ends of the tryptophan operon: evidence that trpE and trpA are the delimiting structural genes. J Bacteriol. 1971 Oct;108(1):615–618. doi: 10.1128/jb.108.1.615-618.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M. Isolation and sequence determination of the 3'-terminal regions of isotopically labelled RNA molecules. Nucleic Acids Res. 1974 May;1(5):653–671. doi: 10.1093/nar/1.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Weissman S., deCrombrugghe B. Termination of transcription in bacteriophage lambda. Heterogeneous, 3'-terminal oligo-adenylate additions and the effects of rho factor. J Biol Chem. 1975 Jun 25;250(12):4755–4764. [PubMed] [Google Scholar]
- Rosenberg M., de Chrombrugghe B., Musso R. Determination of nucleotide sequences beyond the sites of transcriptional termination. Proc Natl Acad Sci U S A. 1976 Mar;73(3):717–721. doi: 10.1073/pnas.73.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C., Lee F., Bertrand K., Squires C. L., Bronson M. J., Yanofsky C. Nucleotide sequence of the 5' end of tryptophan messenger RNA of Escherichia coli. J Mol Biol. 1976 May 15;103(2):351–381. doi: 10.1016/0022-2836(76)90317-x. [DOI] [PubMed] [Google Scholar]
- Winkler M. E., Roth D. J., Hartman P. E. Promoter- and attenuator-related metabolic regulation of the Salmonella typhimurium histidine operon. J Bacteriol. 1978 Feb;133(2):830–843. doi: 10.1128/jb.133.2.830-843.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]



