Abstract
Background:
Securing a diagnosis of ovarian cancer and establishing means to predict outcomes to therapeutics remain formidable clinical challenges. Early diagnosis is particularly important since survival rates are markedly improved if tumour is detected early.
Methods:
Comprehensive miRNA profiles were generated on presurgical plasma samples from 42 women with confirmed serous epithelial ovarian cancer, 36 women diagnosed with a benign neoplasm, and 23 comparably age-matched women with no known pelvic mass.
Results:
Twenty-two miRNAs were differentially expressed between healthy controls and the ovarian cancer group (P<0.05), while a six miRNA profile subset distinguished presurgical plasma from benign and ovarian cancer patients. There were also significant differences in miRNA profiles in presurgical plasma from women diagnosed with ovarian cancer who had short overall survival when compared to women with long overall survival (P<0.05).
Conclusion:
Our preliminary data support the utility of circulating plasma miRNAs to distinguish women with ovarian cancer from those with a benign mass and identify women likely to benefit from currently available treatment for serous epithelial ovarian cancer from those who may not.
Keywords: diagnostics, prognostics, plasma miRNAs, ovarian cancer, biomarkers
Each year, the American Cancer Society estimates over 20 000 new cases of ovarian cancer will be diagnosed and ∼15 500 women died of the disease (Seigel et al, 2012). Although 6% of all cancer death in women is caused by ovarian cancer, there has been relatively little improvement in survival rates over the past decade (Seigel et al, 2012). Five-year survival is critically dependent on the ovarian cancer stage at diagnosis; if diagnosed and treated while localised (stages I and II), 5-year survival rates can reach over 90%. However, most ovarian cancer cases are diagnosed as advanced disease (stages III and IV) where 5-year survival is <30% (Piver et al, 1992; Berkenblit and Cannistra, 2005; Jemal et al, 2010).
Disease outcome is significantly higher with early diagnosis, however, currently there is no non-invasive method to accurately detect early-stage ovarian cancer (Partridge et al, 2009; Andersen et al, 2010). This was recently re-confirmed by the US Preventative Services Task Force that annual screenings with transvaginal ultrasound and measurement of the cancer antigen, CA-125 serum levels does not significantly increase survival rates in asymptomatic women with no genetic risk mutations. In fact, they concluded that screening for ovarian cancer can lead to unnecessary death from surgical complications associated with a false positive result (Moyer, 2012). Other biomarkers such as mesothelin and human epididymis 4 also have limited diagnostic utility (Andersen et al, 2010; Dutta et al, 2010). There clearly exists an unmet need for a non-invasive diagnostic tool with sufficient sensitivity and specificity to positively impact survival rates in ovarian cancer.
MiRNAs are small (19–25 nt), non-coding RNAs that regulate gene expression post-transcriptionally by binding in the 3′ untranslated region (3′UTR) of their specific messenger RNAs and interfering with translation (Bartel, 2009). The presence and expression levels of specific tissue miRNAs have been associated with different stages in ovarian cancer and clinical outcome. Iorio et al (2007) were the first to compare genome-wide miRNA expression profiles from both ovarian cancer and normal ovary tissue. Approximately 30 miRNAs were found to be differentially expressed between normal and cancerous tissue. Since then several others have confirmed the dysregulation of miRNAs in ovarian cancer tissue (Dahiya and Morin, 2010; Creighton et al, 2012).
More recently, it has become apparent that miRNAs circulate in the peripheral blood in several compartments. They may be found within exosomes in the plasma or free of cellular material and bound to proteins such as Argonaut 2 (Arroyo et al, 2011). Thus, they offer a potential biomarker for a wide range of diseases (Devaux et al, 2012; Weiland et al, 2012; Rege et al, 2013; Shen et al, 2013; Zhu et al, 2013). Indeed, there are several reports of circulating miRNAs associated with ovarian cancer and correlated with detection, severity of disease, and response to treatment (Taylor and Gercel-Taylor, 2008; Resnick et al, 2009; Häusler et al, 2010), although the methods for sample preparation and miRNA detection vary between studies. In this study, using an ultracentrifugation method, we focussed on miRNAs that are circulating freely in the plasma unbound to cellular material as potential biomarkers for ovarian cancer detection and outcome.
Materials and methods
Samples
We collected peripheral blood samples before treatment from women being evaluated for a suspicious pelvic mass. We enrolled 42 women with confirmed serous epithelial ovarian cancer and 36 women diagnosed with a benign neoplasm. Additional blood samples were collected from women diagnosed with ovarian cancer >2 weeks post surgery and >24 months post chemotherapy. Blood samples were also collected from 23 healthy female volunteers. The characteristics of study subjects are given in Table 1. The Institutional Review Board of the North Shore–Long Island Jewish Health System approved these studies.
Table 1. Subject summary.
| Healthy controls | Benign mass | Ovarian cancer | |
|---|---|---|---|
| Number of subjects |
23 |
36 |
42 |
|
Age at enrollment (years) | |||
| Mean | 36 | 52 | 64 |
| Range |
18–61 |
34–76 |
31–87 |
|
Ovarian cancer stage |
n/a |
n/a |
|
| IIA |
|
|
3 |
| IIC |
|
|
3 |
| IIIA |
|
|
1 |
| IIIB |
|
|
12 |
| IIIC |
|
|
21 |
| IV | 2 | ||
miRNA extraction and profiling
Whole blood, collected in EDTA tubes, was centrifuged at 4000 r.p.m. for 10 min at room temperature within 5 h of venipuncture. Plasma was removed, aliquoted, and stored at −80 °C. One millilitre of thawed plasma was ultracentrifuged at 100 000 g for 60 min at 4 °C to remove cell debris, microvessicles, and exosomes. Total small RNA from the supernatant was isolated using the mirVana PARIS kit (Ambion, Grand Island, NY, USA) according to the manufacturer's instruction. MiRNA quality was assessed using Agilent's 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Total miRNA profiles were generated using ABI Taqman OpenArray MicroRNA pools A and B to measure the expression of 754 known miRNAs. Briefly, total small RNA isolated from 1 ml of purified plasma was divided equally for each sample and used with TaqMan Megaplex RT primer pools A or B to generate cDNA which was subsequently amplified using the corresponding Megaplex PreAmp Primers (pools A or B, respectively) following the manufacturer's instructions. Real-time PCR was performed on the Taqman Open Array MicroRNA plates using the Applied Biosystem Open Array Real-Time PCR system. Data were processed using the OpenArray Real-Time qPCR Analysis software and exported for analysis using the Applied Biosystems DataAssist Software.
Statistical analysis
Data analysis was done with the R programming language. A cutoff for Ct values at 30 was used. MiRNAs with Ct values higher than 30 were considered as not detected. Data were normalised using a mean-centering restricted (MCR) strategy as described by Wylie et al (2011). The MCR method is a modification of the traditional delta Ct method and uses miRNAs which are expressed in all samples for data normalisation. The mean of these fully expressed miRNAs in a given sample is subtracted from all miRNAs in that sample.
For the comparison of presurgical plasma samples, four miRNAs (miR-320, -720, -1274b, and U6 RNA) were present in all samples and were used for normalisation. For the comparison of presurgical with postsurgical and postchemotherapy samples, 14 miRNAs (miR-16, miR-19b, miR-24, mirR-146a, mmu-miR-451, U6 rRNA, miR-106a, miR-126, miR-320, miR-191, miR-17, miR-483-5p, miR-1274b, and miR-720) were fully expressed and used for data normalisation. After data were normalised, statistical analysis was performed via custom scripts based on the R/Bioconductor package LIMMA (Linear Models for Microarray) (Smyth, 2005).
Comparisons between experimental groups were performed using a moderated t-test from LIMMA and P-values were adjusted for multiple testing via the Benjamini–Hochberg method. Fold changes of each miRNA were calculated by the equation 2 −ΔΔCt where the comparative cycle threshold (ΔΔCt) is defined as the difference between ΔCt (cancer/experimental) minus ΔCt (control) as previously described (Schmittgen and Livak, 2008). A Benjamini–Hochberg adjusted P-value of <0.05 was considered as statistically significant in comparisons between different groups for each miRNA (Benjamini and Hochberg, 1995). As an additional stringency requirement, miRNAs were further considered only if they had at least a 2-fold difference in expression and were present in at least 75% of the samples in each group unless otherwise specified.
Area under the curve (AUC) analysis was performed with the R/Bioconductor package ROC (Carey and Redestig; www.bioconductor.org).
Results
Plasma miRNA expression levels in ovarian cancer and cancer-free subjects
Using plasma isolated miRNAs, we generated comprehensive miRNA expression profiles on presurgical plasma samples from 42 women with confirmed serous epithelial ovarian cancer, 36 women diagnosed with a benign neoplasm, and 23 healthy women with no known pelvic mass (Table 1).
We analysed all miRNAs for differential expression between control plasma and presurgical plasma from confirmed ovarian cancer patients. Twenty-two of the significant miRNAs met our criteria of being expressed in at least 75% samples of each group and demonstrating a fold change of at least 2. These are shown in Figure 1 and Supplementary Table 1. The majority of miRNAs (19 out of 22) were underexpressed in plasma from cancer patients compared with controls. In plasma samples from women without a known pelvic mass (healthy controls) compared with presurgical plasma samples from women diagnosed with ovarian cancer, miR-106a, miR-126, miR-146a, miR-150, miR-16, miR-17, miR-19b, miR-20a, miR-223, miR-24, and miR-92a had at least a 10-fold elevation in expression, while miR-106b, miR-191, miR-193a-5p, miR-30b, miR-30a- 5p, miR-30c, miR-320, and miR-328 were significantly elevated but to a lesser degree (adj. P<0.05; AUC>0.8). Three miRNAs, miR-1274a, miR-625-3p, and miR-720, were elevated only in cancer samples (adj. P<0.05); however, they were not useful discriminators between control and cancer samples (AUC<0.8).
Figure 1.
Comparison of plasma miRNA levels between subject groups. Comparisons of plasma miRNA levels from subjects diagnosed with ovarian cancer, a benign mass, or healthy controls. Only miRNAs that had at least a 2-fold change in expression present in at least 75% of each group were used in the analysis. P<0.05 (cancer subjects n=42, benign subjects n=36, healthy controls n=23). Abbreviation: NS=not significant.
There were only six miRNAs (miR-106b, miR-126, miR-150, miR-17, miR-20a, and miR-92a, adj. P<0.05) that distinguished benign presurgical plasma from that of cancer subjects. In addition to the 22 miRNAs that were significantly differentially expressed and present in at least 75% of each group, we list 161 miRNAs that were well expressed (|ΔCt|>3) in at least two subjects of either benign or ovarian cancer subjects (Supplementary Table 2).
Differences in miRNA expression between presurgical benign samples and normal controls compared with those observed between controls and ovarian cancer subjects showed differential expression of 17 overlapping miRNAs (Figure 1; Supplementary Table 1). There are 10 miRNAs that distinguish plasma of healthy control subjects from presurgical plasma samples from women with ovarian cancer or a benign neoplasm. In the comparison of healthy controls with cancer plasma samples there are five miRNAs, miR-1274a, miR-30b, miR-30c, miR-625-3p, and miR-720 that are differentially expressed. MiR-139-5p, miR-142-3p, miR-484, miR-486, and miR-660 are higher in healthy controls when analysed against benign plasma samples.
Presurgical plasma miRNAs associated with long-term outcome in patients with ovarian cancer
We compared presurgical plasma miRNA profiles of women who turned out to have ovarian cancer, and for whom we had outcome data (N=26). Significant differences in miRs-720, miR-20a, miR-223, miR-126_3p, and miR-1290 expression were found in women who had short overall survival (SOS) (<2 years, N=19) when compared to women with long overall survival (LOS) (>4 years, N=7) (Table 2).
Table 2. Comparison of miRNAs in presurgical plasma of LOS vs SOS subjects.
| ID | P-value | Adj. P-value | Fold change | AUC |
|---|---|---|---|---|
| miR-720 |
0.002 |
0.079 |
−4.57 |
0.13 |
| miR-20a |
0.03 |
0.3 |
−2.63 |
0.15 |
| miR-223 |
0.03 |
0.3 |
4.474 |
0.73 |
| miR-126-3p |
0.016 |
0.24 |
4.818 |
0.74 |
| miR-1290 | 0 | 0.004 | 14.566 | 0.87 |
Abbreviations: AUC=area under the curve; LOS=long overall survival; SOS=short overall survival.
MiR-720 and miR-20a were elevated in women who died of their disease <2 years after diagnosis. Women who survived >4 years after diagnosis have elevated levels of miR-223, miR-126-3p, and miR-1290 in their presurgical plasma (Table 2). However, after correction for multiple testing, only the level of miR-1290 was significantly different between the two groups (adj. P-value<0.05, AUC of 0.87). Age and clinical stage at diagnosis were comparable for both groups analysed (Table 3).
Table 3. Ovarian cancer subject summary and miR-1290 levels.
| Subject ID | Age Dx | Cancer stage | ΔCt | Survival (years) |
|---|---|---|---|---|
|
UND | ||||
| 1 |
53 |
IA |
5.9 |
>1 but <4 |
| 2 |
51 |
IIA |
5 |
>1 but <4 |
| 3 |
79 |
IIIB |
−3.2 |
>1 but <4 |
| 4 |
59 |
IIIB |
−3.4 |
>1 but <4 |
| 5 |
51 |
IIIB |
1.3 |
>1 but <4 |
| 6 |
63 |
IIIB |
0.7 |
>1 but <4 |
| 7 |
67 |
IIIB |
3.8 |
>1 but <4 |
| 8 |
63 |
IIIB |
5.4 |
>1 but <4 |
| 9 |
57 |
IIIC |
−2.8 |
>1 but <4 |
| 10 |
74 |
IIIC |
−1.7 |
>1 but <4 |
| 11 |
50 |
IIIC |
3.4 |
>1 but <4 |
| 12 |
58 |
IIIC |
5.2 |
>1 but <4 |
| 13 |
78 |
IIIC |
4.5 |
>1 but <4 |
| 14 |
50 |
IIIC |
6.8 |
>1 but <4 |
| 15 |
86 |
IV |
5.9 |
> 1 but <4 |
| 16 |
60 |
IV |
ND |
>1 but <4 |
|
LOS | ||||
| 17 |
57 |
IA |
3.2 |
>4 |
| 18 |
51 |
IIC |
2.6 |
>4 |
| 19 |
31 |
IIC |
4.4 |
>4 |
| 20 |
70 |
IIIB |
−0.6 |
>4 |
| 21 |
65 |
IIIB |
2.4 |
>4 |
| 22 |
62 |
IIIC |
−1.8 |
>4 |
| 23 |
66 |
IIIC |
3.7 |
>4 |
|
SOS | ||||
| 24 |
57 |
IIC |
ND |
<2 |
| 25 |
55 |
IIIA |
ND |
<2 |
| 26 |
69 |
IIIB |
3.4 |
<2 |
| 27 |
75 |
IIIB |
5.2 |
<2 |
| 28 |
60 |
IIIB |
7 |
<2 |
| 29 |
69 |
IIIB |
ND |
<2 |
| 30 |
58 |
IIIC |
1.8 |
<2 |
| 31 |
66 |
IIIC |
2.5 |
<2 |
| 32 |
54 |
IIIC |
4.7 |
<2 |
| 33 |
77 |
IIIC |
7.4 |
<2 |
| 34 |
83 |
IIIC |
7.4 |
<2 |
| 35 |
79 |
IIIC |
6.5 |
<2 |
| 36 |
85 |
IIIC |
6.7 |
<2 |
| 37 |
65 |
IIIC |
8.7 |
<2 |
| 38 |
76 |
IIIC |
8.8 |
<2 |
| 39 |
75 |
IIIC |
ND |
<2 |
| 40 |
54 |
IIIC |
ND |
<2 |
| 41 |
87 |
IIIC |
ND |
<2 |
| 42 | 72 | IIIC | ND | <2 |
Abbreviations: LOS=long overall survival; ND=not detected; SOS=short overall survival; UND=undeclared survival outcome.
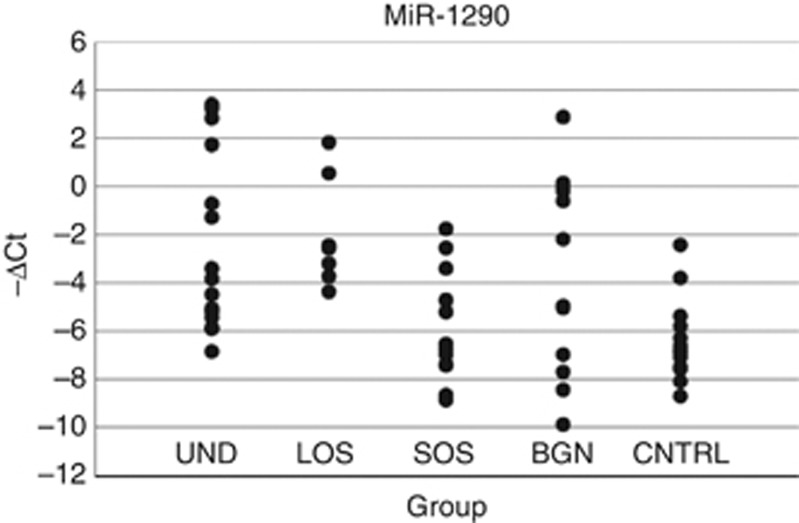
The expression levels of miR-1290 detected in all samples included in this study are shown in Figure 2 and Table 3. MiR-1290 was present in all of the samples from LOS subjects (7 out of 7) tested but was detected in only 12 out of 19 SOS, 15 out of 16 UND, 13 out of 23 control, and 12 out of 36 benign subjects. In this data set, miR-1290 was calculated to have a sensitivity of 63.3% and 100% specificity for long-term survival in subjects with ovarian cancer (e.g., considering only people with ovarian cancer) with a PPV and a PNV of 100% and 75%, respectively. In addition to the differentially expressed miRNAs that were found in at least 50% of the samples, we list 71 miRNAs that were found to be significantly expressed (|ΔCT|⩾3) in at least 2 individuals of either the LOS or SOS subject group (Supplementary Table 3).
Figure 2.
Level of miR-1290 in subject groups. Individual levels of miR-1290 detected in plasma samples from subjects with ovarian cancer, a benign mass, or healthy controls. Expressed as −(ΔCt), see also Supplementary Table 2. Abbreviations: BGN=benign mass (n=32); CNTRL=healthy control (n=23); LOS=long overall survival >4 years (n=7); SOS=short overall survival <2 years (n=19); UND=undeclared survival (n=16).
Changes of miRNA expression in plasma of ovarian cancer patients after surgical treatment and chemotherapy
Five LOS subjects had presurgical and postchemotherapy plasma samples available for analysis. There were seven miRNAs that were differentially expressed between matched presurgical and postchemotherapy samples. These changes are summarised in Figure 3, Supplementary Table 4, and Supplementary Figure 1. MiR-1274a, miR-1274b, and miR-1290 displayed consistent decreases in plasma levels after treatment, whereas miR-19b, -25, -195, and -16 were more highly expressed in postchemotherapy samples when compared with their respective presurgical plasma sample (adj. P<0.05).
Figure 3.
Presurgical vs postchemotherapy plasma miRNAs in LOS subjects. MiRNAs demonstrating significant differences between presurgical and postchemotherapy plasma samples of women with long overall survival (n=5). Expressed as −(ΔCt), see also Supplementary Figure 1 and Supplementary Table 4.
The comparison of presurgical with immediate postsurgical plasma samples, measured before chemotherapy, did not show any significant differences in miRNA expression in paired analyses from the same subjects (n=9; data not shown).
Discussion
To address several unanswered questions regarding the utility of circulating miRNAs as biomarkers in ovarian cancer, we analysed blood samples from women diagnosed with ovarian cancer, a benign neoplasm, or healthy control samples. A number of considerations were taken into account before analysing miRNAs from plasma. For example, the use of circulating miRNAs as biomarkers of cancer have come under increased scrutiny, due to lack of standardised miRNA isolation methods and the possibility of contamination of RBC, lymphoid and myeloid-derived miRNAs. Pritchard et al (2012) reported the potential for confounding factors related to miRNA contamination from lysed cells in previously reported studies including ovarian cancer. Therefore, to minimise the contribution of miRNAs from lysed PBMCs, RBCs, platelets, and other cellular debris, plasma was isolated within 5 h of blood draw. Second, after an initial low speed centrifugation, the plasma was removed and ultracentrifuged leaving only protein bound miRNA in the supernatant. It has been suggested that the majority of disease-related miRNAs found in circulation are vesicle independent originating from the lysis of affected cells (Turchinovich et al, 2011). We confirmed the absence of cellular debris including microvessicles and exosomes in the supernatant by electron microscopy (data not shown). The stable presence of microvessicle and exosome-free miRNAs in circulation is dependent upon being bound to Ago 2 to protect them from degradation (Arroyo et al, 2011).
Previous studies have identified circulating miRNAs in serum (Resnick et al, 2009, Chung et al, 2013), exosomes (Taylor and Gercel-Taylor, 2008), and whole blood (Häusler et al, 2010) from ovarian cancer subjects. Resnick et al (2009) identified eight miRNAs in the serum of 28 ovarian cancer patients. Mir-21,-92, -93, -126, and -29a were elevated in presurgical serum of cancer patients while miR-155, -127, and -99b were lower in cancer patients compared with healthy controls. More recently, Cheung et al (2013) found miR-132, -26a, -145, and let7b to be underexpressed in serum of ovarian cancer patients. In a study by Taylor and Gercel-Taylor (2008), it was found that exosomes isolated from serum of ovarian cancer expressed higher levels of miR-21, -141, -200a, 200b, 200c, 203, 205, and -214 when compared to patients with benign ovarian disease. Häusler et al (2010) measured miRNA levels in 24 patients with relapsed ovarian cancer. In comparison with controls, they found four miRNAs that remained significantly expressed after adjustment for multiple testing; miR-30c1-3p, 342-3p, 181a-3p, and 450b-5p. A comparison of circulating miRNAs identified to distinguish control and/or benign from cancer subjects from previous studies including our own, failed to identify common miRNAs expressed in the same subject group. Interestingly, Resnick et al (2009) found miR-126 to be elevated in serum from cancer patients while we found more robust levels of miR-126 in plasma samples from healthy control and benign mass subjects. This discrepancy among ours and previous studies may be attributable to the source of miRNAs (whole blood, plasma, serum, or exosomes) from primary ovarian cancer or relapse patients, and/or the platform (qPCR or microarray based) used for analysis.
While we recognise that some highly expressed miRNAs relevant to ovarian cancer may go undetected due to degradation, tissue sequestration, or microvessicle/exosome containment, those that we have identified may be seminal to detection and outcome of the disease. To the best of our knowledge, this is the first serous epithelial ovarian cancer study to specifically focus on circulating, protein bound miRNAs in plasma, free of cellular debris, microvessicles, and exosomes.
Initially, we set out to determine whether specific circulating miRNAs could be used to detect the presence of ovarian cancer in subjects being evaluated for a pelvic mass. We assessed the expression patterns of 754 miRNAs in presurgical plasma samples from 42 women with confirmed serous epithelial ovarian cancer compared with 36 plasma samples collected from women who turned out to have a benign pelvic mass at surgery. We also compared both of these groups with age-matched healthy subjects without evidence of pelvic pathology. Interestingly, both cancerous and benign neoplasms were generally associated with lower levels of a group of 22 circulating miRNAs, as shown in Figure 1. A much smaller subset of six miRNAs were significantly different between groups of women with cancer vs a benign neoplasm suggesting that benign neoplasms are associated with a circulating miRNA profile with some similarity to ovarian cancer. Of these, miR-106b, miR-150, and miR-126 have been previously reported to have decreased expression in ovarian cancer tissue compared with non-cancerous ovarian tissue (Iorio et al, 2007; Dahiya et al, 2008; Zhang et al, 2008). There does not seem to be an obvious overlap in gene targets/pathways of these three miRNAs to explain their lowered expression in ovarian cancer. We also found significantly elevated expression of miR-17, miR-20a, and miR-92a of the miR-17-92 cluster in presurgical plasma from women with a benign neoplasm compared to women with ovarian cancer. The role and the function of these miRNAs in cancer are difficult to identify since their function is both cell type and context specific (Olive et al, 2010; Concepcion et al, 2012).
In 2011, the TCGA (Cancer Genome Atlas Research Network, 2011) published and made available genetic, epigenetic, and expression data on 489 high-grade serous ovarian cancer tissue samples. We downloaded and analysed level 3 miRNA microarray expression data of women with ovarian cancer who had not been previously treated for their disease (i.e., adjuvant, chemotherapy). The six miRNAs (miR-106b, miR-126, miR-150, miR-17, miR-20a, and miR-92a) we found to be elevated in presurgical plasma from benign compared with cancer patients were well expressed in ovarian cancer tissue from this data set, however, without comparable miRNA expression data from samples with benign ovarian disease the significance of miRNA expression of miRNAs in ovarian cancer tissue could not be determined.
To take into account the possibility that highly expressed miRNAs in a small number of subjects could also distinguish benign from ovarian cancer subjects, we relaxed our miRNA inclusion criteria to include miRNAs that had a |ΔCt| of >3 and were present in at least two subjects of either the benign or the ovarian cancer group. One hundred and sixty-one samples met this modified criteria, of these 21 were present in presurgical plasma from subjects with a benign mass and 22 were present in presurgical plasma from women subsequently diagnoses with ovarian cancer (Supplementary Table 2). The relevance of these uniquely expressed miRNAs in either benign or ovarian cancer presurgical plasma will need to be evaluated in a larger data set.
We also examined the utility of presurgical circulating plasma miRNA in predicting outcome of those subjects who turned out to have ovarian cancer at operation. Using follow-up data available on 26 ovarian cancer subjects in our study, we compared presurgical plasma miRNA profiles of women who died within 2 years of diagnosis (but survived at least 6 months post surgery) from those who survived 4 years or more. Although 5-year survival is standard, we chose to include subjects with at least 4-year survival for our LOS group to include enough subjects for meaningful analysis. We found distinct differences between these two groups. There were five miRNAs (miR-720, miR-20a, miR-223, miR-126-3p, and miR-1290) that were differentially expressed between LOS and SOS subjects, however, only miR-1290 remained significant after multiple testing adjustments (adj. P=0.004) (Table 2).
There were additional 71 miRNAs that were significantly present (|ΔCt|⩾3) but were detectable in <50% of the LOS or SOS group (Supplementary Table 3). Nineteen of these miRNAs were only found in presurgical plasma samples from SOS while four were present only in LOS subjects. Due to the small number of subjects further analysis could not be performed to determine significance. Using our criteria for LOS and SOS we analysed TCGA miRNA microarray level 3 data for the corresponding tissue miRNA associated with outcome. None of the miRNAs available for analysis were significant.
The expression of miR-1290 may directly influence outcome in women with ovarian cancer and its expression is independent of age and cancer stage at diagnosis. One highly scored potential gene target of miR-1290 is epithelial membrane protein 2 (EMP2) (Supplementary Table 5). EMP2 is a tetraspan protein that facilitates transmembrane activity of specific integrins and can act as an oncogene in a number of cancers (Freije et al, 2004; Fu et al, 2010; Habeeb et al, 2010) and its elevated expression has been associated with poor outcome in patients with endometrial cancer (Wadehra et al, 2006). EMP2 is also expressed in ovarian cancer although unlike with endometrial cancer, the level of expression was not associated with patient survival. In vitro and in vivo treatment of ovarian cancer cells with an engineered bivalent antibody against EMP2 inhibited cell growth and resulted in cell death (Fu et al, 2010). It is conceivable that elevated expression of miR-1290 observed in the LOS subjects led to decreased expression of EMP2 and consequently resulted/contributed to their long-term cancer-free status. Finally, a comparison of presurgical and postsurgical plasma samples from matched individuals did not show any significant differences. This is most likely explained by the timing of ascertainment which was ∼2 weeks post surgery and may have been too soon to detect relevant changes. However, analysis of presurgical and postchemotherapy plasma (cured) samples from LOS subjects showed a dramatic change in miRNA profiles. Although based on a relatively small number of matched presurgical and postchemotherapy sample pairs (5), these miRNAs (miR-1274a, miR-1274b, miR-1290, miR-19b, miR-25, miR-195, and miR-16) maintained their significance even after multiple testing corrections (adj. P<0.05). Interestingly, miR-1290 whose elevated expression was predictive of survival in presurgical plasma is decreased in long overall survivors (>4 years) of ovarian cancer suggesting that it may be induced/regulated in the presence of ovarian cancer. Although suppressed in presurgical plasma miR-19b, miR-25, miR-195, and miR-16 are elevated in the LOS subjects. In particular, there is a 10-fold elevation of miR-16 in LOS postchemotherapy plasma samples when compared with presurgical plasma samples. A recent paper by Pouliot et al (2012) showed that transfection of a miR-16 mimic into cisplatin-resistant cells resulted in a 5-fold increase in cisplatin sensitivity although the mechanism is undefined. Cisplatin is a common chemotherapy used to treat ovarian cancer among others and one could envision that chemosensitivity maintained by elevated levels of miR-16 during the chemotherapy contributes to a favourable outcome in our LOS subjects. To confirm effects of miR-1290 and miR-16 to support long-term cancer-free status in ovarian cancer, confirmation and validation in a larger multi-site cohort will be needed. Given the general nature of miRNAs associated with cancers, it is highly likely these miRNAs have a pivotal role in cell proliferation and/or metastasis but their specific role in affecting ovarian cancer outcome, whether directly or indirectly, remains to be determined. Further studies will be needed to confirm the extent that circulating levels of miRNAs are reflective of disease status.
Acknowledgments
We are indebted to all of the patients and their families who contributed to this study. We also gratefully thank The Great Atlantic Pacific Tea Company, Betta and Jerry Clair, Manhasset Women's Coalition Against Breast Cancer, Moms Who Kick and Partner's Council for Women's Health.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, Bergan L, Thornquist MD, Scholler N, Kim N, O'Briant K, Drescher C, Urban N. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 2010;102:26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy. Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Berkenblit A, Cannistra SA. Advances in the management of epithelial ovarian cancer. J Reprod Med. 2005;50:426–438. [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey V, Redestig H.for C++ language enhancements. ROC: utilities for ROC, with array focus. R package version 1.34.0 http://www.bioconductor.org .
- Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, Lee KW. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18:262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson E, Sheridan R, Xiao W, Spellman PT, Getz G, Wheeler DA, Perou CM, Gibbs RA, Sander C, Hayes DN, Gunaratne PH, Cancer Genome Atlas Research Network Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7:e34546. doi: 10.1371/journal.pone.0034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih IeM, Zhang Y, Wood W, 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17:F77–F89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, Corsten MF, Schroen B, Lair ML, Heymans S, Wagner DR. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–567. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- Dutta S, Wang FQ, Phalen A, Fishman DA. Biomarkers for ovarian cancer detection and therapy. Cancer Biol Ther. 2010;9:668–677. doi: 10.4161/cbt.9.9.11610. [DOI] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- Fu M, Maresh EL, Soslow RA, Alavi M, Mah V, Zhou Q, Iasonos A, Goodglick L, Gordon LK, Braun J, Wadehra M. Epithelial membrane protein-2 is a novel therapeutic target in ovarian cancer. Clin Cancer Res. 2010;16:3954–3963. doi: 10.1158/1078-0432.CCR-10-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeeb O, Goodglick L, Soslow RA, Rao RG, Gordon LK, Schirripa O, Horvath S, Braun J, Seligson DB, Wadehra M. Epithelial membrane protein-2 expression is an early predictor of endometrial cancer development. Cancer. 2010;116:4718–4726. doi: 10.1002/cncr.25259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler SF, Keller A, Chandran PA, Ziegler K, Zipp K, Heuer S, Krockenberger M, Engel JB, Hönig A, Scheffler M, Dietl J, Wischhusen J. Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010;103:693–700. doi: 10.1038/sj.bjc.6605833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Moyer VA. Screening for ovarian cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann Intern Med. 2012;157:900–904. doi: 10.7326/0003-4819-157-11-201212040-00539. [DOI] [PubMed] [Google Scholar]
- Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge E, Kreimer AR, Greenlee RT, Williams C, Xu JL, Church TR, Kessel B, Johnson CC, Weissfeld JL, Isaacs C, Andriole GL, Ogden S, Ragard LR, Buys SS, PLCO Project Team Results from four rounds of ovarian cancer screening in a randomized trial. Obstet Gynecol. 2009;2009;113:775–782. doi: 10.1097/AOG.0b013e31819cda77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piver MS, Malfetano J, Baker TR, Hempling RE. Five-year survival for stage IC or stage I grade 3 epithelial ovarian cancer treated with cisplatin-based chemotherapy. Gynecol Oncol. 1992;46:357–360. doi: 10.1016/0090-8258(92)90232-8. [DOI] [PubMed] [Google Scholar]
- Pouliot LM, Chen YC, Bai J, Guha R, Martin SE, Gottesman MM, Hall MD. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72:5945–5955. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rege SD, Geetha T, Pondugula SR, Zizza CA, Wernette CM, Ramesh Babu J. Noncoding RNAs in neurodegenerative diseases. ISRN Neurol. 2013;30:375852. doi: 10.1155/2013/375852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shen J, Stass SA, Jiang F. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett. 2013;329:125–136. doi: 10.1016/j.canlet.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Smyth G.2005Linear models for microarray data Bioinformatics and Computational Biology Solutions Using R and BioconductorGentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) pp397–420.Springer: New York [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadehra M, Natarajan S, Seligson DB, Williams CJ, Hummer AJ, Hedvat C, Braun J, Soslow RA. Expression of epithelial membrane protein-2 is associated with endometrial adenocarcinoma of unfavorable outcome. Cancer. 2006;107:90–98. doi: 10.1002/cncr.21957. [DOI] [PubMed] [Google Scholar]
- Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- Wylie D, Shelton J, Choudhary A, Adai AT. A novel mean-centering method for normalizing microRNA expression from high-throughput RT-qPCR data. BMC Res Notes. 2011;4:555. doi: 10.1186/1756-0500-4-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Bützow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Pan W, Qian Y. MicroRNA in immunity and autoimmunity. J Mol Med (Berl) 2013;91:1039–1050. doi: 10.1007/s00109-013-1043-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.