Abstract
Pituitary tumor transforming gene (PTTG), the index mammalian securin, is abundantly expressed in several tumors and regulates tumor growth and progression. Molecular mechanisms elucidating PTTG regulation and actions remain elusive. Here, we provide evidence that PTTG acts as a STAT3 target gene. Total STAT3 and Tyr705 phosphorylated STAT3 were concordantly expressed with PTTG in human colorectal tumors (n=97 and n=95 respectively, P<0.001). STAT3 specifically bound the human PTTG promoter and induced PTTG transcriptional activity (2-fold) as assessed by chromatin immunoprecipitation and luciferase reporter assays. STAT3 transfection increased PTTG mRNA and protein abundance 2-fold in HCT116 human colon cancer cells, and induction was further enhanced (3-fold) by constitutively active STAT3 (STAT3-C), while strongly abrogated by dominant negative STAT3 (STAT3-DN). Attenuating PTTG expression by siRNA in STAT3 HCT116 stable transfectants suppressed cell growth and colony formation in vitro, and PTTG cell knockout also constrained activated STAT3-induced explanted murine tumor growth in vivo. STAT3 increased HCT116 cell migration and invasion up to 5-fold, whereas cell mobility was abolished by STAT3-DN (>85%). Impairing PTTG expression by siRNA also strongly suppressed STAT3-faciliated cell migration and invasion by up to 90%. Knocking out PTTG in STAT3-C HCT116 stable transfectants strongly decreased tumor metastases in nude mice, indicating the requirement of PTTG for STAT3-promoted metastasis. These results elucidate a mechanism for tumor cell PTTG regulation, whereby STAT3 induces PTTG expression to facilitate tumor growth and metastasis; and further support the rationale for targeting PTTG to abrogate colorectal cancer growth.
Keywords: STAT3, PTTG, Transformation
Introduction
Pituitary tumor transforming gene (PTTG) and signal transducer and activator of transcription 3 (STAT3) are both involved in tumor transformation and metastatic progression (1, 2). PTTG, the vertebrate securin (3), mediates sister chromatid separation during mitosis (4). PTTG also facilitates cell-cycle progression, maintains chromosomal stability (5), responds to DNA damage (6, 7), and mediates cell transformation in vitro and tumor formation in vivo (4, 8). PTTG is highly expressed in several human tumors including colorectal (9), pituitary (10, 11), thyroid (12), breast (13) and esophageal cancers (14). PTTG overexpression correlates with tumor invasiveness, differentiation, recurrence and prognosis (15, 16), and PTTG has been identified as a key signature gene associated with tumor metastases (17). PTTG abundance promotes lymph node metastases in esophageal carcinomas through modulating metastasis-related factors (18). PTTG exerts oncogenic functions by disrupting genetic stability, altering oncoprotein expression and regulating growth factors. Overexpressed PTTG results in abnormal mitosis and chromosomal instability (19). PTTG activates c-Myc (20) and cyclin D3 (21) to facilitate cell proliferation and also increases basic fibroblast growth factor (b-FGF) and VEGF expression to induce angiogenesis (22), and induces IL-8 to function in metastasis (23). Factors inducing PTTG expression include estrogen, insulin, b-FGF and epidermal growth factor (EGF) (2). PTTG is also regulated by beta-catenin/TCF (14) and Rb/E2F1 pathways (24), which both play important roles in tumorigenesis. Nevertheless, proximal regulatory mechanisms enabling tumor PTTG abundance remain elusive. In this study, we present evidence supporting PTTG as a STAT3 target.
STAT3 acting as a transcription factor, responds to cytokines, growth factors and hormones by JAK kinase phosphorylation, followed by dimerization and nuclear translocation, to regulate target gene transcription (25). STAT3 regulates cell proliferation, survival and differentiation, and is also involved in tumorigenesis, angiogenesis, metastasis, and tumor-promoting inflammation (26). Constitutive STAT3 activation results in NIH3T3 cell transformation and tumor formation in nude mice (27). Overexpression and/or constitutive tumor STAT3 activation are associated with poor prognosis, and occur in a wide variety of cancers including colorectal (28), leukemia, myelomas, melanomas, breast, prostate, pancreatic, ovarian, and head and neck cancers (1). Inhibition of constitutive STAT3 activation, by inhibiting tyrosine kinase signaling, is associated with cell growth suppression and induction of cell death (29-31). Several genes involved in tumorigenesis have been identified as STAT3 targets, including c-Myc (32), cyclin D1 (33), vascular endothelial growth factor (VEGF) (34) and matrix metalloproteinase-2 (MMP-2) (35).
In this study, we present evidence that PTTG, behaving as a STAT3 target gene, acts to mediate STAT3-induced cell transformation and motility. Concordant colorectal cancer STAT3 and PTTG overexpression is accompanied by STAT3 binding to the PTTG promoter and induction of PTTG. Attenuating PTTG inhibited colon cancer cell growth and colony formation in vitro, and constrained STAT3-induced tumor growth in vivo. Furthermore, PTTG inhibition by siRNA attenuated STAT3-promoted cell motility in vitro, and strongly decreased STAT3-facilitated tumor metastasis in vivo. These results elucidate a mechanism for PTTG regulation, whereby STAT3 induces PTTG to promote cell transformation and motility, supporting the PTTG role in tumorigenesis and providing a rationale for PTTG targeting to abrogate colorectal cancer growth.
Results
Total STAT3 and Tyr705 phosphorylated STAT3 are concordantly expressed with PTTG
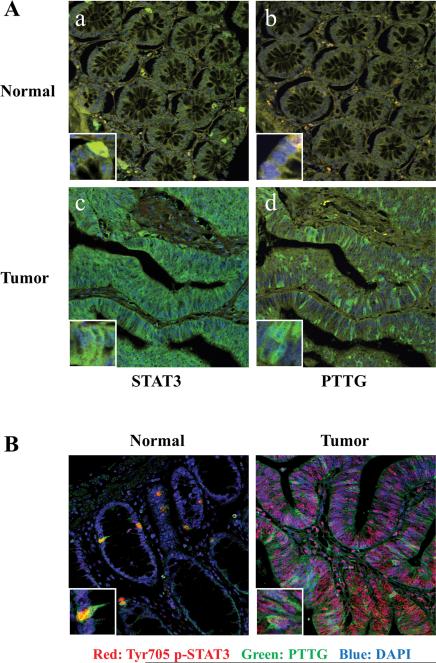
As both STAT3 and PTTG are upregulated in several cancers, we assessed confocal immunofluorescence patterns of STAT3 and PTTG in 97 colorectal tumors and 9 normal tissues. Normal tissue STAT3 and PTTG immunoreactivity was weak or undetectable, whereas STAT3 and PTTG expression was abundant in tumor specimens (Fig. 1A). Upregulated immunoreactive STAT3 and PTTG were detected in 82% (80 of 97) and 74% (72 of 97) of colorectal tumors respectively (Table 1). Sixty-six tumors with strong STAT3 expression were also immunoreactive for PTTG, and eleven cases with unchanged STAT3 levels did not exhibit increased PTTG immunoreactivity. Taken together, 79% of tumor samples exhibited a concordant pattern of STAT3 and PTTG expression (P<0.001, Pearson's χ2-test).
Figure 1.
Concordant STAT3/phospho-STAT3 and PTTG expression in human colorectal tumors. (A) STAT3 and PTTG are concordantly overexpressed in 97 colorectal tumor specimens. Images a-b show weak staining of STAT3 and PTTG in normal tissues; pictures c-d exhibit abundant STAT3 and PTTG expression in tumor specimens. Green signal: STAT3 or PTTG staining; blue signal: DAPI nuclear staining; yellow signal: autofluorescence background. (B) Tyr705 phospho-STAT3 and PTTG are concordantly overexpressed in 95 colorectal tumor specimens. Left image depicts discrete nuclear phospho-STAT3 and PTTG staining in normal tissue. Right image exhibits phospho-STAT3 and PTTG overexpression in tumor specimen. Green signal: PTTG staining; red signal: phospho-STAT3 staining; blue signal: DAPI nuclear staining. Protein immunoreactivity was assessed by fluorescence immunehistochemistry. Picture size is 375μM *375μM; Insert size is 30μM *30μM.
Table 1.
STAT3 and PTTG expression in 97 human colorectal tumors
| PTTG |
Total | |||
|---|---|---|---|---|
| Overexpression | No change | |||
|
STAT3
|
Overexpression | 66 (68%) | 14 | 80 (82%) |
| No change | 6 | 11 (11%) | 17 | |
| Total | 72 (74%) | 25 | 97 | |
STAT3 and PTTG immunoreactivity were assessed by confocal microscopy in 97 human colorectal tumor and 9 normal specimens (Figure 1A). Correlation was analyzed using Pearson chi-square test, P<0.001.
As STAT3 is activated by phosphorylation at Tyr705, we co-stained Tyr705 phospho-STAT3 and PTTG in 95 colorectal tumors and 9 normal tissues. Discrete nuclear phospho-STAT3 and PTTG staining was detected in a small percentage of normal cells; in contrast, most tumor tissues strongly stained with a high percentage of positive cells (Fig. 1B). Upregulated phospho-STAT3 and PTTG expression were observed in 64% and 73% of tumor tissues respectively. Remarkably, 71 of 95 tumors (77%), including 53 cases showing both overexpressed (56%) and 18 cases showing both unchanged (19%), exhibited concordant expression of phospho-STAT3 and PTTG (P<0.001, Pearson's χ2-test) (Table 2).
Table 2.
Tyr705 phospho-STAT3 and PTTG expression in 95 human colorectal tumors
| PTTG |
Total | |||
|---|---|---|---|---|
| Overexpression | No change | |||
|
p-STAT3
|
Overexpression | 53 (56%) | 8 | 61 (64%) |
| No change | 16 | 18 (19%) | 34 | |
| Total | 69 (73%) | 26 | 95 | |
Tyr705 phospho-STAT3 and PTTG immunoreactivity were assessed by confocal microscopy in 95 human colorectal tumor and 9 normal specimens (Figure 1B). Correlation was analyzed using Pearson chi-square test, P<0.001.
IL-6 induces STAT3 phospharylation and PTTG expression, whereas STAT3 inhibitor attenuates STAT3 phosphorylation and PTTG expression
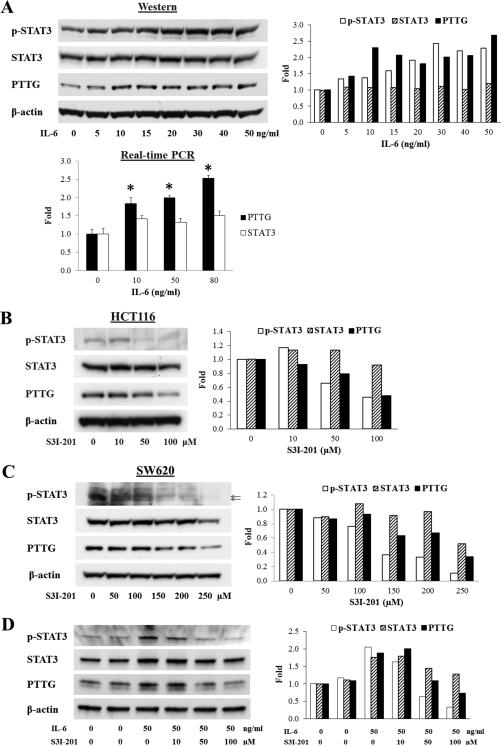
As STAT3 is activated by IL-6, we serum-starved HCT116 cells and subsequently treated with IL-6 for 16 hours. IL-6 dose-dependently (0-50 ng/ml) increased STAT3 Try705 phosphorylation and PTTG expression as measured by Western blotting. Total STAT3 expression did not change as obviously as did phospho-STAT3 (Fig. 2A). IL-6 also increased PTTG mRNA expression as assessed by real-time PCR (Fig. 2A). Moreover, a specific STAT3 inhibitor S3I-201 inhibited STAT3 phosphorylation and also attenuated PTTG expression in HCT116 (Fig. 2B) and SW620 (Fig. 2C) colon tumor cells. When cells were serum-starved and subsequently treated with IL-6 and S3I-201 for 16 hours, IL-6 induction of phospho-STAT3 and PTTG were dose-dependently attenuated (Fig.2D).
Figure 2.
IL-6 induces STAT3 phosphorylation and PTTG expression, whereas STAT3 inhibitor attenuates STAT3 phosphorylation and PTTG expression. (A) IL-6 induces STAT3 phosphorylation and increases PTTG mRNA and protein expression in HCT116 cells. (B-C) S3I-201 attenuates STAT3 phosphorylation and PTTG expression in HCT116 and SW620 cells. (D) S3I-201 suppresses IL-6 effects in HCT116 cells. Right panels show quantitative results of Western blots. Intensity was quantified by Image-J and normalized to β-actin. Experiments were repeated twice. * p<0.05.
STAT3 binds human PTTG promoter and actives PTTG transcription
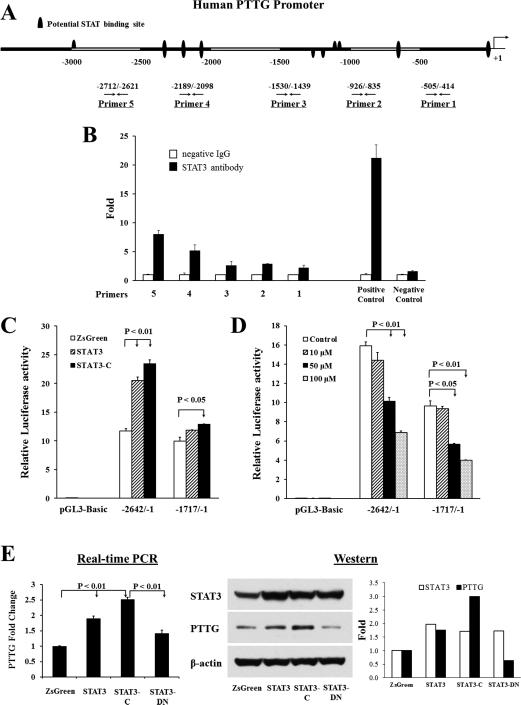
We screened the human PTTG promoter with Genomatix MatInspector and detected several STAT binding sites (Fig. 3A). Accordingly, we performed chromatin immunoprecipitation (ChIP) to identify STAT3 binding to the PTTG promoter. Equal amounts of sonicated HCT116 chromatin DNA were incubated with IgG control or STAT3 antibody respectively. Protein G bead captured chromatin DNA was amplified as template, and five pairs of PTTG promoter primers used for real-time PCR (Fig. 3A), with primer 1 being closest to the ATG translation initiation site and primer 5 furthest (36). Human c-Fos promoter primers were used as positive controls and α-satellite repeat primers as negative controls. Anti-STAT3 immunoprecipitated DNA with the enriched STAT locus was strongly amplified by primers 4 and 5, indicating specific STAT3 binding to the PTTG promoter around these two primer regions (Fig. 3B). Similar results were obtained by using Tyr705 phospho-STAT3 antibody (not shown).
Figure 3.
STAT3 binds the PTTG promoter, activates PTTG transcription and induces PTTG expression. (A) Schematic representation of potential STAT binding motifs on the human PTTG promoter, and five primers designed for chromatin immunoprecipitation. (B) STAT3 binds the PTTG promoter. Normalized inputs of HCT116 chromatin DNA were pulled down by STAT3 or negative IgG antibodies. DNA template was amplified by real-time PCR using primers 1-5. Human c-Fos promoter primers were used as positive controls and α-satellite repeat primers as negative controls. White bars: incubation with negative control IgG; black bars: incubation with STAT3 antibody. Each negative IgG control was normalized to unit “1”. Real-time PCR reaction was performed in triplicate and ChIP experiments repeated twice. (C) STAT3 activates PTTG transcription. HCT116 cells were co-transfected pGL3-Basic, PTTG promoter −2642/−1 or –1717/−1, together with empty vector pIRES2-ZsGreen1 (marked as ZsGreen, white bar), wild type STAT3 (grey bar) or STAT3-C (black bar), as described in Material and Methods, and luciferase activity measured. (D) STAT3 inhibitor S3I-201 suppresses PTTG transcription. HCT116 cells were transfected with pGL3-Basic, PTTG promoter −2642/−1 or –1717/−1, and treated by increasing amounts of S3I-201. Five ng of pRL-TK plasmid were co-transfected to normalize transfection efficiency. Transfections were performed in triplicate, and experiments were repeated three times. (E) STAT3 induces PTTG mRNA and protein expression. HCT116 cells were transfected with pIRES2-ZsGreen1 (marked as ZsGreen), STAT3, STAT3-C or STAT3-DN, and selected for stable transfectants. PTTG mRNA was assessed by real-time PCR (left), and STAT3 and PTTG protein were measured by Western blotting (right). Real-time PCR results are presented as mean ± SE. Western blots were quantified by Image-J and normalized to β-actin. Experiments were repeated three times.
To ascertain whether STAT3 regulates PTTG transcription, we employed two different PTTG promoter plasmids, −2642/−1 and −1717/−1 cloned in pGL3-Basic vector (36), to measure promoter activity by a dual luciferase reporter assay in response to STAT3. The −2642/−1 construct contains multiple STAT binding motifs shown in Figure 3A, whereas the shorter −1717/−1 construct is devoid of the STAT binding sites that exhibited strong binding affinity in ChIP assays. We also cloned two different STAT3 expression plasmids: wild type STAT3 and constitutively active STAT3 (STAT3-C) (27) in pIRES2-ZsGreen1 vector. Figure 3C shows that STAT3 induced PTTG promoter transcriptional activity. Co-transfection of the PTTG promoter (−2642/−1) with STAT3 or STAT3-C plasmid resulted in ~2-fold induction of promoter activity compared to empty vector pIRES2-ZsGreen1. The shorter PTTG promoter (−1717/−1), which lacks STAT binding sites close to ChIP primers 4 and 5 (Fig. 3A), exhibited modest responses to STAT3 and STAT3-C (Fig. 3C). This result implies that STAT3 binding motifs located between −2642 bp and −1717 bp are required for STAT3-induced PTTG transcriptional activity.
Furthermore, we treated PTTG promoter-transfected HCT116 cells with STAT3 inhibitor S3I-201 and measured luciferase activity. As shown in Figure 3D, transcriptional activity of two PTTG promoters (−2642/−1 and −1717/−1) were both suppressed (up to 66% at 100 μM) by S3I-201, indicating that STAT3 is required in PTTG transactivation. To ensure specificity, we performed the same experiment using another STAT3 inhibitor, Stattic, with similar results (data not shown).
STAT3 induces PTTG mRNA and protein expression
As STAT3 binds and activates the PTTG promoter, we tested STAT3 regulation of endogenous PTTG expression in HCT116 stable transfectants. We cloned a dominant negative STAT3 (STAT3-DN) in pIRES2-ZsGreen1 vector, by replacing tyrosine 705 to phenylalanine. We respectively transfected empty vector pIRES2-ZsGreen1, STAT3, STAT3-C and STAT3-DN in HCT116 cells and selected stable transfectants, all of which expressed green fluorescent protein ZsGreen, and sorted before experiments to ensure expression. PTTG mRNA expression was measured by real-time PCR using β-actin as an internal control. As shown in Figure 3E, overexpressed STAT3 doubled PTTG mRNA levels. This induction was further enhanced by STAT3-C (~2.5-fold) and was specifically attenuated by STAT3-DN. Cell lysates analyzed by Western blotting verified that PTTG protein was altered similarly to mRNA (Fig. 3E). Up-regulated STAT3 induced PTTG protein expression 1.8-fold and was further enhanced by STAT3-C to 3-fold. Transfection of STAT3-DN abolished STAT3-induced PTTG protein expression.
Attenuating PTTG expression decreases STAT3 stable cell growth and transformation
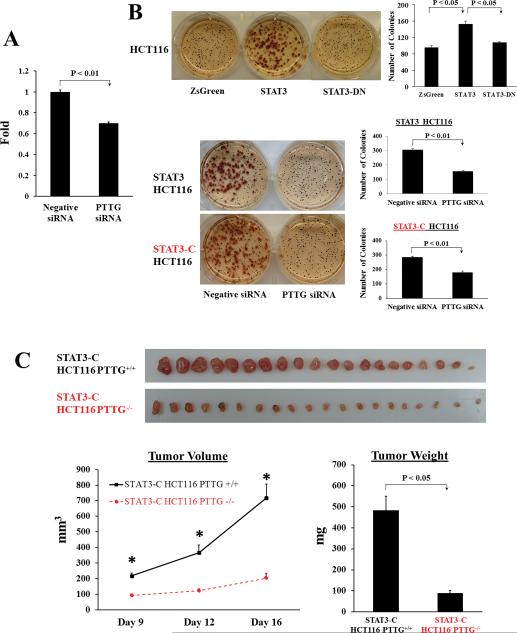
As PTTG promotes cell proliferation and mediates cell transformation, we examined whether inhibiting PTTG would constrain STAT3-facilitated cell growth and transformation. We transfected PTTG siRNA and negative controls in STAT3-overexpressing HCT116 stable cells, and measured cell proliferation using WST-1 cell proliferation reagent. Compared to negative siRNA controls, PTTG siRNA transfected cells exhibited ~30% reduction of WST-1 absorbance after 72 hours (Fig. 4A), demonstrating that attenuating PTTG expression constrains stable HCT116 transfectant growth.
Figure 4.
Attenuating PTTG expression decreases STAT3-promoted cell proliferation and transformation in vitro and xenografted tumor growth in vivo. (A) PTTG siRNA decreases STAT3 stable transfectant proliferation. STAT3 HCT116 stably transfected cells were transfected with PTTG siRNA and negative controls respectively. Cell proliferation was measured using premixed WST-1 cell proliferation reagent. (B) PTTG siRNA inhibits STAT3-faciliated colony formation. Compared to pIRES2-ZsGreen1 controls, STAT3 overexpression increased colony forming ability of HCT116 cells, whereas STAT3-DN overexpression did not exhibit a difference (upper panel). Transfecting PTTG siRNA in STAT3 or STAT3-C stable cells decreased colony formation compared to negative controls (lower panel). Colonies larger than 50 cells (>0.3 mm) were counted manually. Quantitative results were shown as mean ± SE. Experiments were repeated three times. (C) Knockout of PTTG in STAT3-C stable transfectants constrains xenografted tumor growth. STAT-C HCT116 PTTG+/+ and STAT-C HCT116 PTTG−/− cells were implanted subcutaneously into nude mice. Tumor dimensions were measured twice weekly and tumor volume calculated as: (Length*Width2)/2. Seventeen days later, mice were sacrificed and excised tumors weighed. Results are presented as mean ± SE, n=20, * p<0.001.
We then performed soft agar colony formation assays to determine PTTG involvement in STAT3-promoted in vitro transformation. Compared to pIRES2-ZsGreen1 controls, STAT3 overexpression enhanced colony forming ability of HCT116 stable transfectants (1.6-fold), whereas STAT3-DN overexpression did not exhibit a difference (Fig. 4B). Furthermore, suppressing PTTG in STAT3 stable cells by siRNA strongly abrogated STAT3-facilitated colony formation (~50%) (Fig.4B). A similar result was shown in STAT3-C stably expressed HCT116 cells (~38% reduction), further supporting the role of PTTG in STAT3-facilitated anchorage-independent HCT116 colony formation.
PTTG knockout in STAT3-C stable cells suppresses xenografted tumor growth
To assess whether blocking PTTG expression constrains tumor growth in vivo, we subcutaneously injected STAT3-C overexpressed HCT116 PTTG+/+ or HCT116 PTTG−/− stable cells in nude mice and examined xenografted tumor growth. Compared to STAT3-C overexpressed PTTG+/+ controls, knocking out PTTG in STAT-C stable cells strongly suppressed tumor growth. As shown in Figure 4C, tumor volumes were higher in STAT-C HCT116 PTTG+/+ than STAT-C HCT116 PTTG−/−, starting from day 9 (218±17.0 vs 92±8.4 mm3, P<0.001). On day 12, the average volume of STAT-C PTTG+/+ tumor was 366±49.3 mm3 vs 122±12.2 mm3 for STAT-C PTTG−/− tumors (67% inhibition, P<0.001). On day 16, average STAT-C PTTG+/+ tumor volume was 718±89.1 mm3 vs 204±29.4 mm3 for STAT-C PTTG−/− tumors (82% reduction, P<0.001). Seventeen days after tumor cell implantation, mice were sacrificed and tumors harvested. The average weight of STAT-C PTTG+/+ tumors was 484±67.8 mg, while STAT-C PTTG−/− tumors weighed 89±14.2 mg (82% reduction, P<0.001). These results indicate that attenuating PTTG expression inhibits tumor growth in vivo.
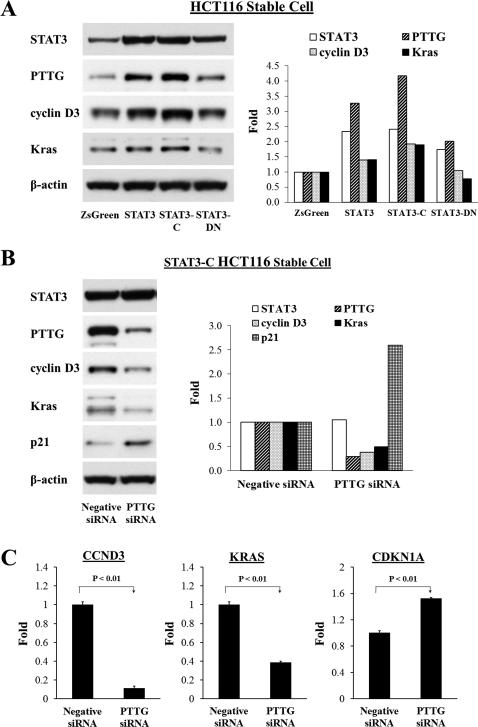
Impairing PTTG expression in STAT3-C stable tranfectants suppresses cyclin D3 and Kras expression
We further studied mechanisms underlying PTTG involvement in STAT3-induced tumor development. By using RT2 Profiler PCR arrays (SABiosciences), we examined alterations of gene expression affected by STAT3-PTTG pathway (data not shown) and further focused on several candidates including CCND3, KRAS and CDKN1A. Induced STAT3 or STAT3-C expression in HCT116 cells elevated cyclin D3 and Kras expression, as measured by Western (Fig. 5A). In contrast, STAT3-DN overexpression attenuated inductions of cyclin D3 and Kras. STAT3 bands were visualized to verify stable exogenous gene expression. Furthermore, we impaired PTTG expression by siRNA in STAT3-C stable cells and showed cyclin D3 and Kras attenuation. Western blot of PTTG confirmed PTTG siRNA suppression (Fig. 5B). In addition, attenuating PTTG in STAT3-C stable transfectants elevated expression of p21, a CDK inhibitor (Fig. 5B). We also measured these gene expressions by real-time PCR and similar results were obtained. As Shown in Figure 5C, inhibiting PTTG expression in STAT3-C stable cells decreased mRNA levels of CCND3 and KRAS, by up to 90%. In contrast, expression of CDKN1A was moderately elevated after PTTG siRNA (~1.5-fold) (Fig. 5C). These results indicate that attenuating PTTG constrained the expression of pro-proliferative factors cyclin D3 and Kras.
Figure 5.
Inhibiting PTTG expression in STAT3-C stably transfected cells attenuates cyclin D3 and Kras expression. (A) Overexpression or activation of STAT3, or STAT3-DN effects on protein expression as measured by Western blotting. (B) Transfection of PTTG siRNA in STAT3-C HCT116 stable cells decreased cyclin D3 and Kras protein expression and increased p21, as assessed by Western blotting. Right panels show quantitative results of Western blots. Intensity was quantified by Image-J and normalized to β-actin. (C) Real-time PCR results verify effects of attenuating PTTG expression in STAT3-C stable cells. Real-time PCR results are presented as mean ± SE. Experiments were repeated three times.
PTTG is involved in STAT3-faciliated cell motility by regulating MMPs expression
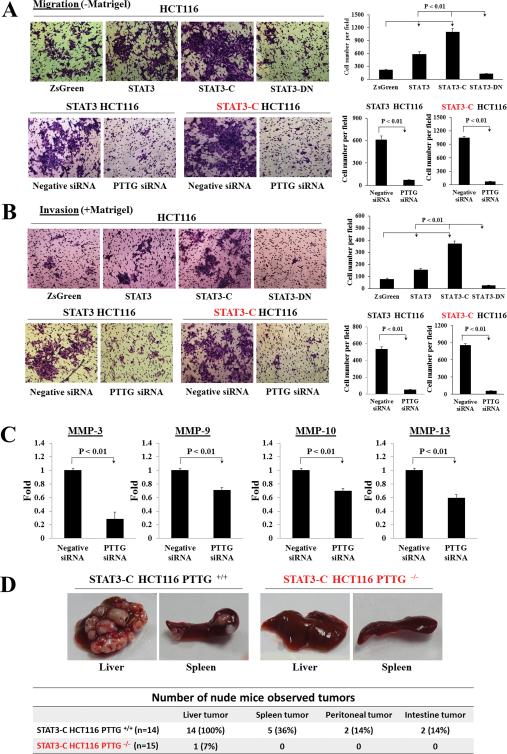
Using pIRES2-ZsGreen1 HCT116 stable transfectants as controls, we employed STAT3, STAT3-C, STAT3-DN HCT116 stable transfectants to measure in vitro cell migration and invasion. Elevated STAT3 expression or activation increased cell migration and invasion abilities (Fig. 6A, 6B). Overexpression of WT STAT3 increased cell migration 2.6-fold and enhanced invasion 2-fold (Fig. 6A-B). Both migration and invasion were further enhanced by activated-STAT3 (STAT3-C) up to 5-fold compared to pIRES2-ZsGreen1 controls, whereas cell mobility was strongly abrogated by STAT3-DN (>88%) (Fig. 6A-B). Moreover, we transfected PTTG siRNA and negative controls to STAT3 or STAT3-C stable transfectants respectively. Attenuating PTTG expression by siRNA strongly suppressed STAT3- or STAT3-C-induced cell migration and invasion by up to 90% (Fig. 6A-B). Impairing PTTG expression in STAT3-C stable transfectants reduced MMP-3 (70% reduction), MMP-9 (30% reduction), MMP-10 (30% reduction) and MMP-13 (40% reduction) expressions as assessed by real-time PCR (Figure 6C), indicating that inhibiting MMPs may underlie the constraint of STAT3-facilitated cell mobility.
Figure 6.
Attenuating PTTG expression decreases STAT3-promoted cell motility in vitro and tumor metastasis in vivo, and regulates MMPs expression. (A-B) In vitro migration (A) and invasion (B) assays of HCT116 stable transfectants. Upper panel: representative migration and invasion images of pIRES2-ZsGreen1 (ZsGreen), STAT3, STAT3-C, STAT3-DN HCT116 stable transfectants. Lower panels: representative migration and invasion pictures of PTTG siRNA transfected STAT3 or STAT3-C stable cells. Negative siRNA was used as control. Cells were counted manually and quantitative results presented as mean ± SE (n=5). Experiments were repeated three times. Picture magnification: 400X. (C) Real-time PCR of MMP-3, MMP-9, MMP-10, MMP-13 mRNA levels in PTTG siRNA transfected STAT3-C HCT116 cells, compared to negative siRNA controls. Results are presented as mean ± SE. Experiments were repeated three times. (D) Summary and representative images of in vivo tumor metastasis. Left: representative photographs of metastatic liver tumors and splenic tumors in STAT-C HCT116 PTTG+/+ implanted nude mice; right panel: representative photographs of liver and spleen exhibiting normal appearance in STAT-C HCT116 PTTG−/− control mice.
PTTG is required for STAT3-promoted tumor metastasis in vivo
To investigate PTTG roles in STAT3-promoted in vivo metastases, we implanted stable STAT3-C overexpressing HCT116 PTTG+/+ or HCT116 PTTG−/− cells into the spleen of nude mice. Mice were sacrificed 5 weeks later when weight loss was observed. As shown in Figure 6D, in the STAT3-C HCT116 PTTG+/+ implanted group (n=14), all 14 mice (100%) developed liver tumors (2-30 metastatic loci) and 5 of 14 mice (36%) harbored splenic tumors. In addition, two mice exhibited a peritoneal tumor, two showed intestine tumors, and one had ascites. In the STAT3-C HCT116 PTTG−/− control group (n=15), only one mouse (6.6%) exhibited small liver tumor foci, while 14 did not exhibit visible metastatic tumor. The incidence of tumor metastasis in the STAT3-C HCT116 PTTG+/+ group (100%) was higher than controls (6.6%) (P<0.01). These results support our in vitro cell motility findings and indicate that PTTG is required for STAT3-promoted tumor metastasis in vivo.
Discussion
Persistent activation or frequent overexpression of STAT3 and PTTG have been reported in several solid tumor types. Both STAT3 and PTTG have been implicated in multiple steps of cancer development and progression, but mechanisms elucidating their coordinated functions were not known. In the present study, we provide evidence that PTTG acts as a STAT3 target gene. Increased expression of STAT3/phosphor-STAT3 and PTTG were concordantly detected in human colorectal cancers (Figure 1). In support of this observation, STAT3 is shown to bind the human PTTG promoter, and activate PTTG transcription, while STAT3 induced PTTG mRNA and protein expression. Moreover, a dominant negative STAT3 mutant, by abolishing Tyr705 phosphorylation, specifically attenuated PTTG induction (Figure 3). This novel regulatory mechanism elucidating induction of intracellular PTTG expression expands our understanding of tumor PTTG abundance, and also extends our knowledge of STAT3 actions in tumor growth.
STAT3, which responds to several cytokines and growth factors, is a mediator of cell proliferation and also plays important roles in cellular transformation and tumor formation (1, 26). As a STAT3 downstream gene, oncogenic PTTG is involved in G2/M mitosis and enables cell proliferation and transformation. Transfecting PTTG siRNA into STAT3 or STAT3-C stable transfectants attenuated cell growth and transformation in vitro (Figure 4). Furthermore, using HCT116 cells devoid of PTTG, depletion of PTTG constrained xenografted tumor growth in vivo. As compared to STAT3-C HCT116 PTTG+/+ controls, xenografted tumors generated from STAT3-C HCT116 PTTG−/− cells exhibited 82% reduction of tumor weight. These results illustrating PTTG actions exemplify PTTG as a potential therapeutic target to abrogate tumor growth.
In addition, mechanisms underlying PTTG actions for STAT3-mediated tumor growth include STAT3 increased cyclin D3 and Kras expression. Attenuating PTTG expression by siRNA constrained expression of these pro-proliferative factors, and also increased tumor suppressor p21 expression. Several direct STAT3 targets are involved in proliferation and cell cycle progression, such as cyclin D1 (33) and c-Myc (32). Moreover, cyclin D3 (21), as well as c-Myc (20) are regulated by PTTG. In this study, we confirmed previous reports, wherein cyclin D3 was induced by STAT3 and suppressed by PTTG siRNA. Importantly, we observed that KRAS was regulated by STAT3-PTTG signaling in these colorectal cancer cells. KRAS is frequently mutated in several cancers (37), and mutant KRAS upregulates fibroblast genes involved in proliferation and angiogenesis (38). Our findings indicate that PTTG siRNA constrains cell transformation and tumor formation in STAT3 HCT116 stable transfectants, associated with decreased cyclin D3 and Kras expression.
STAT3 may be required for cancer cell migration and invasion (39-42), while overexpressed PTTG correlates with metastasis and poor survival in several cancers, and is a key signature gene predicting metastasis (17). Here we show that both migratory and invasive activities of HCT116 cells were induced by STAT3 overexpression and further enhanced by persistent STAT3 activation, whereas cell mobility was abrogated by dominant negative STAT3. Moreover, siRNA-mediated inhibition of endogenous PTTG expression suppressed STAT3-induced cell motility. Remarkably, PTTG depletion in STAT3-C HCT116 cells strongly abrogated tumor metastasis in vivo, suggesting that PTTG is required for metastasis, and attenuating PTTG expression, could be a novel approach to constrain STAT3-faciliated tumor progression.
MMPs, a family of zinc-dependent endopeptidases, contribute to tumor invasion and angiogenesis (43). High levels of distinct MMPs are associated with metastasis, and several cancer MMPs have been reported as STAT3 target genes (35, 44, 45), while PTTG also increases MMP-2 expression (46). Our results here suggest a role for PTTG in STAT3-facilitated tumor invasion, whereby STAT3 regulates PTTG expression to induce MMP expression and promote cell motility.
In summary, we provide evidence that PTTG acts as a STAT3 target gene. STAT3 induces PTTG expression and promotes cell transformation and motility. As attenuated PTTG constrained tumor development and progression, these findings expand our knowledge on regulatory mechanisms underlying tumor PTTG abundance and indicate PTTG as a potential therapeutic target to abrogate colorectal cancer growth.
Materials and methods
Immunofluorescence and Confocal Microscopy
Tissue-arrays of human colorectal cancer specimens were obtained from US Biomax (Cat #BC051110, Rockville, MD), and each pathologically confirmed. Slides were deparaffinized, hydrated, antigen retrieved and blocked as described (24). Slides were hybridized with antibodies against STAT3 (1:600, Cell Signaling #9139, Danvers, MA), phospho-STAT3 (1:300, Cell Signaling #9131) or PTTG (1:250, Abcam DCS-280, Cambridge, MA) at 4°C overnight. Alexa Fluor antibodies (Molecular Probes, Carlsbad, CA) were used as secondary antibody. Slides were mounted with ProLong Gold Antifade Reagent with DAPI (Life technologies, Grand Island, NY), and nuclei dyed by DAPI with blue fluorescence.
Samples were imaged with a Leica TCS/SP spectral confocal scanner (Leica Microsystems, Mannheim, Germany) in dual emission mode to separate autofluorescence from specific staining. For STAT3 and PTTG staining, a spectral window from 500 to 550 nm wavelength detected Alexa 488 emission appeared green. A second window from 560 nm to 620 nm detected autofluorescence contribution colored red. The two images were merged, so autofluorescence appears yellow, and true signals appear green. For double staining, PTTG stained with Alexa 488 were colored in green, and phospho-STAT3 stained with Alexa 568 were imaged with a 540-nm HeNe laser and colored in red.
Evaluation and Statistical Analysis
Immunofluorescence slides were independently examined by two blinded observers. Specimens staining for STAT3, Tyr705 phospho-STAT3 or PTTG were calculated as a percentage of positively stained cells. The average score of nine normal colon tissues was used as basal control, and compared with all tumors (scaled as 40% for STAT3, 19% for phospho-STAT3 and 4% for PTTG). STAT3 expression in tumor tissues was evaluated as: 1) no change, if score is ≤60%; 2) overexpression, if score is >60%. Tumor phospho-STAT3 expression was evaluated as: 1) no change, if score is ≤30%; 2) overexpression, if score is >30%. Tumor PTTG expression was evaluated as: 1) no change, if score is ≤10%; 2) overexpression, if score is >10%. STAT3, phospho-STAT3 and PTTG expression were correlated using Pearson Chi-Square Test with statistical software SPSS10.0 and P <0.05 was considered significant.
Cell Culture, Transfection and Stable Cell Selection
HCT116 cells were obtained from the American Tissue Culture Collection (ATCC, Manassas, VA). HCT116 PTTG+/+ and HCT116 PTTG−/− cells were kindly provided by Dr. Bert Vogelstein, Johns Hopkins University (Baltimore, MD). Generation of HCT116 PTTG+/+ and PTTG−/− cells is described (5). Cell transfection was performed in 70%-80% confluent cells using Lipofectamine 2000 (Life technologies) according to the manufacturer's protocol. G418 (0.5 mg/ml) was added for 14 days to select stable cells. Stable HCT116 transfectants expressed green fluorescent protein ZsGreen. A mass population of green stable transfectants was sorted by MoFlo Cell Sorter to ensure expression prior to in vitro and in vivo experiments.
Chromatin Immunoprecipitation (ChIP)
Ten million cells were cross-linked and lysed using ChIP-IT express kit (Active Motif, Carlsbad, CA). Chromatin was sonicated to 200-800 bp length fragments with eight rounds of 10-second pulses using 25% power. Normalized inputs of sheared chromatin DNA were incubated with 4 μg negative control IgG, STAT3 (Cell Signaling #9139) or phospho-STAT3 antibody (Cell Signaling #9131) overnight at 4°C. Real-time PCR reactions were amplified using precipitated immunocomplexes as template, and the PTTG promoter primers described (24). Human c-Fos promoter primers were used as positive controls (Cell Signaling #4663) and α-satellite repeat primers (Cell Signaling #4486) as negative controls.
Plasmids and siRNA
PTTG promoters, −2642/−1 and −1717/−1, were cloned into pGL3-Basic luciferase reporter vector (Promega, Madison, WI) as described (36). STAT3 wild type expression plasmid was amplified from MHS1011-76652 clone (Open Biosystems, Lafayette, CO) using TaKaRa LA Taq (Clontech, Mountain View, CA), and cloned into the pIRES2-ZsGreen1 vector (Clontech). The following primers were used: Forward: 5'-CCGCTCGAGA CCATGGCCCA ATGGAATCAG CTACAGCA-3'; Reverse: 5'-TCCCCGCGGT CACATGGGGG AGGTAGCGCA CTCCGA-3'. The STAT3-C construct (27) was made by site-directed mutagenesis (QuikChange II Site-Directed Mutagenesis Kit, Agilent Technologies) using primers 5'-GGGCTATAAG ATCATGGATT GTACCTGCAT CCTGGTGTCT CCACTGG-3' and 5'-CCAGTGGAGA CACCAGGATG CAGGTACAAT CCATGATCTT ATAGCCC-3'. The STAT3-DN construct was mutated by primers 5'-CCAGGTAGCG CTGCCCCATT CCTGAAGACC-3' and 5'-GGTCTTCAGG AATGGGGCAG CGCTACCTGG-3', to replace tyrosine at position 705 to phenylalanine. Plasmids were sequenced by Sequetech. Pre-designed PTTG siRNA (ID#4390824) and negative control siRNA (Cat#AM4611) were obtained from Ambion (Grand Island, NY).
Reporter Assay
Cells were split into 24-well plates, and each well co-transfected with 200 ng luciferase vector pGL3-Basic as control, PTTG promoter −2642/−1 or −1717/−1, together with 800 ng pIRES2-ZsGreen1 as control, STAT3 or STAT3-C plasmids. pRL-Tk (Promega) encoding Renilla luciferase was used as an internal control (5 ng/well) to assess transfection efficiency. For STAT3 inhibitors S3I-201 and Stattic (EMD Millipore, Billerica, MA) treatment, cells were transfected with 1000 ng pGL3-Basic or PTTG promoter (−2642/−1 or −1717/−1), and subsequently treated with STAT3 inhibitors. After 24 hours, whole-cell lysate was collected for reporter detection by the Dual Luciferase Reporter System (Promega). Reactions were measured using an Orion Microplate Luminometer (Berthold Detection System). Transfections were performed in triplicate, and repeated three times to assure reproducibility.
RNA Extraction and Real-time PCR
Total RNA was isolated using TRIZOL Reagent (Life technologies). Two micrograms of total RNA were used to synthesize cDNA with SuperScript II Reverse Transcriptase (Life technologies). Real-time PCR was amplified in 20 μl reaction mixtures (100 ng template, 0.5 μM of each primer, 10 μl 2X SYBR GREEN Master Mix (Life technologies)) using the following parameters: 95°C for 1 min, followed by 40 cycles of 95°C for 20 seconds, 60°C for 40 seconds. β-actin was used as internal control. Real-time PCR primers were designed as follows. PTTG Forward: 5’-TGATCCTTGA CGAGGAGAGAG-3’; Reverse: 5’-GGTGGCAATT CAACATCCAGG-3’. KRAS Forward: 5’-TTGAACTAGC AATGCCTGTG-3’; Reverse: 5’-ACCAATTAGA AGGTCTCAACTG-3’. CCND3 Forward: 5’-CAGATGTCAC AGCCATAC-3’; Reverse: 5’-GATGGGTAGG ACCAGATC-3’. CDKN1A Forward: 5’-GCTCTACATC TTCTGCCTTA GTC-3’; Reverse: 5’-ACCTCTCATT CAACCGCCTAG-3’. MMP-3 Forward: 5’-GGTCTCTTTC ACTCAGCCAA CAC-3’; Reverse: 5’-CAGGCGGAAC CGAGTCAGG-3’. MMP-13 Forward: 5’-CCTTGATGCC ATTACCAGTCTC-3’; Reverse: 5’-TCAATACGGT TGGGAAGTTCTG-3’. β-actin Forward: 5’-CATGTACGTT GCTATCCAGGC-3’; Reverse: 5’-CTCCTTAATG TCACGCACGAT-3’. MMP-9 and MMP-10 primers were purchased from SABioscienses (#PPH00152E and # PPH00896B, Valencia, CA).
Western Blot
Western blots were performed as described (24). Primary antibodies used were: STAT3: 1:1000, Cell Signaling #9139; phospho-STAT3: 1:1000, Cell Signaling #9131; PTTG: 1:500, Abcam DCS-280; cyclin D3: 1:500, Santa Cruz sc-182; cK Ras: 1:1000, Abcam ab84573; β-actin: 1:20000, Millipore.
Cell Proliferation and Soft Agar Colony Formation Assay
STAT3 HCT116 stable transfectants were separately transfected with PTTG siRNA or negative control. After 24 hours, cells (104 cells/well in 100 μl medium) were plated in flat-bottomed 96-well plates. Each group (PTTG siRNA or negative control) consisted of eight parallel wells. After 72 hours, premixed WST-1 cell proliferation reagent (Clontech) was added (1:10) and incubated for 4 h at 37 C in a humidified atmosphere maintained at 5% CO2, after which absorbance was measured at 450 nm.
STAT3 or STAT3-C HCT116 stable transfectants were separately transfected with PTTG siRNA or negative control. One thousand cells were plated in the top layer containing 0.3% agarose, and the bottom support layer comprised 0.6% agarose. Cells were stained 10 days later with 0.2% iodonitrotetrazolium chloride (Life technologies) and photographed.
In Vitro Migration and Invasion Assay
Cell migration and invasion assays were performed in 6.5 mm Transwells (Corning #3422, Corning, NY). Cells (2*105) were suspended in 100 μl serum-free medium, added to the upper chamber and the lower chamber was filled with complete medium with 10% serum. Cells were migrated at 37 C for 48 hours. After removing non-migrated cells, membranes were fixed in methanol and stained with 0.05% crystal violet. Migrated cells were photographed and quantified in 5 random fields per membrane. Each sample was assayed in triplicate. A similar system, with Matrigel-coated membranes, was used for assessing invasion.
Subcutaneously Xenografted Tumor Growth
Animal protocols were approved by the Cedars-Sinai Institutional Animal Care and Use Committee. Forty female nude mice (5- to 6-week-old, obtained from Jackson Laboratory) were randomly separated into two groups. One group was injected subcutaneously with STAT-C HCT116 PTTG+/+ stable transfectants and the second group injected with STAT-C HCT116 PTTG−/− stable transfectants (106 cells/100μl Matrigel per animal), into the left lumbar area to generate tumors. Tumor size was measured by caliper measurements twice a week when tumors became visible, and volume calculated with the formula (Length * Width2) /2. Seventeen days after cell injection, animals were euthanized and excised tumors weighed.
Experimental metastasis in vivo
Animal protocols were approved by the Cedars-Sinai Institutional Animal Care and Use Committee. STAT-C HCT116 PTTG+/+ or STAT-C HCT116 PTTG−/− stable cells (106 cells/50μl PBS per animal) were injected into the spleen of 6-week-old female nude mice. Each group included 14-15 animals. Mice were weighed once a week and metastasis development monitored. Five weeks later, mice were sacrificed when weight loss or any adverse effect was observed. Liver, spleen and other organs were visibly observed for tumor metastasis. Tumors were harvested and metastatic loci counted.
Acknowledgments
The authors are grateful to Dr. Bert Vogelstein at Johns Hopkins University (Baltimore, MD) for kindly providing HCT116 PTTG+/+ and HCT116 PTTG−/− cells; and thank Patricia Lin at the Cedars-Sinai Flow Cytometry Core. This work was supported by National Institutes of Health Grant CA75979 (to S.M.) and the Doris Factor Molecular Endocrinology Laboratory.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 2.Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 3.Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- 4.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 5.Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 6.Romero F, Multon MC, Ramos-Morales F, Dominguez A, Bernal JA, Pintor-Toro JA, et al. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 2001;29:1300–1307. doi: 10.1093/nar/29.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero F, Gil-Bernabe AM, Saez C, Japon MA, Pintor-Toro JA, Tortolero M. Securin is a target of the UV response pathway in mammalian cells. Mol Cell Biol. 2004;24:2720–2733. doi: 10.1128/MCB.24.7.2720-2733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD, et al. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol Endocrinol. 1999;13:156–166. doi: 10.1210/mend.13.1.0225. [DOI] [PubMed] [Google Scholar]
- 9.Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Horwitz GA, Heaney AP, Nakashima M, Prezant TR, Bronstein MD, et al. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab. 1999;84:761–767. doi: 10.1210/jcem.84.2.5432. [DOI] [PubMed] [Google Scholar]
- 11.Filippella M, Galland F, Kujas M, Young J, Faggiano A, Lombardi G, et al. Pituitary tumour transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas: a clinical and immunohistochemical study. Clin Endocrinol (Oxf) 2006;65:536–543. doi: 10.1111/j.1365-2265.2006.02630.x. [DOI] [PubMed] [Google Scholar]
- 12.Boelaert K, McCabe CJ, Tannahill LA, Gittoes NJ, Holder RL, Watkinson JC, et al. Pituitary tumor transforming gene and fibroblast growth factor-2 expression: potential prognostic indicators in differentiated thyroid cancer. J Clin Endocrinol Metab. 2003;88:2341–2347. doi: 10.1210/jc.2002-021113. [DOI] [PubMed] [Google Scholar]
- 13.Ogbagabriel S, Fernando M, Waldman FM, Bose S, Heaney AP. Securin is overexpressed in breast cancer. Mod Pathol. 2005;18:985–990. doi: 10.1038/modpathol.3800382. [DOI] [PubMed] [Google Scholar]
- 14.Zhou C, Liu S, Zhou X, Xue L, Quan L, Lu N, et al. Overexpression of human pituitary tumor transforming gene (hPTTG), is regulated by beta-catenin /TCF pathway in human esophageal squamous cell carcinoma. Int J Cancer. 2005;113:891–898. doi: 10.1002/ijc.20642. [DOI] [PubMed] [Google Scholar]
- 15.Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008;15:721–743. doi: 10.1677/ERC-08-0012. [DOI] [PubMed] [Google Scholar]
- 16.Panguluri SK, Yeakel C, Kakar SS. PTTG: an important target gene for ovarian cancer therapy. J Ovarian Res. 2008;1:6. doi: 10.1186/1757-2215-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 18.Yan S, Zhou C, Lou X, Xiao Z, Zhu H, Wang Q, et al. PTTG overexpression promotes lymph node metastasis in human esophageal squamous cell carcinoma. Cancer Res. 2009;69:3283–3290. doi: 10.1158/0008-5472.CAN-08-0367. [DOI] [PubMed] [Google Scholar]
- 19.Yu R, Lu W, Chen J, McCabe CJ, Melmed S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology. 2003;144:4991–4998. doi: 10.1210/en.2003-0305. [DOI] [PubMed] [Google Scholar]
- 20.Pei L. Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. J Biol Chem. 2001;276:8484–8491. doi: 10.1074/jbc.M009654200. [DOI] [PubMed] [Google Scholar]
- 21.Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim CS, Ying H, Willingham MC, Cheng SY. The pituitary tumor-transforming gene promotes angiogenesis in a mouse model of follicular thyroid cancer. Carcinogenesis. 2007;28:932–939. doi: 10.1093/carcin/bgl231. [DOI] [PubMed] [Google Scholar]
- 23.Hamid T, Malik MT, Kakar SS. Ectopic expression of PTTG1/securin promotes tumorigenesis in human embryonic kidney cells. Mol Cancer. 2005;4:3. doi: 10.1186/1476-4598-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou C, Wawrowsky K, Bannykh S, Gutman S, Melmed S. E2F1 induces pituitary tumor transforming gene (PTTG1) expression in human pituitary tumors. Mol Endocrinol. 2009;23:2000–2012. doi: 10.1210/me.2009-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasse J, Hemmann U, Schwartz C, Schniertshauer U, Heesel B, Landgraf C, et al. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol Cell Biol. 1997;17:4677–4686. doi: 10.1128/mcb.17.8.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17:1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 30.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 34.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 35.Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 36.Zhou C, Tong Y, Wawrowsky K, Bannykh S, Donangelo I, Melmed S. Oct-1 induces pituitary tumor transforming gene expression in endocrine tumors. Endocr Relat Cancer. 2008;15:817–831. doi: 10.1677/ERC-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wicki A, Herrmann R, Christofori G. Kras in metastatic colorectal cancer. Swiss Med Wkly. 2010;140:w13112. doi: 10.4414/smw.2010.13112. [DOI] [PubMed] [Google Scholar]
- 38.Horsch M, Recktenwald CV, Schadler S, Hrabe de Angelis M, Seliger B, Beckers J. Overexpressed vs mutated Kras in murine fibroblasts: a molecular phenotyping study. Br J Cancer. 2009;100:656–662. doi: 10.1038/sj.bjc.6604882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. Embo J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, Inghirami G, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 41.Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 42.Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. Faseb J. 1999;13:781–792. [PubMed] [Google Scholar]
- 44.Song Y, Qian L, Song S, Chen L, Zhang Y, Yuan G, et al. Fra-1 and Stat3 synergistically regulate activation of human MMP-9 gene. Mol Immunol. 2008;45:137–143. doi: 10.1016/j.molimm.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Tsareva SA, Moriggl R, Corvinus FM, Wiederanders B, Schutz A, Kovacic B, et al. Signal transducer and activator of transcription 3 activation promotes invasive growth of colon carcinomas through matrix metalloproteinase induction. Neoplasia. 2007;9:279–291. doi: 10.1593/neo.06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik MT, Kakar SS. Regulation of angiogenesis and invasion by human Pituitary tumor transforming gene (PTTG) through increased expression and secretion of matrix metalloproteinase-2 (MMP-2). Mol Cancer. 2006;5:61. doi: 10.1186/1476-4598-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]