Abstract
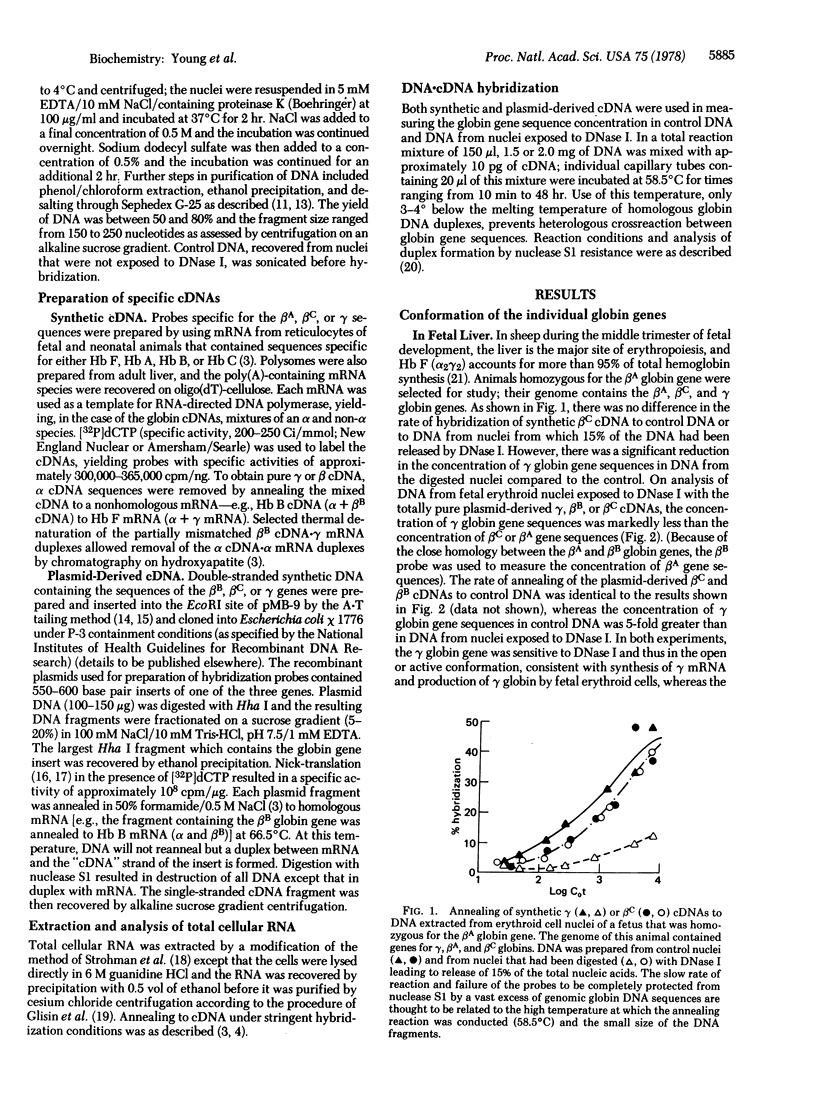
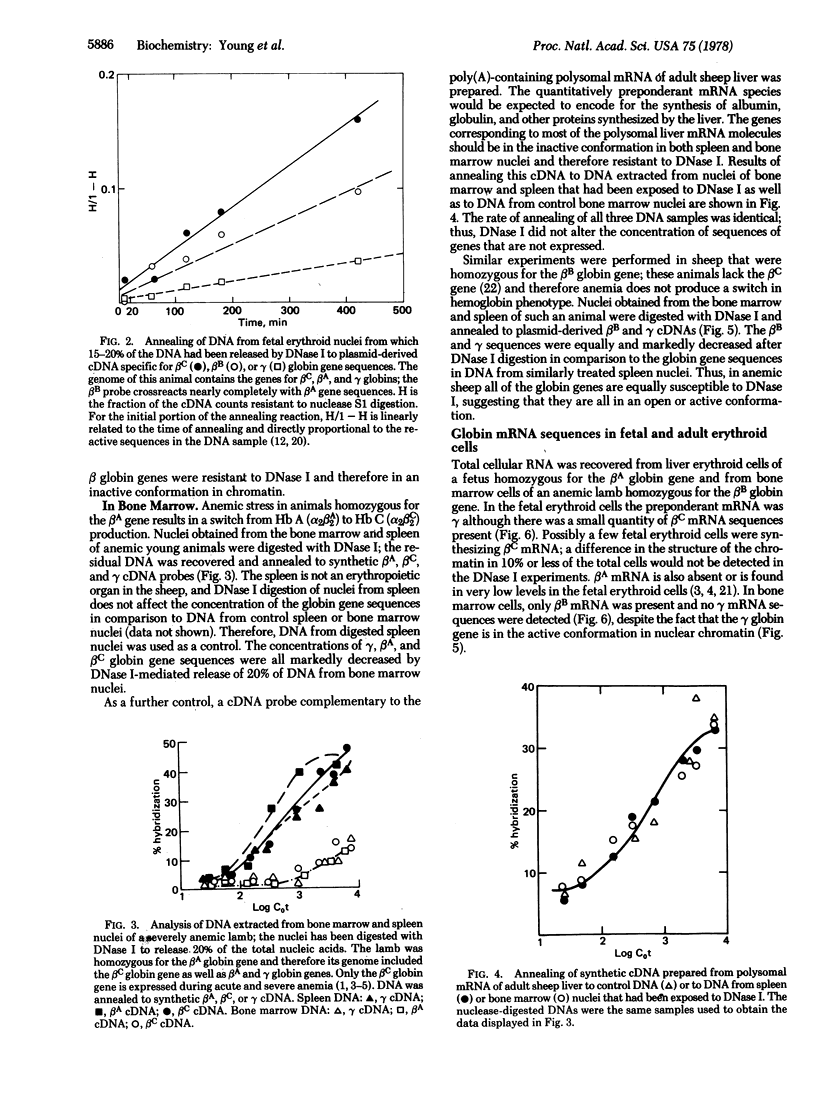
Differential expression of the closely linked γ, βA (or βB), and βC globin genes in sheep results in the production of fetal hemoglobin (Hb F, α2γ2) during gestation and the adult hemoglobins (Hb A, α2β2A, and Hb B, α2β2B) after birth. Erythropoietic stress in certain animals leads to production of Hb C (α2β2C). The molecular mechanism of differential expression of these genes in nuclei of fetal and adult erythroid cells has been investigated by analysis of their susceptibility to digestion by DNase I (genes that are in the conformation associated with active transcription are sensitive to this nuclease). The concentration of globin gene sequences in DNA from control and DNase I-digested nuclei was determined by annealing to synthetic DNAs and analogous cDNA probes derived from recombinant plasmids containing one of the sheep globin genes. In nuclei from sheep fetal liver erythroid cells, the γ genes but not the β genes were digested by DNase I; the γ locus was open but the βA or βC loci was closed, consistent with synthesis of only Hb F by these cells. DNase I digestion of nuclei from bone marrow of anemic sheep making only Hb C or Hb B resulted in equivalent digestion of the β and γ gene sequences, although γ mRNA was not detected in these cells. Digestion by DNase I did not decrease the globin gene sequence concentration in residual DNA of spleen nuclei. As a further control, DNA from digested bone marrow and spleen nuclei were shown to anneal equally well to a cDNA prepared from liver polysomal mRNA. Differential expression of the γ and β globin genes in sheep fetal erythroid cell appears to be based on differences in chromatin structure. The γ globin gene remains in the active conformation in adult erythroid cells; failure of γ mRNA to accumulate in these cells probably reflects transcriptional or post-transcriptional regulation.
Keywords: pancreatic DNase I, complementary DNA, recombinant plasmids, messenger RNA, erythroid cells
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker J. E., Pierce J. E., Kefauver B. C., Nienhuis A. W. Hemoglobin switching in sheep and goats: induction of hemoglobin C synthesis in cultures of sheep fetal erythroid cells. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5078–5082. doi: 10.1073/pnas.74.11.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz E. J., Geist C. E., Steggles A. W., Barker J. E., Nienhuis A. W. Hemoglobin switching in sheep and goats. Preparation and characterization of complementary DNAs specific for the alpha-, beta-, and gamma-globin messenger RNAs of sheep. J Biol Chem. 1977 Mar 25;252(6):1908–1916. [PubMed] [Google Scholar]
- Benz E. J., Jr, Barker J. E., Pierce J. E., Turner P. A., Nienhuis A. W. Hemoglobin switching in sheep: commitment of erythroid stem cells to expression of the betaC-globin gene and accumulation of betaC-globin mRNA. Cell. 1978 Jul;14(3):733–740. doi: 10.1016/0092-8674(78)90255-6. [DOI] [PubMed] [Google Scholar]
- Benz E. J., Jr, Steggles A. W., Geist C. E., Nienhuis A. W. Hemoglobin switching in sheep. Quantitation of betaA- and betaC-mRNA sequences in nuclear and cytoplasmic RNA during the HbA to HbC switch. J Biol Chem. 1978 Jul 25;253(14):5025–5032. [PubMed] [Google Scholar]
- Benz E., Jr, Turner P., Barker J., Nienhuis A. Stability of the individual globin genes during erythroid differentiation. Science. 1977 Jun 10;196(4295):1213–1214. doi: 10.1126/science.860136. [DOI] [PubMed] [Google Scholar]
- Billing R. J., Bonner J. The structure of chromatin as revealed by deoxyribonuclease digestion studies. Biochim Biophys Acta. 1972 Oct 27;281(3):453–462. doi: 10.1016/0005-2787(72)90462-5. [DOI] [PubMed] [Google Scholar]
- Deisseroth A., Nienhuis A., Turner P., Velez R., Anderson W. F., Ruddle F., Lawrence J., Creagan R., Kucherlapati R. Localization of the human alpha-globin structural gene to chromosome 16 in somatic cell hybrids by molecular hybridization assay. Cell. 1977 Sep;12(1):205–218. doi: 10.1016/0092-8674(77)90198-2. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Weintraub H. M. An altered subunit configuration associated with the actively transcribed DNA of integrated adenovirus genes. Cell. 1977 Nov;12(3):783–794. doi: 10.1016/0092-8674(77)90277-x. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Paddock G. V., Wall R., Salser W. A general method for cloning eukaryotic structural gene sequences. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3146–3150. doi: 10.1073/pnas.73.9.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Levy W B., Wong N. C., Dixon G. H. Selective association of the trout-specific H6 protein with chromatin regions susceptible to DNase I and DNase II: possible location of HMG-T in the spacer region between core nucleosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2810–2814. doi: 10.1073/pnas.74.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Kee S. G., Efstratiadis A., Kafatos F. C. Amplification and characterization of a beta-globin gene synthesized in vitro. Cell. 1976 Jun;8(2):163–182. doi: 10.1016/0092-8674(76)90001-5. [DOI] [PubMed] [Google Scholar]
- Miller D. M., Turner P., Nienhuis A. W., Axelrod D. E., Gopalakrishnan T. V. Active conformation of the globin genes in uninduced and induced mouse erythroleukemia cells. Cell. 1978 Jul;14(3):511–521. doi: 10.1016/0092-8674(78)90237-4. [DOI] [PubMed] [Google Scholar]
- Nienhuis A. W., Benz E. J., Jr Regulation of hemoglobin synthesis during the development of the red cell (third of three parts). N Engl J Med. 1977 Dec 29;297(26):1430–1436. doi: 10.1056/NEJM197712292972604. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panet A., Cedar H. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell. 1977 Aug;11(4):933–940. doi: 10.1016/0092-8674(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977 Feb;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Vidali G., Boffa L. C., Allfrey V. G. Selective release of chromosomal proteins during limited DNAase 1 digestion of avian erythrocyte chromatin. Cell. 1977 Oct;12(2):409–415. doi: 10.1016/0092-8674(77)90117-9. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]


