Abstract
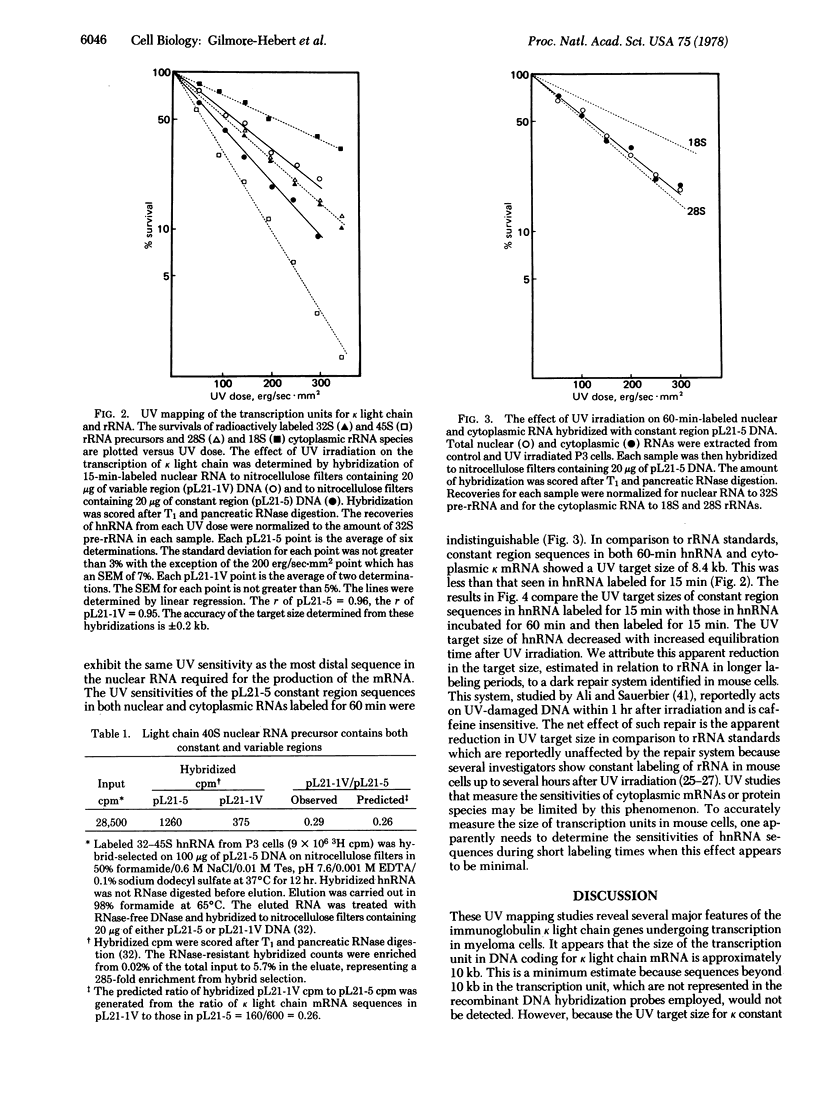
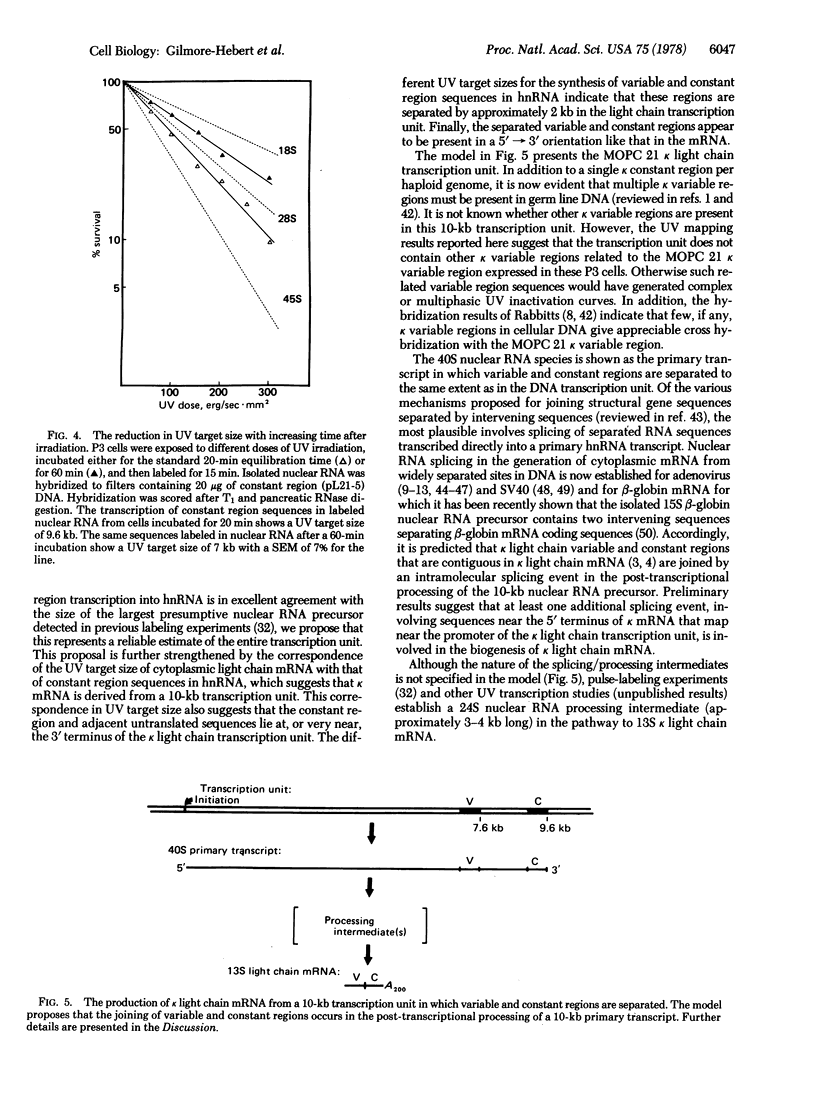
UV transcription mapping with recombinant DNA probes containing immunoglobulin kappa light chain mRNA sequences has been used to determine the size of the transcription unit coding for kappa light chain m RNA and to establish the arrangement of variable and constant regions in this transcription unit. In relation to ribosomal RNA standards, the transcription of kappa light chain constant region sequences into nuclear RNA exhibits a UV target size of 9.6 kbases (kb). The kappa light chain variable region exhibits a UV target size of 7.6 kb indicating that it is separated by approximately 2.0 kb from the constant region in the kappa light chain transcription unit. The size of the primary transcript (i.e., the direct, unprocessed RNA product of transcription) predicted from the constant region target size concurs with our previous pulse-labeling results which showed that the largest presumptive nuclear RNA precursor to kappa light chain mRNA is approximately 10 kb. In addition, the UV target size of cytoplasmic kappa mRNA is indistinguishable from the target size of constant region sequences in nuclear RNA. These results suggest that the kappa light chain transcription unit is copied directly into a 10-kb nuclear RNA precursor in which the kappa variable and constant regions are separated by approximately 2 kb. Accordingly, it is proposed that the joining of immunoglobulin kappa light chain variable and constant regions occurs in the post-transcriptional processing of this large nuclear RNA precursor into kappa light chain mRNA.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ali R., Sauerbier W. Effects of ultraviolet irradiation and postirradiation incubation on heterogenous nuclear RNA size in murine cells. Biophys J. 1978 Jun;22(3):393–411. doi: 10.1016/s0006-3495(78)85495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y., Dhar R., Laub O., Horowitz M., Khoury G. Novel mechanism for RNA maturation: the leader sequences of simian virus 40 mRNA are not transcribed adjacent to the coding sequences. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3686–3690. doi: 10.1073/pnas.74.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Ultraviolet mapping of the adenovirus 2 early promoters. Cell. 1977 Sep;12(1):45–55. doi: 10.1016/0092-8674(77)90184-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brack C., Tonegawa S. Variable and constant parts of the immunoglobulin light chain gene of a mouse myeloma cell are 1250 nontranslated bases apart. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5652–5656. doi: 10.1073/pnas.74.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Mandel J. L., Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977 Nov 24;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- Carlson J., Ott G., Sauerbier W. Transcriptional organization of the Drosophila melanogaster ribosomal RNA genes. J Mol Biol. 1977 May 15;112(2):353–357. doi: 10.1016/s0022-2836(77)80151-4. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Cory S., Adams J. M. A very large repeating unit of mouse DNA containing the 18S, 28S and 5.8S rRNA genes. Cell. 1977 Aug;11(4):795–805. doi: 10.1016/0092-8674(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Dunn A. R., Hassell J. A. A novel method to map transcripts: evidence for homology between an adenovirus mRNA and discrete multiple regions of the viral genome. Cell. 1977 Sep;12(1):23–36. doi: 10.1016/0092-8674(77)90182-9. [DOI] [PubMed] [Google Scholar]
- Gilmore-Hebert M., Wall R. Immunoglobulin light chain mRNA is processed from large nuclear RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):342–345. doi: 10.1073/pnas.75.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R., Sauerbier W. A radiological analysis of the transcription units for heterogeneous nuclear RNA in cultured murine cells. Cell. 1976 Dec;9(4 Pt 2):775–783. doi: 10.1016/0092-8674(76)90140-9. [DOI] [PubMed] [Google Scholar]
- Goldberg S., Nevins J., Darnell J. E. Evidence from UV transcription mapping that late adenovirus type 2 mRNA is derived from a large precursor molecule. J Virol. 1978 Mar;25(3):806–810. doi: 10.1128/jvi.25.3.806-810.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg S., Schwartz H., Darnell J. E., Jr Evidence from UV transcription mapping in HeLa cells that heterogeneous nuclear RNA is the messenger RNA precursor. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4520–4523. doi: 10.1073/pnas.74.10.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg S., Weber J., Darnell J. E., Jr The definition of a large viral transcription unit late in Ad2 infection of HeLa cells: mapping by effects of ultraviolet irradiation. Cell. 1977 Apr;10(4):617–621. doi: 10.1016/0092-8674(77)90094-0. [DOI] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett P. B., Sauerbier W. The transcriptional organization of the ribosomal RNA genes in mouse L cells. J Mol Biol. 1975 Jan 25;91(3):235–256. doi: 10.1016/0022-2836(75)90378-2. [DOI] [PubMed] [Google Scholar]
- Higuchi R., Paddock G. V., Wall R., Salser W. A general method for cloning eukaryotic structural gene sequences. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3146–3150. doi: 10.1073/pnas.73.9.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L., Campbell J. H., Elgin S. C. The organization, expression, and evolution of antibody genes and other multigene families. Annu Rev Genet. 1975;9:305–353. doi: 10.1146/annurev.ge.09.120175.001513. [DOI] [PubMed] [Google Scholar]
- Hozumi N., Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. T., Ford J. Sequence arrangement of the 5' ends of simian virus 40 16S and 19S mRNAs. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4982–4985. doi: 10.1073/pnas.74.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. The rabbit beta-globin gene contains a large large insert in the coding sequence. Cell. 1977 Dec;12(4):1097–1108. doi: 10.1016/0092-8674(77)90172-6. [DOI] [PubMed] [Google Scholar]
- Klessig D. F. Two adenovirus mRNAs have a common 5' terminal leader sequence encoded at least 10 kb upstream from their main coding regions. Cell. 1977 Sep;12(1):9–21. doi: 10.1016/0092-8674(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Lewis J. B., Anderson C. W., Atkins J. F. Further mapping of late adenovirus genes by cell-free translation of RNA selected by hybridization to specific DNA fragments. Cell. 1977 Sep;12(1):37–44. doi: 10.1016/0092-8674(77)90183-0. [DOI] [PubMed] [Google Scholar]
- McGrogan M., Raskas H. J. Two regions of the adenovirus 2 genome specify families of late polysomal RNAs containing common sequences. Proc Natl Acad Sci U S A. 1978 Feb;75(2):625–629. doi: 10.1073/pnas.75.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C., Brownlee G. G., Cartwright E. M., Jarvis J. M., Proudfoot N. J. Sequence analysis of immunoglobulin light chain messenger RNA. Nature. 1974 Nov 29;252(5482):354–359. doi: 10.1038/252354a0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E. Groups of adenovirus type 2 mRNA's derived from a large primary transcript: probable nuclear origin and possible common 3' ends. J Virol. 1978 Mar;25(3):811–823. doi: 10.1128/jvi.25.3.811-823.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H. A molecular hybridization approach for the determination of the immunoglobulin V-gene pool size. Immunol Rev. 1977;36:29–50. doi: 10.1111/j.1600-065x.1977.tb00381.x. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H., Forster A. Evidence for noncontiguous variable and constant region genes in both germ line and myeloma DNA. Cell. 1978 Feb;13(2):319–327. doi: 10.1016/0092-8674(78)90200-3. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Edgell M. H., Leder P. Immunoglobulin light-chain structural gene sequences cloned in a bacterial plasmid. Nature. 1978 Feb 9;271(5645):582–585. doi: 10.1038/271582a0. [DOI] [PubMed] [Google Scholar]
- Svasti J., Milstein C. The complete amino acid sequence of a mouse kappa light chain. Biochem J. 1972 Jun;128(2):427–444. doi: 10.1042/bj1280427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Polsky F. I., Seidman J. G., Leder A., Edgell M. H., Leder P. A comparison of two cloned mouse beta-globin genes and their surrounding and intervening sequences. Cell. 1978 Jun;14(2):237–245. doi: 10.1016/0092-8674(78)90110-1. [DOI] [PubMed] [Google Scholar]
- Tilghman S. M., Curtis P. J., Tiemeier D. C., Leder P., Weissmann C. The intervening sequence of a mouse beta-globin gene is transcribed within the 15S beta-globin mRNA precursor. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1309–1313. doi: 10.1073/pnas.75.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M., Tiemeier D. C., Seidman J. G., Peterlin B. M., Sullivan M., Maizel J. V., Leder P. Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene. Proc Natl Acad Sci U S A. 1978 Feb;75(2):725–729. doi: 10.1073/pnas.75.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S., Hozumi N., Matthyssens G., Schuller R. Somatic changes in the content and context of immunoglobulin genes. Cold Spring Harb Symp Quant Biol. 1977;41(Pt 2):877–889. doi: 10.1101/sqb.1977.041.01.097. [DOI] [PubMed] [Google Scholar]
- Valenzuela P., Venegas A., Weinberg F., Bishop R., Rutter W. J. Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):190–194. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall R., Gilmore-Hebert M., Higuchi R., Komaromy M., Paddock G., Strommer J., Salser W. Recombinant DNA clones constructed from immunoglobulin kappa light chain messenger RNA. Nucleic Acids Res. 1978 Sep;5(9):3113–3128. doi: 10.1093/nar/5.9.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall R., Lippman S., Toth K., Fedoroff N. A general method for the large-scale isolation of polysomes and messenger RNA applied to MOPC 21 mouse myeloma tumors. Anal Biochem. 1977 Sep;82(1):115–129. doi: 10.1016/0003-2697(77)90140-3. [DOI] [PubMed] [Google Scholar]
- Weinstock R., Sweet R., Weiss M., Cedar H., Axel R. Intragenic DNA spacers interrupt the ovalbumin gene. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1299–1303. doi: 10.1073/pnas.75.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Ziff E., Fraser N. Adenovirus type 2 late mRNA's: structural evidence for 3'-coterminal species. J Virol. 1978 Mar;25(3):897–906. doi: 10.1128/jvi.25.3.897-906.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]


