SUMMARY
Misfolded proteins are often cytotoxic, unless cellular systems prevent their accumulation. Data presented here uncover a novel mechanism by which defects in secretory proteins lead to a dramatic reduction in their mRNAs and protein expression. When mutant signal sequences fail to bind to the signal recognition particle (SRP) at the ribosome exit site, the nascent chain instead contacts Argonaute2 (Ago2) and the mutant mRNAs are specifically degraded. Severity of signal sequence mutations correlated with increased proximity of Ago2 to nascent chain, and mRNA degradation. Ago2 knockdown inhibited degradation of the mutant mRNA, while overexpression of Ago2 or knockdown of SRP54 promoted degradation of secretory protein mRNA. The results reveal a previously unappreciated general mechanism of translational quality control, in which specific mRNA degradation preemptively regulates aberrant protein production (RAPP).
INTRODUCTION
Misfolded and mistargeted proteins are often cytotoxic, unless cellular systems prevent their accumulation (Stefani and Dobson, 2003). Cells have strict quality-control systems in the cytosol and in the endoplasmic reticulum (ER) to distinguish properly folded proteins from misfolded (Buchberger et al., 2010; Ellgaard and Helenius, 2003). Secretory and membrane proteins that are not transported to their proper cellular destinations are either degraded by the ER associated degradation (ERAD) pathway (Brodsky, 2012; Smith et al., 2011) or form inclusions (Kopito, 2000). When proteins are successfully translocated from the cytosol to the ER, but are not transported further due to misfolding in stress conditions, the unfolded protein response (UPR) is activated. UPR involves activation of stress response genes, general down-regulation of protein synthesis, and degradation of misfolded ER proteins (Schroder and Kaufman, 2005; Walter and Ron, 2011). Another mechanism that prevents accumulation of misfolded proteins in the ER during UPR is Regulated Ire1-Dependent Decay (RIDD) (Hollien et al., 2009; Hollien and Weissman, 2006). This mechanism is activated under conditions of general ER stress and reduces the quantity of many mRNAs that encode secretory proteins. Thus, these processes are general responses to stress conditions and have not evolved to specifically triage an individual mutant protein that misfolds. Whether cells can specifically prevent the synthesis of “unwanted”, and potentially hazardous proteins derived from mutated or foreign mRNAs is only poorly understood. If mutant proteins do not fold properly or fail to be delivered to the appropriate cellular compartment, they can be removed by the proteasome system (Ward et al., 1995). Such a post-translational mechanism, however, does not prevent continued de novo production of the mutant gene product. Currently known systems of mRNA quality control, nonsense mediated decay (NMD), nonstop decay (NSD), and no-go decay (NGD), detect only profound defects, including premature stop codons, lack of a natural stop codon, or stalled ribosomes (Doma and Parker, 2007; Shoemaker and Green, 2012). These quality control mechanisms are not known to involve sensors that detect misfolded nascent chains or target mRNA templates whose products are not properly folded/targeted because of missense mutations.
We hypothesized that when an individual protein misfolds or fails to be correctly sorted to the appropriate compartment due to alterations in its protein sequence, processes that specifically reduce its translation could provide an incisive means of reducing potential toxicity caused by the accumulation of misfolded conformers. Here we demonstrate the existence of such a mechanism for specific mRNA degradation in response to the synthesis of defective proteins.
RESULTS
Mutations in the Signal Sequence of a Secretory Protein Cause a Defect in Protein Transport and Lead to Decreased Production of Mutant Proteins
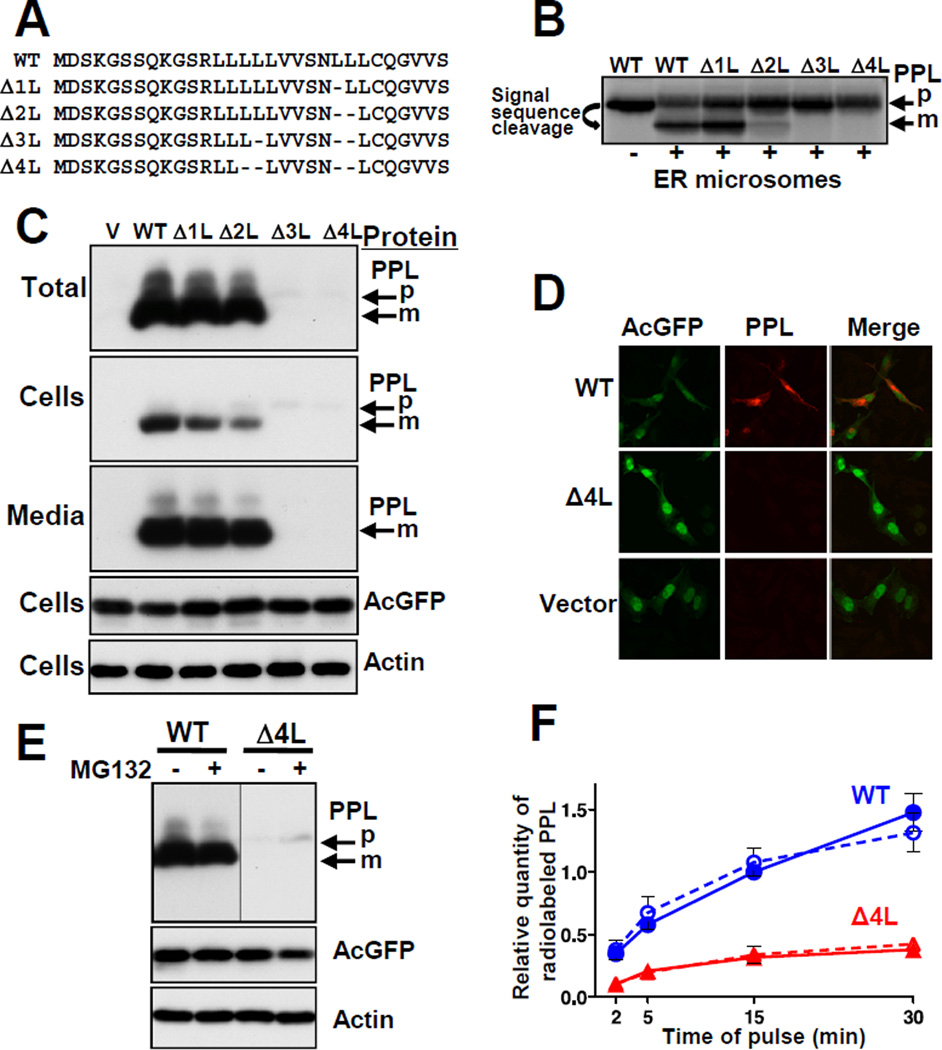
Secretory proteins contain N-terminal signal sequences (von Heijne, 1983). The signal sequence is co-translationally recognized by SRP (Walter et al., 1981). The ribosome-nascent chain-SRP complex is targeted to its receptor in the ER membrane where the nascent chain is translocated into ER lumen by the Sec61p/TRAM translocon, and finally the signal sequence is removed by signal peptidase (Alder and Johnson, 2004; Egea et al., 2005; Rapoport, 2007; Wild et al., 2004). Preprolactin (PPL) is a secretory protein that contains a prototypical signal sequence. We constructed mutant PPLs with deletions within the signal sequence hydrophobic core (Figure 1A) and assessed protein translocation. In vitro-translated wild-type (WT) PPL was efficiently translocated into ER microsomes as detected by processing of the precursor (signal sequence cleavage) (Figure 1B). Deletion of leucines from the signal sequence reduced translocation, with the number of deleted leucines directly correlating with the degree of the transport defect.
Figure 1. Deletions in the PPL Signal Sequence Lead to Defects in Protein Transport and Expression.
(A) Signal sequences of WT and mutated PPLs, deletions indicated by dashes. (B) WT and mutant PPLs were synthesized in a rabbit reticulocyte translation system in vitro in the presence or absence of ER microsomes, analyzed by SDS-PAGE, and detected by autoradiography. Positions of mature PPL (m) and precursor (p) are shown. (C and D) WT and mutant PPLs were transiently expressed in HeLa Tet-On cells (cells with empty vector (V) were controls) and detected by Western blot or by immunofluorescence, respectively. AcGFP, expressed from the same plasmids, and endogenous actin were controls. (E) Effect of the proteasome inhibitor MG132 on the level of WT and Δ4L PPLs (detection by Western blot). (F) Pulse-labeling analysis of translated WT (blue circles) and Δ4L (red triangles) PPLs in the presence (dashed lines, n=4) and absence (solid lines, n=5) of MG132 (mean ± SEM). See also Figure S1.
WT PPL expressed in cell culture was efficiently secreted, reflected by the presence of its mature form in the cells and media (Figure 1C). However, the Δ3L and Δ4L mutants did not mature and their protein levels were dramatically decreased. The decrease directly correlated with the number of deleted leucines. In contrast to WT PPL, the Δ4L mutant was not detected by immunocytochemistry (Figure 1D). Notably, levels of AcGFP expressed from an independent promoter on the same plasmid, and of endogenous actin, were not affected (Figures 1C and 1D). Thus, mutations in the signal sequence lead to specific reductions in the quantities of the mutant proteins. The simplest explanation is that mutant proteins are degraded by the proteasome. However, inhibition of the proteasome with MG132 produced only a marginal change (Figure 1E). Hence, reduction in the level of the proteins cannot be explained by proteasomal degradation. Alternatively, the decrease in mutant protein levels could be caused by a reduction in their translation. Pulse-labeling of WT and Δ4L proteins showed that translation of the mutated protein was dramatically impaired while turnover was not affected (Figures 1F and S1), suggesting that the lower level of mutant translation may be caused by reduction in mutant mRNA.
Deletions in the Signal Sequence of Secretory Proteins Cause a Reduction in Their mRNA Levels
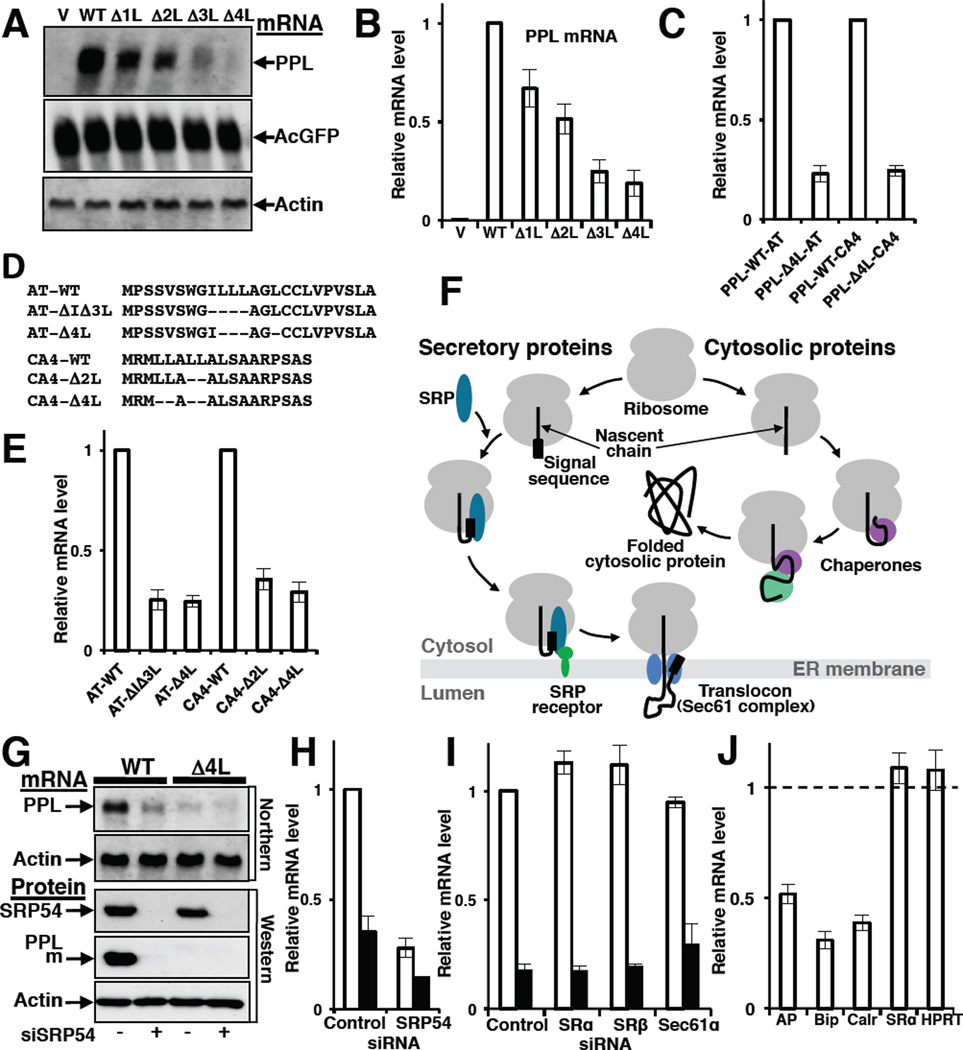
Using two independent techniques, Northern blot and quantitative real-time PCR (qPCR), we found that levels of PPL mRNA decreased as the number of deleted leucines in the signal sequence increased (Figures 2A and 2B). This effect was specific for mutant PPLs, as neither AcGFP nor actin mRNAs levels changed (Figure 2A). The same effect was observed in two other cell lines (Figure S2), indicating that the process of mutant mRNA elimination is independent of the cell type. We also constructed hybrid proteins containing WT PPL or mutated PPL Δ4L signal sequences fused to mature sequences of two other secretory proteins, α1-antitrypsin (AT) or carbonic anhydrase IV (CA4), and measured their mRNA levels (Figure 2C). The mRNA levels of the hybrid proteins with mutated signal sequences were reduced, indicating that the presence of aberrant signal sequence was sufficient to trigger mRNA depletion regardless of the sequence of the mature secretory protein. These data suggested the existence of a specific mechanism for decreasing mutant mRNA levels.
Figure 2. Defects in the Signal Sequence or in a Targeting Factor, SRP, Lead to Decreased Levels of a Secretory Protein mRNA.
(A and B) Effect of deletions in signal sequence on PPL mRNA levels. PPL and control mRNAs were analyzed by Northern blot (A) or qPCR (B; n=9, mean ± SD) 42 h after transfection of HeLa Tet-On with WT and mutated PPL plasmids. (C) Presence of a mutated PPL signal sequence (PPL- Δ4L) in hybrid proteins containing the mature part of a1-antitrypsin (AT) or carbonic anhydrase IV (CA4) is sufficient to trigger mRNA depletion. mRNA levels measured by qPCR (n=3, mean ± SD) are shown relatively to mRNA levels of corresponding hybrid proteins containing WT PPL signal sequence (PPL-WT). (D) Deletions (indicated by dashes) in the natural signal sequences of secretory proteins AT or CA4. (E) Deletions in the hydrophobic core of natural signal sequences of AT or CA4 proteins lead to a decrease in their mRNA levels. Graph shows mRNA levels measured by qPCR (n=3, mean ± SD). (F) Scheme shows differences in biogenesis of secretory and cytosolic proteins. When the signal sequence of a secretory protein emerges from the ribosomal tunnel it is recognized by SRP, and the complex is targeted to SRP receptor and finally to a translocon in ER membrane. Nascent chains of cytosolic proteins do not have signal sequences, however, their nascent chains are recognized by ribosome-associated chaperones and that help them fold in the cytosol. (G–H) SRP depletion causes a reduction in the level of secretory protein mRNA. Detection of PPL, and actin mRNAs (Northern blot), SRP54, PPL and actin proteins (Western blot) in HeLa Tet-On cells transfected with siRNA for SRP54 and WT or Δ4L PPL plasmids as indicated (G). Quantification of WT PPL (open bars) or Δ4L mutant (black bars) mRNAs by qPCR in independent sets of SRP54 knockdown experiments (n=3, mean ± SD) (H). (I) Knockdown of SRP receptor subunits SRα and SRβ, or translocon component, Sec61α, does not affect PPL mRNA level (qPCR, n=3, mean ± SD). WT PPL (open bars), Δ4L mutant (black bars). (J) SRP depletion causes reduction in endogenous mRNA levels of secretory and ER proteins. Measured by qPCR (n=3, mean ± SD) mRNA levels in SRP depleted cells are shown relatively those treated with control siRNA (taken as 1). Intestinal alkaline phosphatase (AP) is a secretory protein, Bip and calreticulin (Calr) are ER lumen proteins. mRNA levels of the cytosolic protein HPRT and the SRP-independent ER membrane-associated protein SRα were not altered. See also Figures S2 and Table S1.
To establish generality of the mechanism we made deletions in the natural signal sequences of AT and CA4 and found that their mRNA levels were also reduced (Figures 2D–E), similar to the reductions of mRNA levels of mutated PPL and hybrid proteins (Figures 2B–C). These results indicate that the effect of mRNA reduction is determined by the defects in the translated signal sequences independently of their amino acid sequences, making the scenario of reduction of mRNA levels due to changes in mRNA secondary structure unlikely. These data demonstrate that the mRNA-depletion mechanism is general and selective.
SRP Depletion Leads to a Decrease in the mRNA Levels of Secretory Proteins
Secretory and cytosolic proteins have different biogenesis pathways. While cytosolic proteins require different ribosome associated chaperones and chaperonins for their proper folding in the cytoplasm, for a secretory protein, the first step in protein transport is recognition of its signal sequence by SRP and formation of the SRP-RNC (Ribosome Nascent Chain) complex (Figure 2F). Since mRNA levels were reduced by mutations that were predicted to alter signal sequence-SRP interactions, the role of SRP in the mRNA quality control process was independently evaluated by decreasing its intracellular concentration. siRNA was used to efficiently decrease SRP in human cell culture. SRP depletion caused a dramatic reduction in mRNA levels (as detected by Northern blot and qPCR) and protein levels of both WT and mutant PPLs (Figures 2G–H). In the next step of biogenesis the SRP-RNC complex is targeted to the SRP receptor at the ER membrane, then the RNC is transferred to a translocon and the nascent chain is co-translationally translocated into the ER lumen, where it is folded with the help of ER chaperones (Figure 2F). Contrary to SRP depletion, no effect on WT or Δ4L mRNA was found when SRP receptor subunits SRα and SRβ or the core component of the translocon, Sec61α, were depleted (Figure 2I). The data clearly demonstrate that mRNA depletion of a secretory protein occurs when its nascent chain loses interaction with SRP, suggesting that selection of mRNA for degradation takes place at the ribosome during translation, when the N-terminal portion of a mutated nascent chain exits the ribosomal tunnel but is not recognized by SRP.
To address the fate of mRNAs of other potential substrates for SRP, endogenous mRNA levels of secretory and ER resident proteins were measured in SRP depleted cells. mRNA levels of the secretory proteins alkaline phosphatase, as well as ER localized proteins Bip and calreticulin, were dramatically reduced in SRP54 depleted cells (Figure 2J). All of these proteins have signal sequences and are transported from the cytosol. By contrast, mRNA levels of the non-secreted cytosolic protein hypoxanthine phosphoribosyltransferase (HPRT) and the SRP-independent ER membrane-associated protein SRα (Andrews et al., 1989; Lakkaraju et al., 2007) were not affected (Figure 2J). The data demonstrate that the loss of targeting factor SRP leads to specific elimination of the mRNAs coding for its substrates. Thus, mRNA depletion correlates with an abnormality in the signal sequence recognition process, including either a mutation in the signal sequence or a decrease in the amount of functional SRP.
Decrease in mRNA Level is due to mRNA Degradation
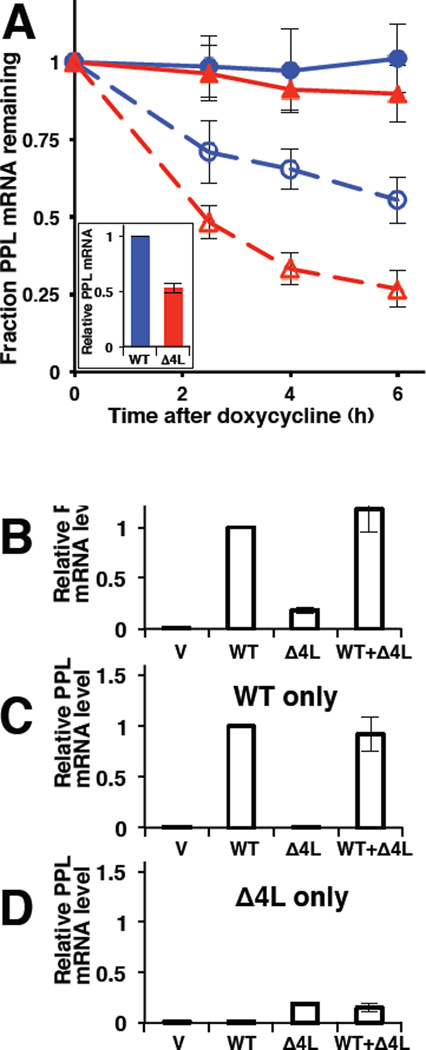
Reduction of steady state mRNA levels could be achieved by either reducing transcription or increasing mRNA degradation. To test whether mutant mRNA degradation was increased, mRNA levels were monitored as a function of time after transcriptional inhibition (Figure 3A). WT and mutant PPL were cloned into the pTRE2hyg vector. This vector contains the inducible Tet-response element and a minimal CMV promoter. In HeLa Tet-Off cells cultured in the absence of the antibiotic, the mRNA is transcribed normally, while the addition of doxycycline inhibits transcription. Doxycycline was added 20 h after transfection to ensure that adequate levels of Δ4L mRNA still remained at the start of the experiment (Figure 3A, inset). The turnover of WT PPL mRNA (observed by qPCR) as a function of time in doxycycline-treated (versus untreated) cells indicates the normal rate of mRNA degradation (Figure 3A). The Δ4L PPL mRNA turned over significantly faster than WT PPL mRNA, indicating that the mutant mRNA was preferentially degraded.
Figure 3. Preferential Degradation of Mutant PPL mRNA.
(A) qPCR analysis of WT (blue circles) or Δ4L (red triangles) PPL mRNAs under conditions of transcription inhibition (doxycycline treatment, dashed lines and open symbols) or mock (solid lines and symbols). The quantity of mRNA at each time point is shown relative to the initial quantity of the respective mRNA species. Doxycycline was added 20 h after transfection to ensure that adequate levels of Δ4L mRNA still remained at the start of the experiment; the initial WT and Δ4L mutant mRNA content was about 2:1 (inset). The quantity of mRNA at each time point is shown relative to the initial quantity of the respective mRNA species. Data are from 3 experiments, where each time point contained 3 repeats in each experiment (mean ± SD). (B to D) qPCR analysis (n=5, mean ± SD) of PPL WT and Δ4L mRNAs using primers amplifying both WT and mutant (B), only WT (C), or only Δ4L (D) in the cultured human cells expressing WT PPL only (WT), Δ4L mutant only (Δ4L), both WT and Δ4L (WT+ Δ4L), or transfected with empty vector (V). mRNA levels are shown relative to WT, except in D, where mRNA levels were scaled to equate the Δ4L level in D to that in B. See also Figure S3.
The Mechanism Discriminates Mutant from Wild-Type mRNAs
To determine whether the mutant protein induces a general cellular response that acts on both mRNAs or acts specifically on the mutant mRNA, WT and Δ4L PPLs were coexpressed. Protein levels were detected by Western blotting (Figure S3) and mRNA levels were determined by qPCR using primers to detect both WT and Δ4L, or specifically the WT or the mutant (Figures 3B–D). The total amount of PPL message in cells coexpressing mutant and WT was equivalent to the sum of the two mRNAs expressed independently. Thus, the presence of WT protein and mRNA did not prevent degradation of mutant mRNA (Figure 3D), nor did the presence of mutant mRNA lead to increased degradation of WT mRNA (Figure 3C). These data indicate that the cellular machinery discriminates between WT and mutant mRNAs and specifically targets mutant mRNA for degradation.
A Functional Signal Sequence Protects mRNA of a Secretory Protein from Degradation
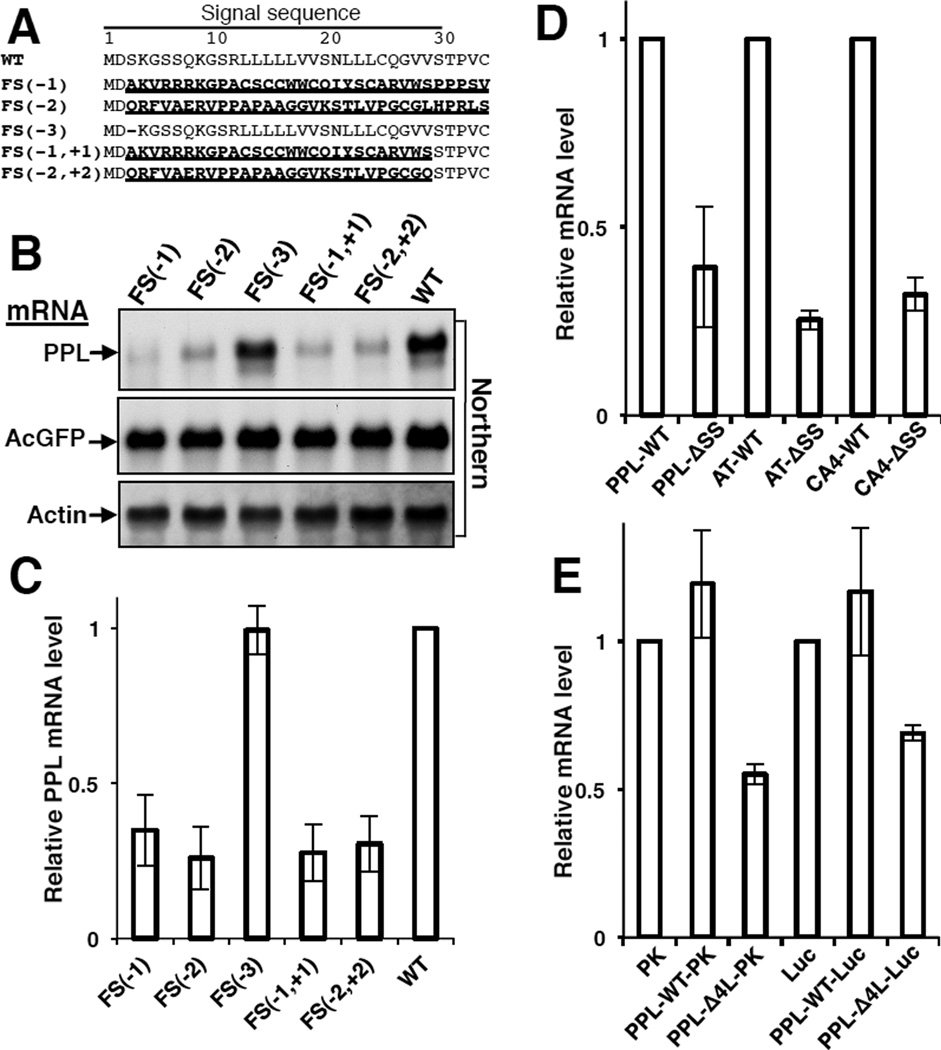
The results of the SRP knockdown experiment on endogenous transcripts suggest that the mechanism does not involve direct recognition of the abnormal mutant mRNA structures or sequences. Instead, the data suggest a link between the translation of abnormal proteins and the degradation of their mRNA. To directly test this hypothesis, PPL constructs were analyzed that contained a deletion of one, two, or three nucleotides at the beginning of the signal sequence to create either frame-shifts in the open reading frame or the deletion of a codon (Figure 4A). The deletions of one or two nucleotides led to dramatic changes in the protein sequence because of a frame shift in the reading frame and only minor changes (removing one or two nucleotides) in mRNA. By contrast, the deletion of 3 nucleotides leads to larger changes in mRNA and smaller changes (deletion of one residue) in the protein. As predicted, the mRNA levels of frame-shift mutants were depressed, while the level of mRNA with a deleted codon was not affected (Figures 4B–C). These results indicate that the translation of the native nascent chain is important. To localize the sequence that is important for protection of mRNA from degradation, additional nucleotide(s) were introduced into frame-shift mutants to restore the PPL reading frame after the signal sequence (Figure 4A). The resultant nascent chains contained dramatic changes in the signal sequence, but no changes in the mature protein. The levels of these mRNAs were low (Figure 4B–C). These results suggested that the signal sequence protects secretory protein’s mRNA from degradation. To test this hypothesis we prepared a PPL construct without a signal sequence, PPL- ΔSS, and found that its mRNA level was decreased (Figure 4D). Similar results were obtained using other two secretory proteins, AT and CA4 lacking signal sequences (Figure 4D). All data together demonstrate that the presence of a functional signal sequence in the nascent chain is important for protection of the mRNA from degradation.
Figure 4. Functional Signal Sequence is Required for Protection of the Secretory Protein mRNA from Degradation.
(A) N-terminal sequences of PPL mutants with amino acid substitutions in bold and underlined, and deletions as dashes. FS(−1), FS(−2), FS(−3) contain deletions of one, two, three nucleotides, correspondingly, in the third codon. FS(−1, +1), FS(−2,+2) contain additional insertion of one or two nucleotides in the codon 29 to restore original reading frame. mRNAs were analyzed by Northern blots (B) or by qPCR (C; n=3, mean ± SD). (D) Deletions of natural signal sequences from PPL, AT, and CA4 lead to decrease in their mRNA levels (qPCR, n=3, mean ± SD). (E) Presence of a mutated signal sequence in hybrid cytosolic proteins leads to their mRNA depletion (qPCR, n=3, mean ± SD).
Presence of a Mutant Signal Sequence in Hybrid Cytosolic Proteins Leads to Their mRNA Depletion
While the lack of a functional signal sequence in secretory proteins leads to significant decrease of their mRNA levels, the mRNAs of cytosolic proteins are stable despite the fact that they do not have signal sequences. The biogenesis of cytosolic proteins is significantly different from that of secretory proteins (Figure 2F). During de novo synthesis of cytosolic proteins, nascent chains interact with a set of ribosome associated chaperones linked to protein synthesis (CLIPS) (Albanese et al., 2006; Pechmann et al., 2013) or ribosome-associated protein biogenesis factors (RPBs) (Raue et al., 2007). Although a sequence consensus for interaction with ribosome bound chaperones has not been reported for cytosolic proteins, it is known that the affinities of ribosome bound chaperones, as well as positioning of nascent chains, are influenced by the amino acid sequence of the nascent polypeptide (Raue et al., 2007). Thus, the N-terminal sequence of nascent chains is likely responsible for specificity of these interactions.
Hybrids of cytosolic proteins luciferase (Luc) or pyruvate kinase (PK) containing WT or Δ4L PPL signal sequences at their N-termini were prepared. While the WT signal sequence did not lead to mRNA depletion of a hybrid protein, the presence of the mutant Δ4L signal sequence led to a significant decrease of mRNAs for both tested hybrid proteins, PK and luciferase (Fig. 4E). The presence of a WT signal sequence may not influence the mRNA level because it is recognized by SRP, and thus the mRNA remains protected. However, an aberrant signal sequence will not be recognized by SRP, nor by normal cytosolic protein partners at the ribosome. This observation indicates that the selection of the mRNA for degradation is conducted at a very early step in translation, when the N-terminal sequence of a nascent chain first appears from the ribosomal tunnel. The experiments clearly demonstrate that cellular quality control machinery can distinguish between nascent chains of a normal cytosolic protein and a hybrid containing mutated signal sequence.
Mutation of the PPL Signal Sequence Shifts its Interaction from SRP to a ~100 kDa Protein
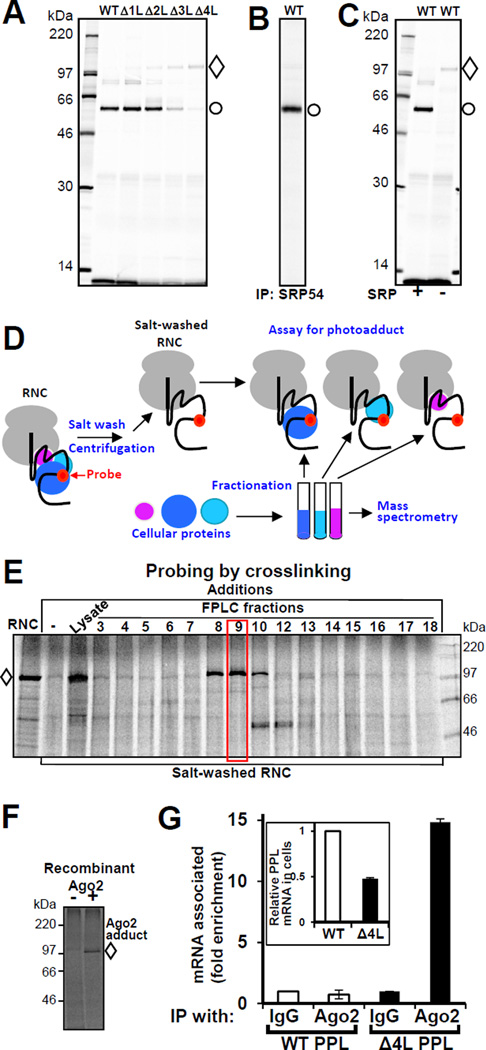
We hypothesized that mutations in the nascent chain might lead to the recruitment of specific interacting factors at the ribosome exit site during translation that trigger mRNA degradation. Site-specific photo-crosslinking (Krieg et al., 1986; McCormick et al., 2003) has been employed to study the interactions of a nascent chain with cellular factors. This approach is based on the tRNA-mediated incorporation of a photoreactive probe into a nascent chain at a specific position(s) during in vitro translation (Figure S4A). Radiolabeled ribosome-nascent chain complexes (RNCs) of WT and mutant PPLs were synthesized in vitro in the presence of SRP using mRNAs truncated after codon 86, so only a short portion of the nascent chain, including the signal sequence, was exposed outside the ribosome. The photoreactive probe was positioned in the middle of the signal sequence. When exposed to UV light, the photoprobes form covalent bonds with molecules in close proximity, and the resulting photoadducts are detected by a shift in nascent chain electrophoretic mobility (Figure S4B). WT PPL forms a photoadduct with a ~50 kDa protein (Figure 5A) that was confirmed by immunoprecipitation to be the SRP54 subunit of SRP (Figure 5B). Deletion of leucines in the mutant PPL decreased the efficiency of photocrosslinking to SRP54 while simultaneously increasing the efficiency of formation of a photoadduct with a ~100 kDa protein (Figure 5A). Consistent with the SRP knockdown experiment (Figure 2 G-H), the adduct to the ~100 kDa protein was also formed with WT PPL when the reaction was not supplemented with SRP (Figure 5C). This behavior implicated the ~100 kDa protein in the initial step of the process that leads to the specific degradation of mutant PPL mRNA or WT PPL mRNA when SRP is limiting.
Figure 5. Ago2 is in Close Proximity to Mutant Nascent Chains and Associated with Mutant mRNA.
(A) Deletions in the PPL signal sequence lead to reduced interaction with SRP54 and increased proximity to a ~100 kDa protein. Photocrosslinking patterns of WT and mutated PPLs are shown. RNCs containing the first 86 residues of WT and mutant PPLs with a photoreactive probe in the signal sequence were produced in vitro in the presence of SRP. Following UV irradiation, samples were analyzed by electrophoresis and autoradiography. The positions of photoadducts containing ~50 kDa (circle) or ~100 kDa (diamond) proteins are shown. (B) Immunoprecipitation (IP) using SRP54 specific antibody demonstrates that the ~50 kDa protein is SRP54. (C) WT PPL crosslinks to the ~100 kDa protein are maximized when lysate is not supplemented with SRP. (D) Scheme of the method for identification of Proteins Interacting with Nascent Chains (iPINCH). (E) Identification of fractions that contain the ~100 kDa protein. Δ4L RNCs were prepared as above and treated with a high salt buffer. The latter were combined with lysate or aliquots of fractionated rabbit reticulocyte ribosome-associated proteins, incubated, UV irradiated and analyzed as in A. The photocrosslinking pattern of the fraction examined by mass spectrometry is boxed. (F) Recombinant Ago2 is in close proximity to mutant nascent chain. Purified recombinant human Ago2 or buffer was added to salt-washed RNCs of Δ4L PPL mutant with photocrosslinking probe. Samples were photocrosslinked and analyzed as above. (G) mRNA of Δ4L PPL is preferentially enriched in Ago2 immunoprecipitates. A short incubation time (20–24 h after plasmid transfection) was used to ensure that significant levels of Δ4L mRNA still remained at the start of the experiment: the WT and Δ4L mutant mRNA content in cell lysates was about 2:1 before IP (inset). Mouse monoclonal Ago2 antibody was used for IP. Data are from two qPCRs, reactions were in triplicates, mean ± SD. See also Figures S4 and Table S2.
Argonaute2 (Ago2) is in Close Proximity to Mutant Nascent Chains During Translation and Associates with Mutant mRNA
To identify proteins interacting with nascent chains we combined site-specific photocrosslinking with protein fractionation and mass spectrometry. This allowed identification of proteins involved in short-lived, transient interactions during translation. The approach includes: in vitro preparation of salt-washed RNC complexes with an incorporated photocrosslinking probe, by the use of modified amber-suppressor tRNA (salt-washing removes ribosome-associated proteins); ii) fractionation of cellular proteins, and selection of fractions containing the protein of interest using the photocrosslinking assay; and iii) identification of proteins in the selected fractions by mass spectrometry (Figure 5D). This method for identification of Proteins Interacting with Nascent Chains (iPINCH) is a valuable tool for identification of partners in transient protein-protein interactions during translation.
RNCs of Δ4L PPL containing the photocrosslinking probe were prepared in vitro translation system. As expected the nascent chain of the mutated protein photocrosslinked the ~100K protein (Figure 5E, lane “RNC”). Washing the RNCs with high salt buffer removed most of the ~100 kDa protein, but upon adding lysate back to salt-washed RNCs, photocrosslinking to the ~100 kDa protein was reconstituted (Figure 5E, lane “Lysate”). A crude reticulocyte preparation of ribosome-associated proteins, which restored photoadduct formation, was fractionated and fractions containing the protein of interest were identified (Figure 5E, fractions 8–10). Analysis by mass spectroscopy revealed that the protein was Argonaute2 (Ago2) (Table S2). Ago2, a member of the argonaute protein family, is involved in gene silencing (Hammond et al., 2001; Hutvagner and Simard, 2008; Martinez et al., 2002; Tolia and Joshua-Tor, 2007). The identification of Ago2 was validated independently by reconstituting the photoadduct using recombinant human Ago2 purified from insect cells (Figure 5F), confirming its close proximity to mutated PPL nascent chain during translation.
We tested whether Ago2 preferentially interacts with the mutant mRNA. RNA immunoprecipitation (RNA-IP) of endogenous Ago2 from human cells expressing WT or mutant PPLs revealed that mutant Δ4L PPL mRNA was enriched in Ago2 immunoprecipitates (Figures 5G, S4C–F).
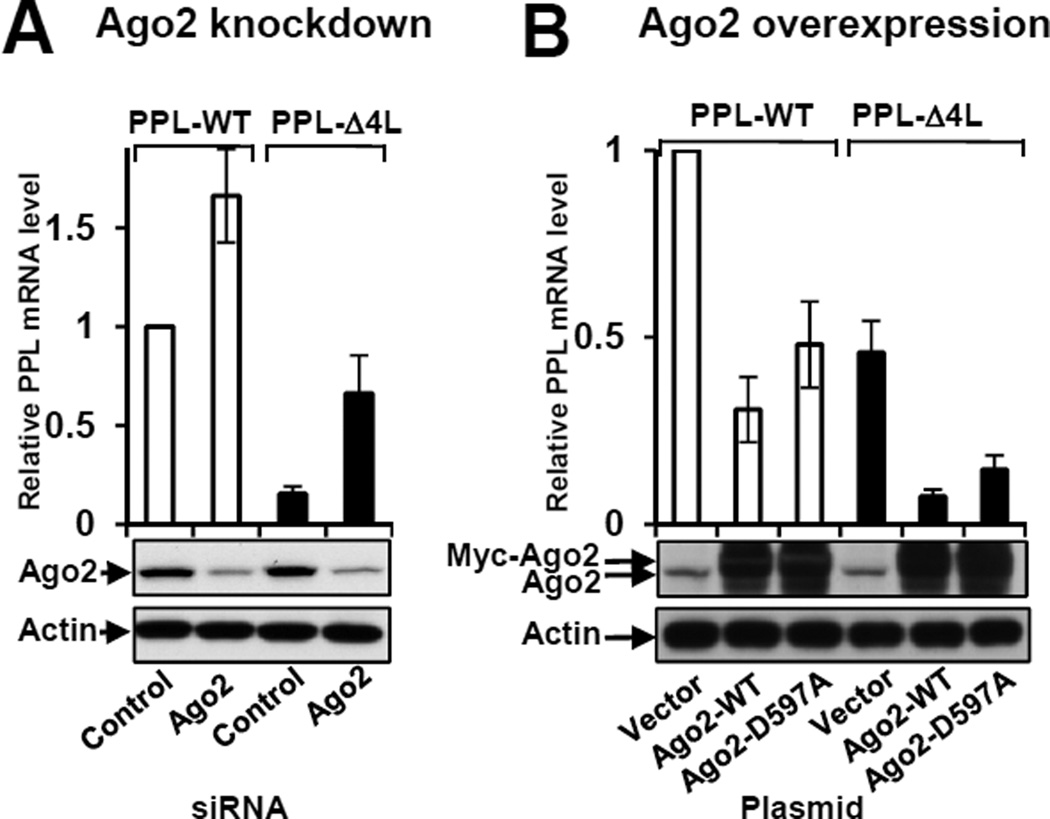
To test functional involvement of Ago2 in the specific reduction of mutant mRNA levels we measured PPL mRNA levels in the cultured cells in which Ago2 was depleted or overexpressed. The reduction of mutant PPL mRNA was significantly inhibited in cells when Ago2 was knocked-down (Δ4L mRNA level increased ~4.4 fold, Figure 6A), while WT mRNA level was elevated in Ago2 knock-down cells to a lesser degree (~1.7 fold). The results suggest that Ago2 is involved in the process of specific mRNA degradation. For Ago2 overexpression experiments, Ago2-containing plasmids were transfected a day before transfection with PPL plasmids. A short incubation time (~24 h after PPL plasmids’ transfections) was used to ensure that significant level of Δ4L mRNA (about 45% of WT PPL mRNA level) still remained in the control (Figure 6B). When Ago2 was over-expressed, the level of Δ4L mRNA was dramatically reduced (~6.2 fold), while WT PPL mRNA level was decreased to a lesser degree (~3.3 fold) (Figure 6B).
Figure 6. Process of mRNA degradation involves Ago2.
Ago2 knockdown inhibited reduction of PPL mRNA (A), while overproduction of recombinant WT or mutant D597A Myc-Ago2 reduced the level of PPL mRNA (B). Bars are PPL mRNA levels from Northern blot (n=3 (but n=2 for D597A), mean ± SEM). WT PPL mRNA levels are open bars, Δ4L mutants are black bars. Depletion or overexpression of Ago2 was confirmed by Western blot, actin was a control. See also Figure S5.
Although Ago2 possesses ribonuclease H activity (slicer activity) and amino acid residues in the active site are known (Liu et al., 2004; Song et al., 2004), not all Ago2-dependent processes require its nuclease activity. To test if Ago2 slicer activity is required for mutant PPL mRNA degradation we conducted experiments with a slicer defective Ago2, containing mutation D597A (Liu et al., 2004; Rivas et al., 2005). Overproduction of Ago2-D597A leads to significant decrease in the level of Δ4L mRNA, and to lesser degree, WT PPL mRNA (Figures 6B). Although the effect of Ago2-D597A was slightly less than that of Ago2-WT, the results demonstrate that overproduction of slicer mutant Ago2 leads to decreased levels of PPL mRNAs. Overproduction of Ago2-WT or mutant Ago2-D597A under conditions when endogenous Ago2 was knocked-down independently confirmed this observation (Figure S5A) and showed that the effects were specific (Figure S5B). These results indicate that Ago2 is involved in the process, but its slicer activity is not required. Interestingly, knock down of other proteins of the argonaute family, Ago1, Ago3, and Ago4, and proteins involved in small RNA biogenesis, Dicer and Drosha, as well as proteins involved in miRNA dependent association with Ago2, TNRC6A, B, C, were without measurable effect on Δ4L PPL mutant mRNA levels (Fig. S5C–H). Modest increases in mRNA levels of WT PPL mRNA in some cases was observed, reflecting involvement of the above proteins in normal PPL mRNA biogenesis, however, the knock-downs did not significantly affect the process of mutant mRNA elimination.
DISCUSSION
Misfolded/mistargeted proteins, as well as their aggregates can be cytotoxic, leading to many diseases (Stefani and Dobson, 2003). However, the fact that protein aggregates do not accumulate in unstressed cells despite their potential continued production indicates the existence of efficient quality control machinery (Kopito, 2000). Cells have developed multiple mechanisms for protection from these “unwanted” products at different levels. One of the protection levels is labeling with ubiquitin and degradation of already synthesized aberrant proteins by the proteasome (Buchberger et al., 2010; Smith et al., 2011). Energy dependent degradation of mutant proteins and their futile translation are expensive processes and cells evolved mechanisms to prevent synthesis of mutant cytotoxic proteins. NMD directing mRNAs with premature stop codons for degradation, NSD acting on mRNAs without a natural stop codon, and NGD acting on mRNAs on stalled ribosomes, are examples of quality control on the template mRNAs with dramatic aberrations (Doma and Parker, 2007; Shoemaker and Green, 2012). However, none of these systems are known to act on mutant mRNA templates that code for proteins with small aberrations in their primary structure. The mechanism reported here fills this gap in quality control by specifically acting on mutant mRNA templates encoding proteins that have lost interactions with their normal binding partners at the ribosome exit site.
In this study we report the discovery of a previously unappreciated mechanism of translational quality control, in which translation of a defective protein leads to specific degradation of its mRNA. This mechanism monitors the absence of normal interactions of nascent chains early during translation and transfers the signal to mRNA degradation machinery that allows cells to prevent synthesis of these “unwanted” aberrant proteins. This process is distinct from known quality control mechanisms. It involves surveillance of newly synthesized nascent chain interactions and triggers mRNA degradation in response to a loss of these normal interactions during translation. This mechanism can discriminate between normal and mutated mRNAs even when they are co-expressed (Figures 3B–D). This specificity is graded, as demonstrated by the strong correlation between decreased efficiency of SRP-nascent chain interaction and mRNA degradation (Figures 2A–B, 5A). Therefore, the system balances the quantity of synthesized protein with decreased ability for targeting and secretion. Notably, this process is not a generalized stress response, but rather is specifically directed to reduce the burden of non-transportable proteins as well as potentially other types of newly synthesized misfolded proteins. Furthermore, as demonstrated by two different approaches, introduction of deletions in signal sequences of several secretory proteins and assessment of mRNA levels of secretory and ER lumen proteins under conditions when SRP was depleted, the mechanism is general (Figures 2).
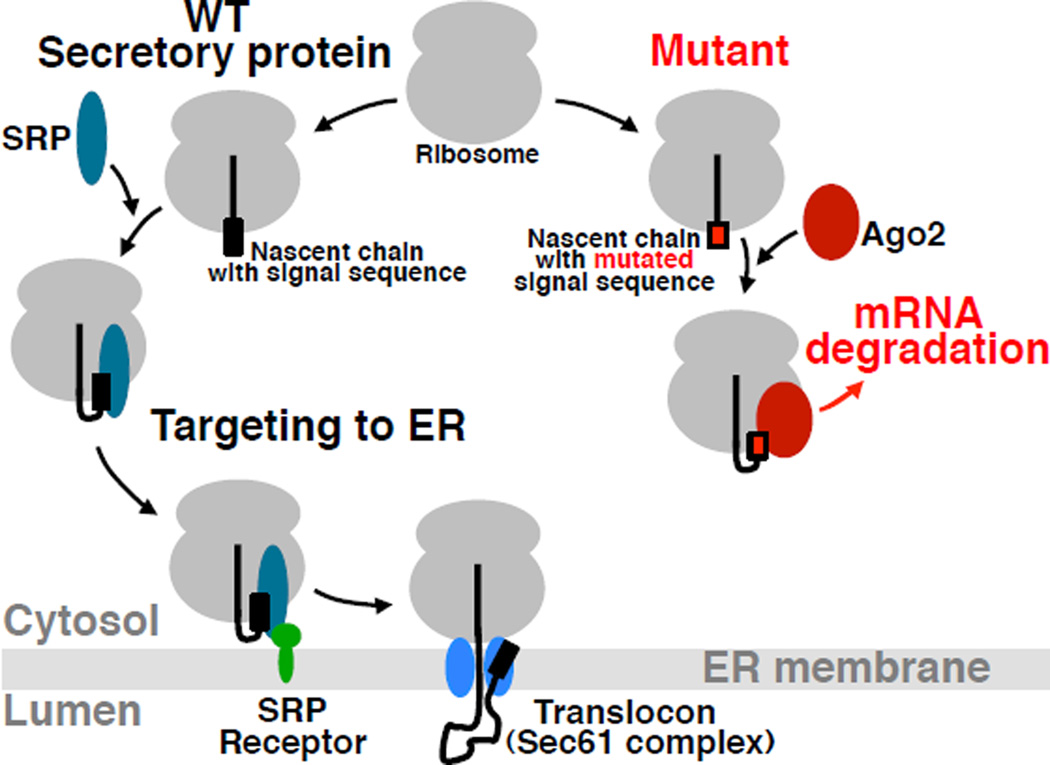
Based on our data, we propose that the loss of co-translational interactions between a nascent chain and factors required for transport and/or folding (SRP in this study, but possibly ribosome-associated chaperones for cytosolic proteins (Hartl et al., 2011; Pechmann et al., 2013; Raue et al., 2007), and other targeting factors for nuclear, mitochondrial, or peroxisomal proteins) can trigger a cellular response that leads to specific elimination of the corresponding mRNA (Figure 7). According to this hypothesis, Ago2 is associated with the ribosome within the reach of the nascent chains that are either defective or unable to bind their normal binding partners after emerging from the translating ribosome and the encoding mRNA. This association correlates with mRNA elimination. Although it is still unknown if Ago2 is a direct sensor, the data demonstrate that Ago2 is functionally involved in the process. This mechanism of Regulation of Aberrant Protein Production (RAPP) would provide high flexibility for regulating the synthesis of aberrant proteins regardless of their sequence and structure, without negative effects on the production of undamaged proteins in the same cell.
Figure 7. Model for Regulation of Aberrant Protein Production (RAPP).
When the nascent chain of a WT secretory protein containing a signal sequence emerges from the ribosome polypeptide exit tunnel during translation it is recognized by SRP. This interaction leads to targeting of the complex to SRP receptor in the ER membrane and finally to the translocon. The nascent chain is co-translationally translocated into the ER lumen, where it is folded with the help of ER chaperones. However, when the nascent chain of a secretory protein contains critical mutations preventing SRP recognition, the unbound nascent chain is exposed outside the polypeptide tunnel. Despite the absence of its interaction with SRP, a defective secretory protein does not typically behave as a cytosolic protein because of the absence of appropriate sequences/conformations/signals required for normal cytosolic nascent chains interactions. Thus, the space normally occupied by SRP at the ribosome remains unoccupied. Ago2 occupies the space and directs mRNA for degradation initiating the RAPP process.
The following observations support this model: 1) Ago2 co-translationally crosslinks to mutant nascent chains that have lost the ability to bind SRP (Figure 5A); 2) in the absence of SRP, nascent WT PPL photocrosslinks instead to Ago2 (Figure 5C), indicating that Ago2 occupies the space when it is not occupied by SRP; 3) Ago2 was found preferentially associated (directly or non-directly) with the mutant mRNA (Figures 5G and S4C–F); 4) overexpression of Ago2 or depletion of SRP in human cells decreases the levels of PPL mRNA, while depletion of Ago2 stabilizes the level of mutant mRNA (Figure 6); 5) increase in Ago2 crosslinking efficiency directly correlates with the number of leucines deleted from the PPL signal sequence, a decrease in efficiency of RNC interaction with SRP, and a decrease in the level of mutant mRNA (Figures 2A–B, and 5A); 6) mRNAs of secretory proteins with defective signal sequences were efficiently degraded despite the absence of homology in signal sequences (Figure 2E); 7) SRP knockdown leads to mRNA degradation of even WT protein, while defects in other downstream components of the protein translocation machinery were without effect (Figure 2 G–I).
Several different scenarios were examined to determine which signals are used for selection for degradation of mRNAs coding aberrant proteins. The above model is the simplest model that explains all the data. Indeed, direct surveillance of mRNA sequence by a quality control mechanism is unlikely (see discussion to Figure 4). A model in which the decreased stability is due to mislocalization of ER-targeted mRNAs, or a mechanism involving surveillance of the overall change in the structure of synthesized aberrant secretory protein as a result of folding in the cytosol, an inappropriate environment, is is inconsistent with the observations that knock down of SRP receptor or translocon protein Sec61α did not effect the mRNA level of secretory proteins, by contrast to SRP knockdown (Figure 2). All these defects in components of the secretory machinery abolish protein translocation, but only SRP knock down leads to mRNA elimination. Recognition of a specific sequence or features of an aberrant protein (e.g. a mutated signal sequence) is inconsistent with data on secretory proteins lacking their signal sequences or with scrambled N-termini (Figure 4). This scenario also cannot explain secretory protein mRNA depletion when SRP was knocked-down (Figure 2G, H, J). Therefore the simplest model is that the quality control system does not recognize specific sequences of a mutant protein or overall changes in its structure, but rather, the absence of normal interactions of nascent chains early in translation.
How does this mechanism discriminate aberrant secretory proteins with defects in signal sequences from cytosolic proteins that are not natural substrates for SRP and therefore do not interact with SRP? In this regard, it is important to note that the presence of signal sequence is not the only feature that distinguishes secretory proteins from cytosolic proteins. The removal of a signal sequence from a secretory protein will not typically make it truly cytosolic (having failed translocation, it will be localized to the cytosol, but it will lose its natural binding partners and will not likely be correctly folded, and will be directed for degradation), just as adding a signal sequence to a cytosolic protein will not typically make it truly secretory (although exceptions are known).
Other factors interact with cytosolic proteins during their translation. A special network of chaperones linked to protein synthesis, CLIPS (Albanese et al., 2006; Pechmann et al., 2013), and ribosome-associated protein biogenesis factors, RPBs (Raue et al., 2007), represent the very complex machinery at the ribosome peptide exit tunnel. Examples include ribosome associated chaperones Hsp70 SSB (Willmund et al., 2013), MPP11 and Hsp70L1 (Otto et al., 2005), chaperonin TRiC as well as numerous others (see for details reviews (Hartl et al., 2011; Pechmann et al., 2013). Thus, in the case of cytosolic proteins, while their nascent chains do not bind SRP, they interact with other factors. These interactions may prevent Ago2 association and thus would not trigger RAPP.
The crosslinking and functional studies described here implicate Ago2 in the process of specific degradation of mRNAs in response to synthesis of mutated nascent chains not capable of efficiently binding SRP. Ago2 is a member of the argonaute protein family. These proteins are highly conserved and divided into 3 subfamilies: Ago-like, PIWI-like, and group 3 (Hutvagner and Simard, 2008; Tolia and Joshua-Tor, 2007). Mammals contain four Ago-like subfamily members, and Ago2 is the only member known to possess RNA cleavage activity (Liu et al., 2004; Meister et al., 2004). Historically, and most commonly known, Ago2 function is associated with non-coding RNAs (miRNAs or siRNAs) and its participation in formation of the RNA-induced silencing complex (RISC) responsible for gene silencing through mRNA cleavage (Hammond et al., 2001; Martinez et al., 2002). Ago2 has also been reported to play a key role in translational repression, but by incompletely understood mechanisms (Djuranovic et al., 2011; Filipowicz et al., 2008). Recently, Ago2 has been associated with multiple additional functions in the cell (Meister, 2013). Ago2 is involved in RNAi mediated regulation of gene expression, miRNA biogenesis, protection from viruses, transcriptional regulation, and even alternative splicing (Ameyar-Zazoua et al., 2012; Cernilogar et al., 2011; Czech and Hannon, 2011; Djuranovic et al., 2011; Fabian et al., 2010; Filipowicz et al., 2008; Hutvagner and Simard, 2008). Although many Ago2 functions are most often associated with its ability to bind small RNAs (Fabian et al., 2010), recent examples indicate that small RNAs are not involved in all Ago2 functions. A PUF-Ago-eEF1A complex responsible for attenuation of translational elongation does not depend on small RNAs (Friend et al., 2012). Ago2 recruitment to chromatin is also RNAi independent (Moshkovich et al., 2011)… Small RNA-independent recruitment to mRNA was also recently demonstrated for one of two argonautes in Drosophila (Pinder and Smibert, 2013). The wide variability in Ago2 reliance on small RNAs and RNase activity may reflect multiple parallel functions in the cell. Experiments with Ago2 knock down and its overproduction demonstrated that it is functionally involved in the RAPP process (Figure 6). Ago2 was found in proximity to mutated nascent chains during their synthesis, with direct correlation between the severity of a mutation in signal sequence and more efficient crosslinking of the nascent chain to Ago2 (Figure 5A). Moreover, we have found that mutant protein mRNA was specifically enriched in Ago2 IP experiments (Figures 5G, S4C–F). Although questions remain about whether Ago2 binds mutant mRNA directly, during translation or at later stages, the data suggest Ago2 is in close proximity to the mutant nascent chain and mRNA at the same time at the ribosome. Association of Ago2 with mutated nascent chains during translation is consistent with earlier observations of Ago2 association with translating ribosomes (Landthaler et al., 2008; Nelson et al., 2004). It was also shown that Ago2 forms a complex containing L5, L11 ribosomal proteins, along with 5S RNA (Ishizuka et al., 2002). Experiments with slicer inactive Ago2, as well as, experiments on Dicer and Drosha knockdowns (Figures 6B and S5) suggest that Ago2’s function in RAPP is small RNA- and slicer-independent.
Although precise role for Ago2, its ability to act as a sensor in the process, as well as other details of the mechanism remain obscure, the data presented here indicate that the stability of mRNA is dictated by the balance of nascent chain interactions with specific cellular factors at the ribosome exit site, thereby providing a means to selectively reduce the load of defective proteins at an early point in translation by specifically degrading their mRNAs. RAPP provides a novel mechanism by which defects in the signal sequence of a secretory protein, or in the secretory machinery, lead to a dramatic reduction in substrate mRNA and thus expression of the protein.
EXPERIMENTAL PROCEDURES
Protein translocation was studied using an in vitro translation/translocation system in the presence of purified canine microsomes or by maturation of the proteins in human cell culture. Protein visualization in the cells was conducted by immunocytochemistry and confocal microscopy. Cellular mRNA levels were analyzed by Northern blot or qPCR, and proteins by Western blot. For the mRNA stability/degradation experiments, WT and mutant PPLs cloned under control of the Tet-response element into pTRE2hyg were expressed in HeLa Tet-Off cells (Clontech) in the presence/absence of doxycycline. SRP54, Ago2, and other protein depletions in human cell culture were achieved by siRNA technique. Site-specific photocrosslinking using tRNA-mediated incorporation of a photoreactive probe into a nascent chain during in vitro translation was conducted as before (Krieg et al., 1986; McCormick et al., 2003). Method for identification of proteins interacting with nascent chains employs: 1) in vitro preparation of salt-washed RNCs containing the photocrosslinking probe in specific position of the nascent chain; 2) fractionation of the active lysates and assay of fractions containing the protein of interest using photocrosslinking with salt-washed RNCs; and 3) identification of proteins in the selected, purified fractions by mass spectrometry. SRP and microsomes for in vitro experiments were purified from canine pancreas. Recombinant human Ago2 was expressed in and purified from cultured insect cells (Ye et al., 2011).
Supplementary Material
Highlights.
Mechanism for regulation of aberrant protein production (RAPP) is described
Mutations in the signal sequence shift proximity from SRP to Ago2
Defective SRP nascent chain interactions trigger degradation of the encoding mRNA
Ago2 is required for the specific degradation of the mutant mRNA
ACKNOWLEDGEMENTS
We thank Xuecheng Ye for recombinant human Ago2 purification; Yiwei Miao and Yuanlong Shao for SRP, microsomes and εANB-Lys-tRNAamb preparation for some experiments; Tara Hessa for introducing some mutations into the PPL gene; Yuh Min Chook for the plasmid containing pyruvate kinase gene. Research reported in this publication was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDKS) of the National Institutes of Health R37DK049835 and CFF grants to P.J.T.; by National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award number R01GM26494 and Robert A. Welch Foundation Chair grant BE-0017 to A.E.J.; grants SFB 746, Forschergruppe 967, and EXC 294 to S.R.; by the Swedish Foundation for Strategic Research, the Swedish Research Council and the Swedish Cancer Foundation to G.v.H. and I.M.N.; the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), Magn. Bergvalls Stiftelse and the Carl Trygger Foundation to I.M.N. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and three tables and can be found with this article online.
NOTE ABOUT CONTRIBUTION
Anna E. Patrick and Zemfira N. Karamysheva contributed equally to this work.
REFERENCES
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- Andrews DW, Lauffer L, Walter P, Lingappa VR. Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J. Cell Biol. 1989;108:797–810. doi: 10.1083/jcb.108.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biology. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Krieg UC, Walter P, Johnson AE. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Luyet PP, Parone P, Falguieres T, Strub K. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp. Cell Res. 2007;313:834–847. doi: 10.1016/j.yexcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- McCormick PJ, Miao Y, Shao Y, Lin J, Johnson AE. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA. 2005;102:10064–10069. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013;49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder BD, Smibert CA. microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep. 2013;14:80–86. doi: 10.1038/embor.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nature Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. (Berl) 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nat. Struct. Mol. Biol. 2004;11:1049–1053. doi: 10.1038/nsmb853. [DOI] [PubMed] [Google Scholar]
- Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat. truct. Mol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.