Abstract
Several models of cell fate determination can be invoked to explain how single retinal progenitor cells (RPCs) produce different cell types in a terminal division. To gain insight into this process, the effects of the removal of a cell fate regulator, Notch1, were studied in newly postmitotic cells using a conditional allele of Notch1 (N1-CKO) in mice. Almost all newly postmitotic N1-CKO cells became rod photoreceptors, whereas wild-type (WT) cells achieved a variety of fates. Single cell profiling of wild-type and N1-CKO retinal cells transitioning from progenitor to differentiated states revealed differential expression of inhibitor of DNA binding factors Id1 and Id3, as well as Notch-regulated ankyrin repeat protein (Nrarp). Misexpression of Id1 and Id3 was found to be sufficient to drive production of Müller glial cells and/or RPCs. Moreover, Id1 and Id3 were shown to partially rescue the production of bipolar and Müller glial cells in the absence of Notch1 in mitotic and newly postmitotic cells. Misexpression of Nrarp, a downstream target gene and inhibitor of the Notch signaling pathway, resulted in the overproduction of rod photoreceptors at the expense of Müller glial cells. These data demonstrate that cell fate decisions can be made in newly postmitotic retinal cells, and reveal some of the regulators downstream of Notch1 that influence the choice of rod and non-rod fates. Taken together, our results begin to address how different signals downstream from a common pathway lead to different fate outcomes.
Keywords: Id, Notch, Nrarp, Cell fate, Postmitotic, Retina, Mouse
INTRODUCTION
The retina is an area of the central nervous system that offers excellent accessibility and a well-characterized anatomy. These attributes have allowed investigation of its development, including lineage analyses (Holt et al., 1988; Turner and Cepko, 1987; Turner et al., 1990; Wetts and Fraser, 1988) and birthdating studies (Rapaport et al., 2004; Young, 1985a). During development, retinal neurons arise in a conserved, temporal sequence from multipotent, cycling RPCs (reviewed by Livesey and Cepko, 2001). Retinal ganglion cells are born first, followed by horizontal cells, cone photoreceptors, rod photoreceptors, amacrine cells, bipolar cells and Müller glia (Sidman, 1961; Wong and Rapaport, 2009; Young, 1985a).
Previous studies have not established how fate decisions are determined in retinal cells. One possibility is that a RPC might decide the fate of daughter cells and then pass on this decision via determinants. In support of this model, heterochronic mixing experiments showed that embryonic RPCs determine the fate of their amacrine cell daughters (Belliveau and Cepko, 1999). Furthermore, recent studies of chick and zebrafish retinal development revealed RPCs that make horizontal cells exclusively, suggesting the inheritance of the horizontal fate from a committed RPC (Godinho et al., 2007; Rompani and Cepko, 2008). In the cerebral cortex, the laminar fate of cortical cells was shown to be determined in the late S or G2 stage of a terminal cell cycle, and thus in the progenitor cell (McConnell and Kaznowski, 1991). Alternatively, even though newly postmitotic cells receive determinants from their progenitors, they may remain uncommitted. For example, when newly postmitotic cells fated to become rod photoreceptor cells were treated with ciliary neurotrophic factor, they began to express bipolar cell markers (Ezzeddine et al., 1997). This suggests that some postmitotic cells are plastic, or at least can change fate in response to signals after exiting cell cycle. It is possible that cell fate determination can occur at different stages in the continuum from RPC to postmitotic daughter, perhaps with different cell types choosing their fates at different points in this continuum.
During retinal development, the Notch signaling pathway regulates both cell cycle exit and cell fate specification (Jadhav et al., 2006; Yaron et al., 2006). Genetic removal of Notch1, or Notch downstream effectors such as Hes1 and RBP-J, during early and late stages of retinal development resulted in precocious cell cycle exit and the overproduction of cone and rod photoreceptors (Jadhav et al., 2006; Riesenberg et al., 2009; Tomita et al., 1996; Yaron et al., 2006; Zheng et al., 2009). Similar outcomes were observed when retinal explants were treated with DAPT, an inhibitor of the protease γ-secretase, which is required to activate Notch signaling (Nelson et al., 2007). These studies provide evidence that Notch signaling maintains RPCs in a cycling state, and determines cell type identity by inhibiting the photoreceptor fate. However, it is unknown whether the timing of Notch1 cell fate regulation is restricted to RPCs only, or whether postmitotic cells still require signal input to achieve non-rod fates. In order to investigate whether cell fate determination was dependent on Notch1 after cell cycle exit, the effects of removal of a Notch1 conditional allele in newly postmitotic cells were analyzed. In addition, single cell microarrays were performed to investigate gene expression changes that might lead to the acquisition of rod and non-rod fates (Mizeracka et al., 2013). Expression of Id1 and Id3, which may be direct targets of Notch signaling (Meier-Stiegen et al., 2010; Reynaud-Deonauth et al., 2002; Ruzinova and Benezra, 2003; Yokota, 2001), was found to be reduced. These factors have been shown to prevent differentiation during neurogenesis in the central nervous system (CNS) (Cai et al., 2000; Lyden et al., 1999). Another gene whose expression levels changed, Nrarp, is a known Notch downstream target gene (Krebs et al., 2001; Pirot et al., 2004) that serves as a feedback inhibitor of the Notch signaling pathway (Lamar et al., 2001; Yun and Bevan, 2003). Functional studies were carried out to assess the role of Id factors and Nrarp in the developing retina. These data begin to address how different fate outcomes arise from the same signaling pathway in RPCs and their newly postmitotic daughter cells.
MATERIALS AND METHODS
Animals
Notch1fl/fl were maintained as homozygotes (Radtke et al., 1999). CD-1 mice were obtained from Charles River Laboratories. All experiments were approved by the Institutional Animal Care and Use Committee at Harvard University.
Misexpression constructs
CAG:Id1 and CAG:Id3 were constructed by PCR amplification from full-length mouse cDNA clones (Matsuda and Cepko, 2004). Each construct was verified by sequencing. The full-length mouse cDNA sequence encoding Nrarp was cloned into the LIA vector at the NotI site (Bao and Cepko, 1997).
Electroporation and infection
In vivo injections of DNA constructs and viruses were performed as previously described, with the exception that an oocyte microinjector (Drummond) and pulled glass pipettes (Dumont/Drummond) were used to deliver 0.2 μl of 5 μg/μl DNA solution or 107 CFU/ml viral stock into the subretinal space of the postnatal mouse eye (Matsuda and Cepko, 2004). In vitro electroporations were performed as previously described (Matsuda and Cepko, 2004).
Viruses used include LIA, LIA-Cre (Bao and Cepko, 1997; Jadhav et al., 2006), BAG (Price et al., 1987), LIA-Id1-2A-Cre and LIA-NRARP. DNA constructs used include CAG:GFP, CAG:Cre, CALNL-GFP (Matsuda and Cepko, 2004), Cralbp:dsRed, Hes1:tdTomato (Matsuda and Cepko, 2007), Chx10:tdTomato (Kim et al., 2008) and CAG:Id1, CAG:Id3. Empty vectors were added to maintain equimolar ratios among DNAs that were co-injected.
Intraperitoneal injections into newborn pups were performed to deliver EdU at 1 μg/μl in PBS, with a total of 10 μl per pup.
Histology and immunohistochemistry
Retinas were fixed and processed for cryosections as described previously (Matsuda and Cepko, 2004; Trimarchi et al., 2007), starting either as wholemounts (fixed for 30 minutes at 4°C with 0.5% glutaraldehyde) or as eyeballs (fixed for 2 hours in 4% PFA at room temperature in PBS, pH 7.4). Primary antibodies used in this study include: chicken anti-GFP (1:2000; Abcam), rabbit anti-Chx10 (1:500; C. L. Cepko’s laboratory), rabbit anti-Id3 (1:500; Abcam) and mouse anti-p27Kip1 (1:50; BD Biosciences Transduction Laboratories). EdU detection and TUNEL staining were performed according to manufacturer’s instructions. X-gal and alkaline phosphatase staining was performed as described previously (Bao and Cepko, 1997; Price et al., 1987). Section in situ hybridization was performed as previously described (Trimarchi et al., 2007).
Microscopy and image analysis
Confocal microscopy to obtain images was performed using a Leica TCS SP5 microscope. Imaris 5.7 software (Bitplane) was used to analyze, quantify and uniformly adjust images.
FACS purification and semi-quantitative PCR
Electroporated retinas were dissociated to single cells via papain treatment (Trimarchi et al., 2007). FACS was performed on a BD Aria II sorter or Accuri C6 Analyzer, gated for GFP and dsRed/tdTomato detection. For semi-quantitative PCR, 3-5×105 GFP+ cells were collected from two dissociated retinas for each sample. After sorting, GFP+ cells were lysed in Trizol (Invitrogen) and stored at -80°C. Phenol-chloroform extractions were performed to isolate total RNA. cDNA was generated using Accuscript High Fidelity (Agilent Technologies) according to manufacturer’s guidelines. Semi-quantitative real-time PCR was performed and gene expression was normalized according to actin expression in each sample. Primers used included: actin, accaactgggacgacatggagaa and tacgaccagaggcatacagggac; Id1, acatgaacggctgctactca and gtggtcccgacttcagactc; Id3, actcagcttagccaggtgga, tcagtggcaaaagctcctct.
Statistical methods
For each condition, three or more retinas were analyzed. Data were grouped together according to category (i.e. percentage of electroporated cells of a particular cell type). A Student’s two-tailed t-test was used to compare differences between control and experimental values for statistical significance.
Microarray data
The raw and processed Affymetrix data files have been deposited in the NCBI Gene Expression Omnibus, Accession Number GSE35682.
RESULTS
Viral mediated loss of Notch1 reveals activity in newly postmitotic cells
In order to determine whether Notch1 signaling plays a role in cell fate specification in newly postmitotic cells, two independent strategies were undertaken. The first strategy takes advantage of the manner in which gammaretroviruses integrate the viral genome and express viral genes. Upon entering a host cell, viral reverse transcriptase creates only a single copy of the viral genome in the cytoplasm. The viral DNA in the pre-integration complex of a gammaretrovirus, which is the type used for lineage tracing, cannot penetrate the nuclear envelope. Thus, integration of the viral DNA into the host genome, which allows for stable marking of a clone, can occur only after the breakdown of the nuclear envelope during M phase (Roe et al., 1997). As the host genome will be 4N at this time, and there is a single copy of the viral genome, only one of the daughter cells of the initially infected cell will inherit the viral genome. In all subsequent cell cycles, the integrated viral genome will be replicated along with that of the host, and thus all of the progeny of the cell with the integrated viral genome will be marked (Fig. 1A,B). Clones can consist of one to thousands of cells following infection of the retina at various times (Fekete et al., 1994; Fields-Berry et al., 1992; Rompani and Cepko, 2008; Turner and Cepko, 1987; Turner et al., 1990). We focused our analysis on one-cell clones, in which viral transgene expression would initiate in a cell that exited the cell cycle, after being generated by the initially infected cell (Fig. 1B).
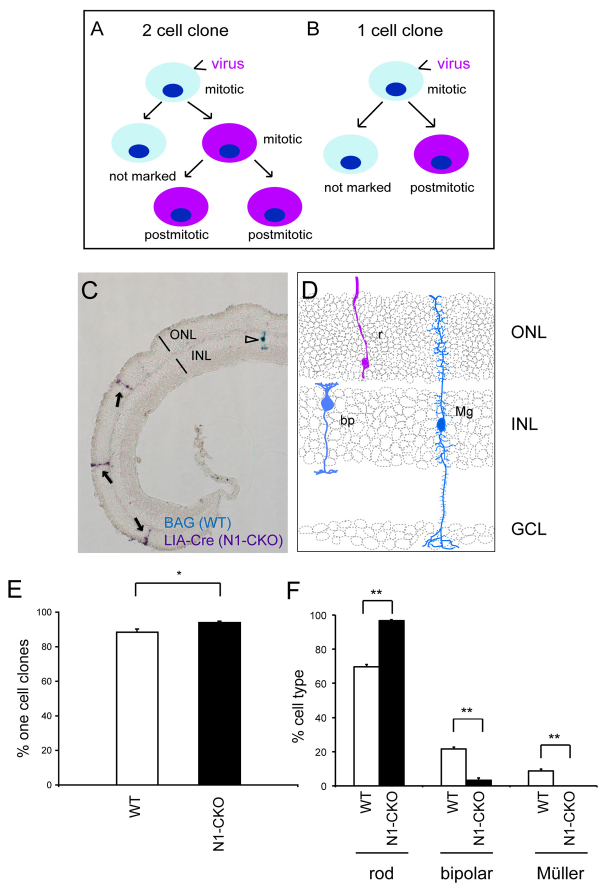
Fig. 1.
Clonal inactivation of Notch1 in newly postmitotic cells. (A,B) Retinal progenitor cells (RPCs) were infected with replication-incompetent gammaretroviruses that integrate into the genome of the host cell only after the breakdown of the nuclear envelope during mitosis. Because integration is necessary for expression, the initially infected RPC does not express the viral genome. When the initially infected RPC divides, one of its daughter cells inherits a single integrated copy of the viral genome and expresses it. A two-cell clone results when a daughter cell that inherited the viral genome divides to make two postmitotic daughters (A), whereas a one-cell clone results when a daughter cell that inherited the viral genome becomes postmitotic (B). (C) Notch1fl/fl retinas were co-infected in vivo at P3 with LIA-Cre and BAG retroviruses. LIA-Cre virus encodes both AP and Cre, and was used to generate Notch1-conditional knockout (N1-CKO) clones (purple cells, arrows). BAG encodes β-galactosidase, but not Cre, to mark wild-type clones (blue cell, arrowhead). An analysis of the size and composition of all clones was performed after P21. (D) Schematic of cell types marked. (E) Percentage of all clones from wild-type and N1-CKO that comprised only one cell. (F) Quantification of cell types found in wild-type or N1-CKO one-cell clones. n=3 retinas per condition, 464 BAG one-cell clones, 600 LIA-Cre one-cell clones. **P<0.01. Error bars indicate s.e.m. in all figures. r, rod; bp, bipolar cell; Mg, Müller glial cell; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
We wanted to remove Notch1 at a time point when the majority of clones produced normally would be one-cell clones. Proliferation wanes in the postnatal rodent retina (Alexiades and Cepko, 1996; Rapaport et al., 2004; Young, 1985a), and many small or one-cell clones have been observed following viral infection at these time points (Fields-Berry et al., 1992; Turner and Cepko, 1987; Turner et al., 1990). In order to deplete postmitotic cells of Notch1, postnatal day 3 (P3) was chosen as a time point for infection of Notch1fl/fl mice with a retrovirus encoding Cre and alkaline phosphatase. As a control, BAG, a virus encoding lacZ, but not Cre, was delivered at the same time to the same retinas. This allowed an assessment of the clone size and the types of cells normally produced by P3 RPCs. Co-infection was unlikely, as the infection rates of both BAG and LIA-Cre were low.
BAG and LIA-Cre retroviruses were co-injected in vivo into the subretinal space of P3 Notch1fl/fl retinas. After the completion of retinal development (P21 or later), retinas were processed via histochemical staining to detect BAG and LIA-Cre clones (Fig. 1C, schematized in D). As predicted by previous studies of retinal proliferation (Young, 1985a), 88.6±1.7% of BAG clones comprised only one cell. These data demonstrate that the vast majority of cells that integrated the viral genome did not re-enter the cell cycle, i.e. were in the process of exiting or were postmitotic when they initiated expression of viral genes (Fig. 1E). Among the one-cell BAG clones, 69.6±1.5% of cells were rods, 21.7±1.0% were bipolar cells and 8.7±1.3% were Müller glial cells (Fig. 1C,F). By contrast, the one-cell clones derived from infection with LIA-Cre resulted in 96.6±0.8% rods, 3.4±0.8% bipolar cells and 0±0% Müller glial cells (Fig. 1C,F). There also was an increase in the frequency of one-cell clones, to an average of 94.3±0.8% when compared with 88.6±1.7% (Fig. 1E). This is likely due to the loss of Notch1 in cells that would normally re-enter the cell cycle to produce two-cell clones. Although this effect on cell cycle likely occurred in RPCs, the number of RPCs affected was too small to create the increase in rods among the one-cell LIA-Cre clones. The overproduction of rods at the expense of other cell types is consistent with Notch1 being required for the acquisition of non-rod fates in newly postmitotic cells.
Loss of Notch1 function in newly postmitotic electroporated N1-CKO cells
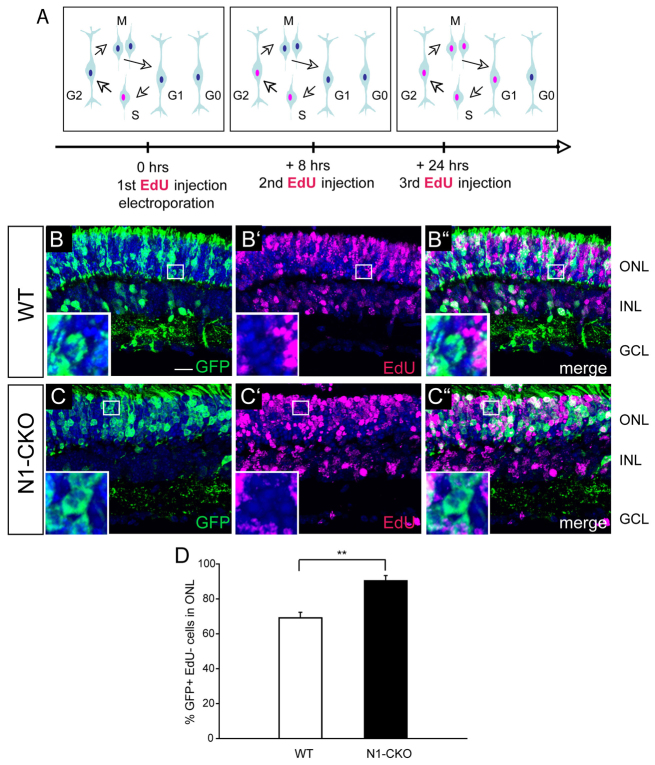
An independent approach was undertaken to assess the role of Notch1 signaling in newly postmitotic cells. Newly postmitotic cells were identified among cells that had been electroporated with a plasmid encoding GFP, but which had not undergone an S-phase. This was accomplished by labeling with EdU, a thymidine analog that is incorporated during S phase (Salic and Mitchison, 2008). A plasmid encoding CAG:Cre, which uses the broadly active promoter CAG to drive Cre expression, and a plasmid, CALNL-GFP, which uses CAG to drive expression of a Cre-responsive GFP reporter (Matsuda and Cepko, 2007), were co-electroporated in vivo into wild-type and Notch1fl/fl retinas at P1. Mitotic cells were labeled with three injections of EdU: immediately after electroporation, 8 hours after electroporation and 24 hours after electroporation. These EdU injections were timed according to the lengths of the cell cycle phases (16 hours for S, minimum of 2.6 hours for G2, 8.5 hours for G1 and 2.5 hours for M) and the overall cell cycle length at P1 (∼30 hours) (Young, 1985b) to ensure that all cycling cells were labeled (Fig. 2A,B′,C′). As previous work suggested that mitotic cells and exiting cells are preferentially electroporated (Matsuda and Cepko, 2004), GFP+ EdU-cells were interpreted as cells that were near the scleral surface where DNA was injected, but were exiting or had recently exited cell cycle, and thus did not take up the EdU label. Retinas were harvested after P14 and the fate of GFP+ EdU-cells was assessed by location in the outer nuclear layer (where only photoreceptors reside) versus inner nuclear layer (where interneurons and Müller glia reside) (Fig. 2B,B′,B′,C,C′,C′). The majority, 90±3.2%, of GFP+ EdU-cells became photoreceptors in Notch1fl/fl retinas when compared with 69±3.0% in wild-type retinas, providing evidence that postmitotic precursors that lost Notch1 became rod photoreceptors at the expense of other cell types (Fig. 2D). These results are in accordance with the viral Cre experiments described above. In addition, this cell fate change did not appear to be due to excessive cell death, as N1-CKO retinas did not show significantly more TUNEL+ cells as compared with wild-type retinas (9.0±4.2 cells per 300×300 μm2 electroporated area in N1-CKO versus 9.2±5.5 in wild type).
Fig. 2.
Depletion of Notch signaling in newly postmitotic cells. (A) Wild-type and Notch1fl/fl retinas were electroporated in vivo at P1 with CAG:Cre and a Cre-responsive GFP reporter, CAG:CALNL-GFP. In order to assess the fate of electroporated newly postmitotic cells, all cells that underwent an S phase during or after the electroporation were labeled with EdU. Based on the length of the cell cycle (∼30 hours) and S phase (∼16 hours) (Young, 1985b), three EdU injections were performed. The first injection was performed at the time of electroporation to label cells in S phase (indicated by magenta nuclei). The second injection was 8 hours after electroporation to label any electroporated cells that had progressed into S phase. The third injection was 24 hours after electroporation to label any cells that had been in G2 at the time of electroporation and had re-entered S phase. (B-C′) Electroporated cells that were not labeled by EdU, and thus had become postmitotic after electroporation, were identified as GFP+ and EdU-. Retinas were harvested after P14, sectioned and stained for GFP (B,C), EdU (B′,C′) and DAPI. Examples of GFP+ EdU-cells are shown at higher magnification in insets. (B-C′) The fate of GFP+ EdU-cells was assessed by location either in the outer nuclear layer (rod photoreceptors) or in the inner nuclear layer (interneurons and Müller glia). (D) Percentage of GFP+ EdU-cells found in the ONL for wild-type or N1-CKO conditions. Scale bar: 50 μm. n=3 retinas per condition. **P<0.01. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Id1 and Id3 expression is reduced in the absence of Notch1
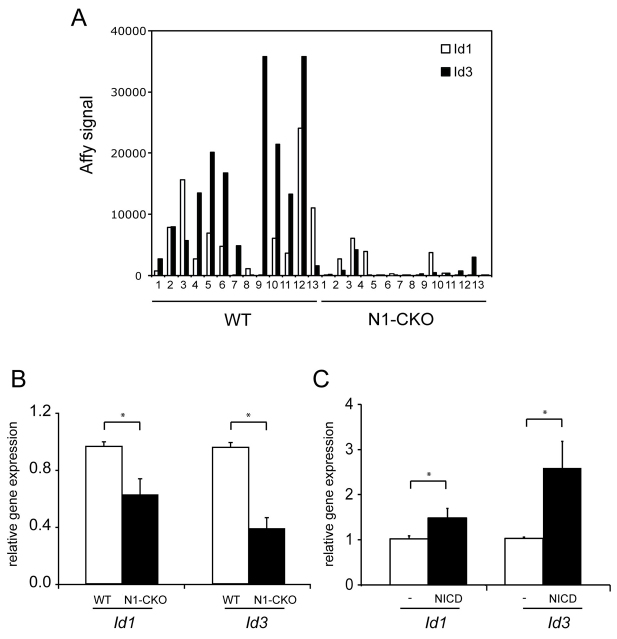
Examination of microarray data from N1-CKO and wild-type cells transitioning from RPCs to newly postmitotic cells led to the discovery that Id1 and Id3 were almost completely absent in N1-CKO cells (Fig. 3A) (Mizeracka et al., 2013). This was confirmed by a semi-quantitative PCR assay on cDNA made from populations of N1-CKO and wild-type cells. Retinas of Notch1fl/fl P0 pups were electroporated in vitro with plasmids encoding CAG:Cre, along with a Cre-responsive GFP reporter, CALNL-GFP. For controls, the retinas of sibling Notch1fl/fl pups were electroporated with CAG:GFP. Electroporated retinas were cultured for 3 days, dissociated to single cells, and GFP+ cells were collected using fluorescence-activated cell sorting (FACS). In accordance with the changes observed by microarray analysis, semi-quantitative qPCR determined that Id1 and Id3 RNA levels were lower in N1-CKO cells when compared with wild-type cells (Fig. 3B). In order to further investigate whether Id1 and Id3 were sensitive to Notch signaling, P0 wild-type retinas were electroporated with an activated form of the Notch receptor (CAG:NICD) and CAG:GFP. GFP+ cells were collected 14 hours later and subjected to qPCR, which showed that both Id1 and Id3 were upregulated following the addition of NICD (Fig. 3C).
Fig. 3.
Analysis of Id1 and Id3 expression following Notch1 manipulation. (A) Microarrays were performed on 13 single wild-type cells and 13 single N1-CKO cells from retinas electroporated at P0 and harvested at P3 (Mizeracka et al., 2013). The Affymetrix signal levels for Id1 and Id3 in each single cell. (B) Notch1fl/fl retinas were electroporated at P0 in vitro with CAG:GFP to mark wild-type cells, or with CAG:Cre and CALNL-GFP, a Cre-responsive reporter, to mark N1-CKO cells. Electroporated retinas were cultured for 3 days and dissociated to single cells. GFP+ cells from wild-type or N1-CKO conditions were collected by FACS and used to prepare cDNA. Samples were subjected to semi-quantitative real-time PCR in order to detect differences in expression of Id1 and Id3 between N1-CKO and wild-type cells. All expression values were normalized to actin expression levels in each sample. *P<0.05. (C) Wild-type retinas were electroporated at P1 in vitro with CAG:GFP for controls, or with CAG:GFP and CAG:NICD, and harvested for qPCR analysis 14 hours later. Differences in expression of Id1 and Id3 between wild-type and NICD cells, normalized to actin. n=3 retinas per condition. *P<0.05.
Detection of Id1 and Id3 RNA by in situ hybridization showed that these genes are expressed throughout the progenitor layer at P1, with expression becoming more restricted to the inner neuroblastic layer, where newly postmitotic amacrine cells are found, and to the ganglion cell layer a few days later (supplementary material Fig. S1A,B,D,E). At P5, when proliferation has almost ceased in the central retina (Young, 1985a), Id1 and Id3 expression was localized to the inner nuclear layer, most likely in newly exited cells that are becoming Müller glial cells (supplementary material Fig. S1C,F). To confirm that Id factors are expressed in newly postmitotic cells, mitotic cells were labeled by three successive EdU injections. The EdU-labeled retinas were stained by anti-Id3 and examined for EdU and Id3 signals. Id3 staining was detected in both EdU+ and EdU-cells, demonstrating that this protein is expressed in both mitotic and postmitotic populations (supplementary material Fig. S2).
Functional analysis of Id1 and Id3 in the developing WT retina
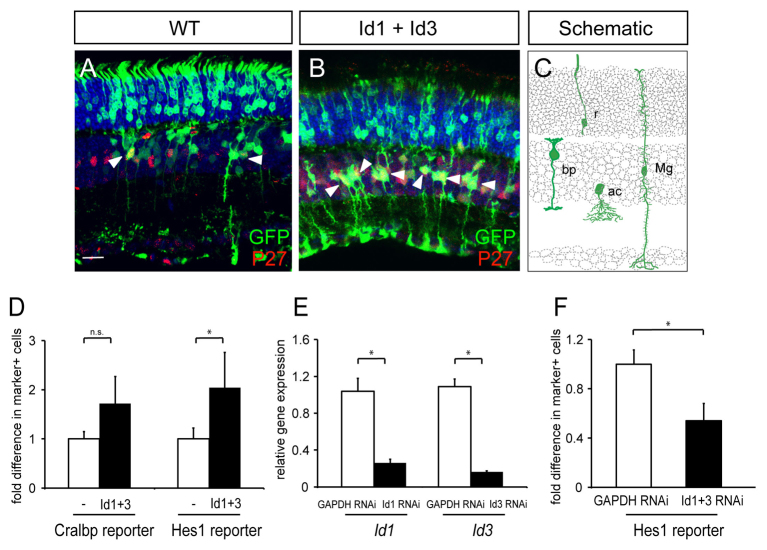
In order to better understand Id function, retinas of wild-type P0 pups were electroporated in vivo with plasmids expressing the Id genes (CAG:Id1, CAG:Id3 or both plasmids) along with CAG:GFP. Retinas were harvested after P14, and the fates of the electroporated cells were assessed by morphology and molecular markers. Misexpression of CAG:Id1 and CAG:Id3 led to the overproduction of cells that exhibited features of both RPCs and Müller glial cells, when compared with wild-type control retinas (Fig. 4A-C). First, the morphology of the induced cells resembled that of RPCs or Müller glial cells, in that their cell bodies were located in the INL, with long processes that extended both into the photoreceptor layer and the GCL (Fig. 4B, schematized in C). Furthermore, these cells were positive for a marker of both RPCs and Müller glial cells, the cyclin-dependent kinase inhibitor p27, by immunohistochemistry (Fig. 4B). Misexpression of Id1 or Id3 alone also resulted in induction of extra RPC/Müller-like cells, but not to the extent observed when both Id factors were misexpressed together (data not shown).
Fig. 4.
Misexpression of Id1 and Id3 in the wild-type postnatal retina. (A,B) Wild-type retinas were electroporated in vivo at P0 with CAG:GFP for controls or CAG:GFP, CAG:Id1 and CAG:Id3. The fates of electroporated cells were analyzed in the mature retina (after P14) by assessment of their location and morphology in the retinal layers. Electroporation of CAG:GFP alone (WT) or CAG:GFP, CAG:Id1 and CAG:Id3 (Id1+Id3) labeled GFP+ photoreceptors, interneurons and Müller glial cells. Immunohistochemistry for p27 (red)-labeled Müller glial cells (arrowheads demarcate examples). Scale bar: 50 μm. (C) Schematic of cell types marked. (D) Combinations of plasmids encoding CAG:GFP, cell type-specific reporters (Cralbp:dsRed to mark Müller glial cells or Hes1:tdTomato to mark RPCs/Müller glial cells), and Id factors were co-electroporated in vitro into P0 wild-type retinas. Retinas were cultured for 11-12 days and dissociated to single cells. FACS was used to determine the percentage of GFP+ cells that were dsRed+ or tdTomato+ for each condition. Fold difference is the percentage of GFP+marker+ cells induced by Id1+Id3 misexpression relative to wild type. n=5 retinas per condition. *P<0.05; n.s., P>0.05. (E) Wild-type retinas were electroporated at P0 in vitro with CAG:GFP and GAPDH:RNAi for controls, CAG:GFP and Id1 RNAi or CAG:GFP and Id3 RNAi. Retinas were harvested for qPCR analysis 3 days later to detect differences in expression of Id1 and Id3, normalized to actin expression. n=2 retinas per condition. *P<0.05. (F) Wild-type retinas were electroporated in vitro at P0 with CAG:GFP, Hes1:tdTomato, and GAPDH RNAi or Id1+3 RNAi. Retinas were cultured for 11-12 days and dissociated to single cells. FACS was used to determine the percentage of GFP+ cells that were tdTomato+ for each condition. Fold difference is the percentage of GFP+Hes:tdTomato+ cells in Id1+3 RNAi conditions relative to GAPDH RNAi. n=5 retinas per condition. *P<0.05. r, rod; bp, bipolar cell; Mg, Müller glial cell; ac, amacrine cell.
In order to quantify a large number of retinal cells transduced by combinations of genes via electroporation, we used FACS of electroporated cells marked by expression of cell type-specific reporters. Wild-type retinas were electroporated in vitro at P0 with combinations of CAG:GFP, CAG:Id1, CAG:Id3 and a cell type-specific reporter. These reporters included regulatory sequences upstream of Cralbp (which marks Müller glial cells at P14) and Hes1 (which marks RPCs at early postnatal stages and Müller glia at later stages) driving dsRed and tdTomato expression, respectively (Matsuda and Cepko, 2007). After electroporation, retinas were cultured in vitro as explants for 11-12 days and dissociated to single cells. FACS was used to count single- (GFP+) and double-positive cells (GFP+dsRed+ or GFP+tdTomato+). Co-expression of CAG:Id1, CAG:Id3, CAG:GFP and Cralbp:dsRed resulted in an increase of marker-positive cells by 72.5% when compared with wild-type retinas electroporated with CAG:GFP and Cralbp:dsRed without the Id genes (Fig. 4D). Co-expression of CAG:Id1 and CAG:Id3, along with CAG:GFP, and Hes1:tdTomato resulted in a robust increase of marker-positive cells, by 104.1%, when compared with wild type (Fig. 4D).
Two RNAi constructs, targeting either Id1 or Id3, were individually tested for efficacy (Fig. 4E), and then electroporated together with CAG:GFP and Hes1:tdTomato at P0 (Fig. 4F). For controls, GAPDH RNAi, which does not have an effect on retinal development (Matsuda and Cepko, 2004), was co-electroporated with CAG:GFP and Hes1:tdTomato. Co-expression of Id1 and Id3 RNAi resulted in a 45.9% decrease in the number of marker-positive cells, consistent with a reduction in RPC/Müller glial cells (Fig. 4F).
Rescue of Notch1 loss-of-function phenotype by Id1 and Id3
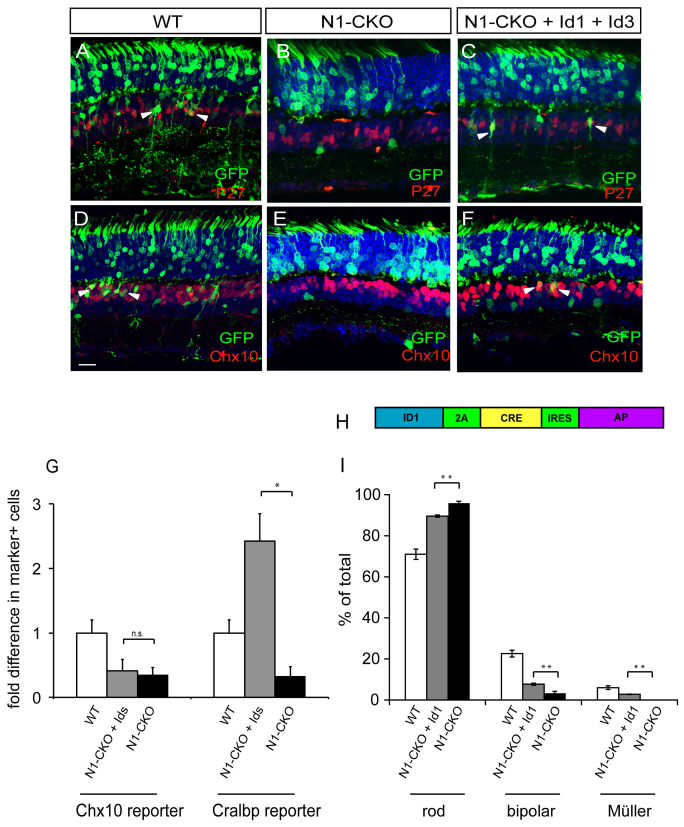
Id1 and Id3 RNA levels were significantly lower in the absence of Notch1, and increased shortly after delivery of NICD. In addition, RPC/Müller-like cells were induced by misexpression of Id1 and Id3 in wild-type retinas. These observations led to the hypothesis that these genes are upregulated by Notch signaling, whereupon they induce non-rod fates. To test this, retinas of Notch1fl/fl animals were electroporated in vivo at P0 with combinations of CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3. Retinas were harvested after P14, sectioned, and stained for GFP and cell type-specific markers. Because Müller glial cells and bipolar cells were the predominant cell types lost in the absence of Notch1 (Jadhav et al., 2006), the production of these cell types was used as a means to assess the extent of rescue by Id1 and Id3. When CAG:Cre and CALNL-GFP were co-expressed in Notch1fl/fl retinas, and immunohistochemistry and morphology were used to assess the fate of the cells, none of the resultant GFP+ cells resembled Müller glial cells or bipolar cells (Fig. 5B,E). Additionally, none of these GFP+ cells expressed the Müller glial marker p27 (Fig. 5B) or the bipolar cell marker Chx10 (Fig. 5E). By contrast, retinas electroporated with CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3 exhibited cells with the morphology of bipolar and Müller glial cells (Fig. 5C,F), although fewer than in wild-type retinas (Fig. 5A,D). In addition to their specific morphologies, these cells expressed p27 or Chx10 (Fig. 5C,F).
Fig. 5.
Misexpression of Id1 and Id3 in N1-CKO retinas. (A-F) Notch1fl/fl retinas were electroporated in vivo at P0 with CAG:GFP, or CAG:Cre and CALNL-GFP, or CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3. The fates of the electroporated cells were analyzed in the mature retina (after P14) using morphology, location and immunohistochemistry. Müller glial cells express p27 and bipolar cells express high levels of Chx10 (red). (A,D) Electroporation of CAG:GFP alone resulted in GFP+ photoreceptors, interneurons and Müller glial cells, some of which were positive for p27 or Chx10 (arrowheads indicate examples). (B,E) Electroporation of CAG:Cre and CALNL-GFP into Notch1fl/fl retinas resulted in GFP+ photoreceptors and some amacrine cells, none of which was positive for p27 or Chx10. (C,F) Electroporation of CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3 resulted in GFP+ photoreceptors, interneurons and Müller glial cells, some of which were positive for p27 and Chx10 (arrowheads demarcate examples). Scale bar: 50 μm. (G) Combinations of plasmids encoding CAG:GFP or CAG:Cre and CALNL-GFP, cell type-specific reporters (Chx10:tdTomato to mark bipolar cells and Cralbp:dsRed to mark Müller glial cells), and Id factors were co-electroporated in vitro into P0 Notch1fl/fl retinas. Retinas were cultured for 11-12 days and dissociated to single cells. FACS was used to determine the percentage of GFP+ cells that were dsRed+ or tdTomato+ for each condition. Fold difference is the percentage of GFP+marker+ cells in N1-CKO conditions, or induced by Id1+Id3 misexpression in N1-CKO conditions, relative to wild type. n=5 retinas per condition. *P<0.05; n.s., P>0.05. (H) The LIA retrovirus was engineered to express Id1, Cre and AP. Notch1fl/fl retinas were infected with LIA-Id1-2A-Cre-IRES-AP at P3 and the fate of one-cell clones was assessed after P21 by histochemical staining. (I) Quantification of cell types found in BAG (WT), LIA-Cre (N1-CKO) (as described in Fig. 1) and LIA-Id1-2A-Cre (N1-CKO+Id1) one-cell clones. n=3 retinas per condition, 464 BAG one-cell clones, 746 LIA-Id2-2A-Cre one-cell clones. **P<0.01.
As the overall percentage of Müller glia and bipolar cells induced by the Ids among all GFP+ cells was relatively slight, we carried out quantitative assays that allowed assessment of the fates of a large number of cells. First, the FACS assay described above was used to quantify the extent of rescue by CAG:Id1 and CAG:Id3 in N1-CKO cells. A Chx10 reporter, to mark bipolar cells (Kim et al., 2008), and a Cralbp reporter, to mark Müller glial cells, were used (Matsuda and Cepko, 2007). For controls, P0 wild-type or Notch1fl/fl retinas were electroporated in vitro with combinations of CAG:GFP and one of the cell type-specific reporters. To assess rescue, Notch1fl/fl retinas were electroporated in vitro with combinations of CAG:Cre, CALNL-GFP, CAG:Id1, CAG:Id3 and a cell type-specific reporter. Removal of Notch1 resulted in a decrease in the number of cells marked by the Chx10 reporter and a decrease in the number of cells marked by the Cralbp reporter (Fig. 5G). Removal of Notch1 and co-expression of CAG:Id1 and CAG:Id3 resulted in a slight rescue of expression of the bipolar reporter and a significant induction in the reporter for Müller glial cells (Fig. 5G).
As not all bipolar and Müller glial cells are labeled by the reporters used above, and to further assess the degree of maturation of rescued cells, a viral approach was undertaken to determine whether Id1 could rescue the cell fates lost when Notch1 was deleted in vivo. Because virally labeled cells can be identified readily by location and morphology, we found that this method was the most unambiguous for assessing retinal cell fates. Furthermore, we assessed whether the rescue occurred in postmitotic cells, as one-cell clones were assayed (as in experiments shown in Fig. 1). A retroviral vector that expressed three genes, Id1, Cre and AP (Id1-2A-Cre-IRES-AP) was constructed (Fig. 5H). This virus was injected in vivo into the subretinal space of P3 Notch1fl/fl pups and retinas were harvested after retinal maturation (P21 or later). LIA-Id1-2A-Cre-IRES-AP-infected cells were detected by histochemical staining for AP activity. Quantification of the identity of cells in one-cell clones showed that Id1 could provide partial and statistically significant rescue of both bipolar cells and Müller glia in postmitotic cells (Fig. 5I).
Analysis of Nrarp function in the developing retina
Similar to Id1 and Id3, Nrarp RNA was reduced in N1-CKO cells assayed by microarray (Mizeracka et al., 2013). Nrarp expression was detected by in situ hybridization in the postnatal retina in areas where both mitotic and postmitotic cells are located (supplementary material Fig. S1G-I). To examine Nrarp function, the FACS assay described above was used to determine whether Nrarp misexpression affected the Hes1:tdTomato reporter. Overexpression of Nrarp resulted in a 72.5% decrease in Hes1 reporter expression, when compared with the reporter in wild type (Fig. 6A). These results suggest that Nrarp can inhibit Notch signaling in the developing retina, as has been shown in other developmental systems (Krebs et al., 2012; Yun and Bevan, 2003).
Fig. 6.
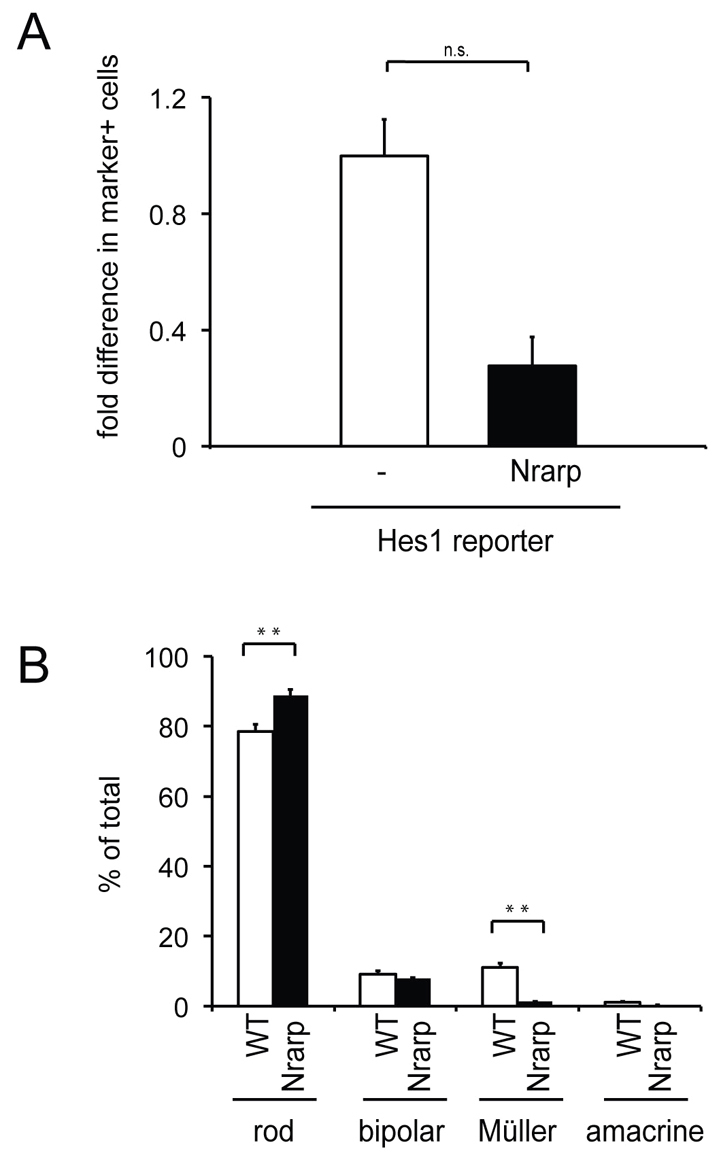
Analysis of Nrarp function in the developing retina. (A) Wild-type retinas were electroporated in vitro at P0 with CAG:GFP for controls, or CAG:GFP and CAG:Nrarp, along with the Hes1:tdTomato reporter. Retinas were cultured for 3 days and dissociated to single cells. FACS was used to determine the percentage of GFP+ cells that were tdTomato+ for each condition. Fold difference is the percentage of GFP+marker+ cells induced by Nrarp misexpression relative to wild type. n=3 retinas per condition. n.s., P>0.05. (B) Wild-type retinas were infected in vivo at P3 with LIA and LIA-Nrarp, and the fate of single cells was assessed after P21 by histochemical staining. Quantification of cell types found in one-cell clones infected with LIA or LIA-Nrarp. n=3 retinas per condition, 718 LIA clones and 672 LIA-Nrarp clones were scored. **P<0.01.
The role of Nrarp in specifying cell fates during postnatal retinal development was examined. Viruses expressing Nrarp and AP (LIA-Nrarp) or expressing AP alone (LIA) were injected separately into the subretinal space of P3 wild-type pups. Retinas were harvested after retinal maturation (P21 or later) and clonal composition was examined. Again, only one-cell clones were examined in order to assess Nrarp function in postmitotic cells. Misexpression of Nrarp led to the increase of rod photoreceptors, no change in bipolar cells, a slight decrease in amacrine cells and a loss of Müller glial cells (Fig. 6B). These results provide evidence that Nrarp inhibits the production of Müller glial cells in postmitotic cells.
DISCUSSION
Numerous studies have determined a role for Notch signaling in regulating neural cell diversity in a variety of developmental systems (Cau and Blader, 2009; Yamamoto et al., 2006). Our study shows that Notch signaling in newly postmitotic cells is required for acquisition of non-rod fates. In the developing retina, diversity may already exist at the progenitor cell level, with molecularly distinct RPCs producing restricted types of postmitotic progeny that respond differentially to Notch signaling. This is similar to ganglion mother cells (GMCs) in the fly ventral nerve cord, which often undergo terminal divisions to give rise to two different cell types in a Notch-dependent manner (Skeath and Doe, 1998; Spana and Doe, 1996; Truman et al., 2010). Recently, we have identified RPCs that express Olig2 and behave like GMCs, in that they undergo terminal divisions and produce specific fates, either two rods or a rod and an amacrine cell in the postnatal period (Hafler et al., 2012). As both rod and amacrine fates are produced under N1-CKO conditions (Jadhav et al., 2006, this study), it is likely that the fates of the progeny of Olig2-expressing RPCs are not dependent on Notch1 signaling. By contrast, although we do not yet have a molecular marker for this population, Olig2-negative RPCs produce the other types of progeny of terminal divisions: a rod and a bipolar cell, or a rod and a Müller glial cell. Our current study shows that the bipolar and Müller glial fates of the daughters of these RPCs are very dependent on Notch signaling, and that this signaling is required in the newly postmitotic state. Thus, we speculate that there are molecularly distinct RPCs whose postmitotic progeny inherit a requirement for Notch1 input to achieve their non-rod fates (Fig. 7A).
Fig. 7.
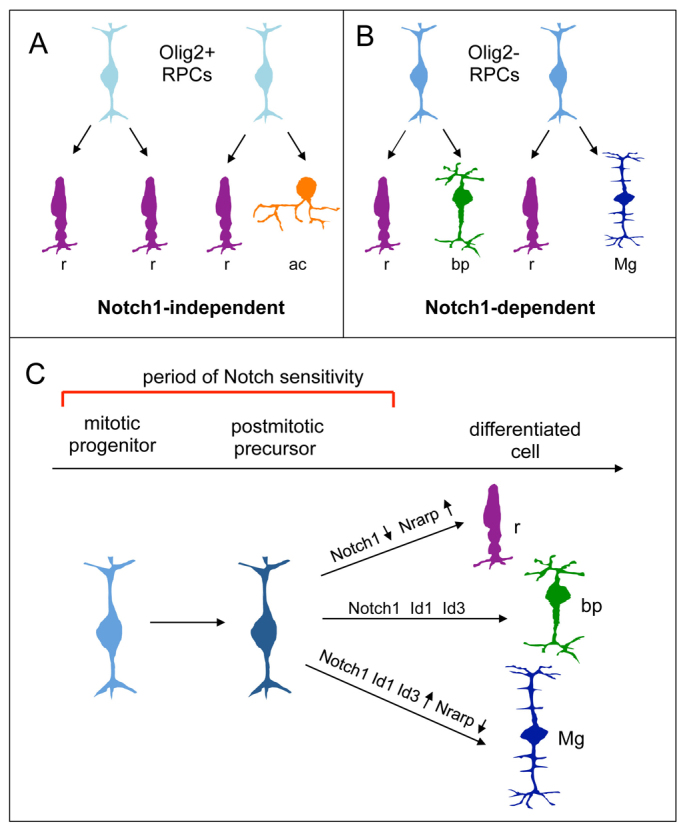
A model of Notch regulation of cell fate decisions in the postnatal retina. Removal of Notch1 from RPCs or their postmitotic progeny results in almost all cells achieving the rod fate at the expense of bipolar and Müller glial cells (this study) (Jadhav et al., 2006; Nelson et al., 2007; Yaron et al., 2006). Postnatal RPCs frequently divide to give rise to two postmitotic cells (Turner and Cepko, 1987). These two daughter cells commonly consist of two rods, as well as a rod and an amacrine cell, a rod and a bipolar cell, and a rod and a Müller glial cell. Previous work has determined that Olig2+ RPCs divide to produce two rods or a rod and an amacrine in a terminal division (Hafler et al., 2012). (A) We hypothesize that the progeny of postnatal Olig2-positive RPCs are Notch1 independent, as rods and amacrines are produced under N1-CKO conditions. (B) Conversely, Olig2-negative RPCs divide to give rise to a rod and a bipolar cell or a rod and Müller glial cell. The progeny of these divisions are Notch1 dependent, as loss of Notch1, even after cell cycle exit, results in a loss of bipolar and Müller glial fates. (C) We speculate that Notch signaling induces high levels of Id1 and Id3, which play a role in specifying the Müller glial fate. Conversely, high levels of Nrarp expression, an inhibitor of Notch signaling, promote the formation of rod photoreceptors and a loss of Müller glial cells. Commitment to the bipolar fate is proposed to require intermediate levels of Notch1 signaling. Mg, Müller glial cell; ac, amacrine cell.
The existence of distinct postnatal RPCs and progeny is consistent with the observation that not all cells respond to the addition of Id factors and Nrarp. The majority of cells take on the rod fate even when Id1 and Id3 are misexpressed. Differences in the intrinsic state of responding cells probably explain an intriguing aspect of development, which is that many common signaling pathways produce distinct effects at different times and places. Here, it appears that even though most RPCs express Notch and its downstream regulators (Trimarchi et al., 2008; Mizeracka et al., 2013), only a subset is responsive to these regulators, and they translate this responsiveness to varying extents.
Ids act downstream of Notch to induce the Müller glial fate
From microarray studies of single wild-type and NI-CKO cells (Mizeracka et al., 2013), we identified Id1 and Id3 as genes that were downregulated in the absence of Notch1. We found that misexpression of these factors in the postnatal wild-type retina resulted in an overproduction of cells that resemble RPC/Müller glial cells. Interestingly, expression of the activated form of the Notch receptor, the NICD, in the postnatal retina resulted in cells that also had characteristics of RPCs and Müller glial cells, with almost all transduced cells taking on this phenotype (Bao and Cepko, 1997; Furukawa et al., 2000). The difference in the penetrance of this effect is likely due to NICD being a more potent regulator of gene expression, and/or only certain types of newly postmitotic cells being sensitive to Id factor expression.
We found that Id factors could partially rescue the Notch loss-of-function phenotype, with the robustness of the rescue dependent upon the assay. The FACS-based assay, which used as a readout the activation of the Cralbp:dsRed reporter, showed that a large number of cells could respond to the addition of the Ids. The viral misexpression assay, which used more stringent morphological criteria for the assessment of fate induction, showed that only a small percentage of cells could take on the Müller glial fate following addition of an Id. Moreover, the viral misexpression assay showed that the induction occurred in newly postmitotic cells and that Ids are sufficient for induction of the Müller glial and bipolar cell fates, even in the absence of other Notch regulated genes. Taken together, these results suggest that Id genes are not only involved in maintaining cells in the progenitor state, but are also directly involved in cell fate specification.
Notch signaling activates expression of Hes family members, which can transcriptionally repress pro-neurogenic bHLH factors (Kageyama et al., 2007). Misexpression of Hes genes can also induce a RPC/Müller glial state and repress the rod and bipolar fates (Furukawa et al., 2000). Similarly, Id factors are also known to maintain cells in an undifferentiated state. They function as dominant negatives by binding pro-neurogenic bHLH transcription factors via a HLH binding domain, thus preventing these factors from activating downstream target genes (Benezra et al., 1990). In addition, Id factors can sustain the expression of Hes1, which also delays the onset of differentiation (Bai et al., 2007). From our microarray studies, we found that NeuroD1, a rod-inducing bHLH transcription factor (Akagi et al., 2004; Cherry et al., 2011; Morrow et al., 1999), was upregulated in the absence of Notch1 (Mizeracka et al., 2013). Potentially, in a postmitotic cell, Notch signal transduction leads to the expression of Hes and Id factors, which then prevent the activity of factors such as NeuroD1 at the level of RNA expression and protein function. We speculate that Id factors only inactivate bHLH transcription factors (and perhaps some non-bHLH transcription factors) that are specific to rod induction.
Nrarp activity during retinal development
Nrarp is a known downstream target and inhibitor of Notch signaling (Lamar et al., 2001; Pirot et al., 2004; Yun and Bevan, 2003). As the role of this protein had not been studied in the mammalian CNS, and as it is one of the genes reduced in expression level following removal of Notch1, we analyzed its role in the developing retina. We found that overexpression of Nrarp resulted in reduced expression of a Notch activity reporter, in keeping with its role as a feedback inhibitor of Notch signaling. Consistent with this role, the effects of Nrarp misexpression resulted in induction of rods and inhibition of Müller glial cell production in newly postmitotic cells. These results suggest that under wild-type conditions, Nrarp expression must be low in order to generate Müller glial cells, whereas high expression may be part of the normal process of rod genesis. Nrarp is thus part of a network that includes bHLH, Hes and Id genes, which regulate the number of Müller glia and rods in the postnatal retina.
Model for Notch activity in the postnatal retina
Our findings demonstrate a role for Notch1 in specifying cell fate in newly postmitotic cells, similar to its activity described during Drosophila neurogenesis. We have identified factors downstream of Notch that mediate this function. We propose that the cells with the highest levels of Notch activity also express high levels of Hes and Id factors, and take on the fate of Müller glial cells, while those with lower levels take on the bipolar fate. Cells with low or no Notch signal, but high Nrarp expression, take on the rod fate (Fig. 7). Notch signaling in newly postmitotic cells may allow those cells to stay in a plastic state, or give them time to execute the non-rod fate specification program(s) inherited from their progenitor cell.
Supplementary Material
Acknowledgments
We thank members of the Cepko, Tabin and Dymecki labs for helpful discussions and advice.
Footnotes
Funding
This work was supported by a National Institutes of Health grant [R01EY09676]. C.L.C. is an Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
K.M. and C.L.C. conceived the project. K.M. and C.R.D. performed the experiments. K.M. and C.L.C. wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.090696/-/DC1
References
- Akagi T., Inoue T., Miyoshi G., Bessho Y., Takahashi M., Lee J. E., Guillemot F., Kageyama R. (2004). Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 279, 28492–28498 [DOI] [PubMed] [Google Scholar]
- Alexiades M. R., Cepko C. (1996). Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev. Dyn. 205, 293–307 [DOI] [PubMed] [Google Scholar]
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., Jing N. (2007). Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell 13, 283–297 [DOI] [PubMed] [Google Scholar]
- Bao Z. Z., Cepko C. L. (1997). The expression and function of Notch pathway genes in the developing rat eye. J. Neurosci. 17, 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau M. J., Cepko C. L. (1999). Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development 126, 555–566 [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. (1990). The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61, 49–59 [DOI] [PubMed] [Google Scholar]
- Cai L., Morrow E. M., Cepko C. L. (2000). Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development 127, 3021–3030 [DOI] [PubMed] [Google Scholar]
- Cau E., Blader P. (2009). Notch activity in the nervous system: to switch or not switch? Neural Dev. 4, 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry T. J., Wang S., Bormuth I., Schwab M., Olson J., Cepko C. L. (2011). NeuroD factors regulate cell fate and neurite stratification in the developing retina. J. Neurosci. 31, 7365–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine Z. D., Yang X., DeChiara T., Yancopoulos G., Cepko C. L. (1997). Postmitotic cells fated to become rod photoreceptors can be respecified by CNTF treatment of the retina. Development 124, 1055–1067 [DOI] [PubMed] [Google Scholar]
- Fekete D. M., Perez-Miguelsanz J., Ryder E. F., Cepko C. L. (1994). Clonal analysis in the chicken retina reveals tangential dispersion of clonally related cells. Dev. Biol. 166, 666–682 [DOI] [PubMed] [Google Scholar]
- Fields-Berry S. C., Halliday A. L., Cepko C. L. (1992). A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89, 693–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Mukherjee S., Bao Z. Z., Morrow E. M., Cepko C. L. (2000). rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells. Neuron 26, 383–394 [DOI] [PubMed] [Google Scholar]
- Godinho L., Williams P. R., Claassen Y., Provost E., Leach S. D., Kamermans M., Wong R. O. L. (2007). Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron 56, 597–603 [DOI] [PubMed] [Google Scholar]
- Hafler B. P., Surzenko N., Beier K. T., Punzo C., Trimarchi J. M., Kong J. H., Cepko C. L. (2012). Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc. Natl. Acad. Sci. USA 109, 7882–7887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. E., Bertsch T. W., Ellis H. M., Harris W. A. (1988). Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1, 15–26 [DOI] [PubMed] [Google Scholar]
- Jadhav A. P., Mason H. A., Cepko C. L. (2006). Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development 133, 913–923 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 [DOI] [PubMed] [Google Scholar]
- Kim D. S., Matsuda T., Cepko C. L. (2008). A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J. Neurosci. 28, 7748–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs L. T., Deftos M. L., Bevan M. J., Gridley T. (2001). The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238, 110–119 [DOI] [PubMed] [Google Scholar]
- Krebs L. T., Bradley C. K., Norton C. R., Xu J., Oram K. F., Starling C., Deftos M. L., Bevan M. J., Gridley T. (2012). The Notch-regulated ankyrin repeat protein is required for proper anterior-posterior somite patterning in mice. Genesis 50, 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar E., Deblandre G., Wettstein D., Gawantka V., Pollet N., Niehrs C., Kintner C. (2001). Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 15, 1885–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey F. J., Cepko C. L. (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2, 109–118 [DOI] [PubMed] [Google Scholar]
- Lyden D., Young A. Z., Zagzag D., Yan W., Gerald W., O’Reilly R., Bader B. L., Hynes R. O., Zhuang Y., Manova K., et al. (1999). Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401, 670–677 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Cepko C. L. (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA 101, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Cepko C. L. (2007). Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. USA 104, 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S. K., Kaznowski C. E. (1991). Cell cycle dependence of laminar determination in developing neocortex. Science 254, 282–285 [DOI] [PubMed] [Google Scholar]
- Meier-Stiegen F., Schwanbeck R., Bernoth K., Martini S., Hieronymus T., Ruau D., Zenke M., Just U. (2010). Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS ONE 5, e11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K., Trimarchi J. M., Stadler M.B., Cepko C.L. (2013) Analysis of gene expression in wild type and Notch1 mutant retinal cells by single cell profiling. Dev. Dyn. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow E. M., Furukawa T., Lee J. E., Cepko C. L. (1999). NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23–36 [DOI] [PubMed] [Google Scholar]
- Nelson B. R., Hartman B. H., Georgi S. A., Lan M. S., Reh T. A. (2007). Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 304, 479–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirot P., van Grunsven L. A., Marine J.-C., Huylebroeck D., Bellefroid E. J. (2004). Direct regulation of the Nrarp gene promoter by the Notch signaling pathway. Biochem. Biophys. Res. Commun. 322, 526–534 [DOI] [PubMed] [Google Scholar]
- Price J., Turner D., Cepko C. (1987). Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. USA 84, 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H. R., Aguet M. (1999). Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547–558 [DOI] [PubMed] [Google Scholar]
- Rapaport D. H., Wong L. L., Wood E. D., Yasumura D., LaVail M. M. (2004). Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474, 304–324 [DOI] [PubMed] [Google Scholar]
- Reynaud-Deonauth S., Zhang H., Afouda A., Taillefert S., Beatus P., Kloc M., Etkin L. D., Fischer-Lougheed J., Spohr G. (2002). Notch signaling is involved in the regulation of Id3 gene transcription during Xenopus embryogenesis. Differentiation 69, 198–208 [DOI] [PubMed] [Google Scholar]
- Riesenberg A. N., Liu Z., Kopan R., Brown N. L. (2009). Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J. Neurosci. 29, 12865–12877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T., Chow S. A., Brown P. O. (1997). 3′-end processing and kinetics of 5′-end joining during retroviral integration in vivo. J. Virol. 71, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompani S. B., Cepko C. L. (2008). Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc. Natl. Acad. Sci. USA 105, 192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzinova M. B., Benezra R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13, 410–418 [DOI] [PubMed] [Google Scholar]
- Salic A., Mitchison T. J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105, 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman R. (1961). Histogenesis of the mouse retina studied with thymidine-3H. In The Structure of the Eye, pp. 487–506 New York, NY: Academic Press; [Google Scholar]
- Skeath J. B., Doe C. Q. (1998). Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, 1857–1865 [DOI] [PubMed] [Google Scholar]
- Spana E. P., Doe C. Q. (1996). Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 [DOI] [PubMed] [Google Scholar]
- Tomita K., Ishibashi M., Nakahara K., Ang S. L., Nakanishi S., Guillemot F., Kageyama R. (1996). Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16, 723–734 [DOI] [PubMed] [Google Scholar]
- Trimarchi J. M., Stadler M. B., Roska B., Billings N., Sun B., Bartch B., Cepko C. L. (2007). Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J. Comp. Neurol. 502, 1047–1065 [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., Stadler M.B., Cepko C.L. (2008). Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE 3, e1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., Moats W., Altman J., Marin E. C., Williams D. W. (2010). Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development 137, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. L., Cepko C. L. (1987). A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136 [DOI] [PubMed] [Google Scholar]
- Turner D. L., Snyder E. Y., Cepko C. L. (1990). Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4, 833–845 [DOI] [PubMed] [Google Scholar]
- Wetts R., Fraser S. E. (1988). Multipotent precursors can give rise to all major cell types of the frog retina. Science 239, 1142–1145 [DOI] [PubMed] [Google Scholar]
- Wong L. L., Rapaport D. H. (2009). Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development 136, 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Tanigaki K., Tsuji M., Yabe D., Ito J., Honjo T. (2006). Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J. Mol. Med. 84, 37–45 [DOI] [PubMed] [Google Scholar]
- Yaron O., Farhy C., Marquardt T., Applebury M., Ashery-Padan R. (2006). Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development 133, 1367–1378 [DOI] [PubMed] [Google Scholar]
- Yokota Y. (2001). Id and development. Oncogene 20, 8290–8298 [DOI] [PubMed] [Google Scholar]
- Young R. W. (1985a). Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205 [DOI] [PubMed] [Google Scholar]
- Young R. W. (1985b). Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 353, 229–239 [DOI] [PubMed] [Google Scholar]
- Yun T. J., Bevan M. J. (2003). Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 170, 5834–5841 [DOI] [PubMed] [Google Scholar]
- Zheng M.-H., Shi M., Pei Z., Gao F., Han H., Ding Y.-Q. (2009). The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol. Brain 2, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.