Abstract
G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptor (GPCR)–mediated increases in the second messenger cyclic adenosine monophosphate (cAMP) activate the mitogen-activated protein kinase (MAPK) extracellular signal–regulated kinase (ERK), and in neuroendocrine cells, this pathway leads to cAMP-dependent neuritogenesis mediated through Rap1 and B-Raf. We found that the Rap guanine nucleotide exchange factor Rapgef2 was enriched from primary bovine neuroendocrine cells by cAMP-agarose affinity chromatography and that it was specifically eluted by cAMP. With loss-of-function experiments in the rat neuronal cell line Neuroscreen-1 (NS-1) and gain-of-function experiments in human embryonic kidney 293T cells, we demonstrated that Rapgef2 connected GPCR-dependent activation of adenylate cyclase and increased cAMP concentration with the activation of ERK in neurons and endocrine cells. Furthermore, knockdown of Rapgef2 blocked cAMP- and ERK-dependent neuritogenesis. Our data are consistent with a pathway involving the cAMP-mediated activation of Rapgef2, which then stimulates Rap1, leading to increases in B-Raf, MEK, and ERK activity.
INTRODUCTION
Cyclic adenosine monophosphate (cAMP) plays a central role as a second (intracellular) messenger for the transduction of signals regulating cellular metabolism, secretion, and plasticity by hormones and other first messengers that act through plasma membrane receptors (1). cAMP-dependent protein kinase A (PKA) was identified as the major downstream effector of intracellular cAMP, acting as a cAMP sensor and in turn transducing cAMP-initiated signaling to further downstream cellular targets (2, 3). Investigations of the cellular actions of PKA led to the working hypothesis that all of the biological effects of cAMP within cells might be PKA-dependent (4). By the 1990s, however, additional cAMP sensors were identified. These include the Rap guanine nucleotide exchange factors (GEFs) Epac1 and Epac2 (5, 6) and the hyperpolarization-activated potassium (HCN) and calcium (CNGC) cyclic nucleotide–gated channels (7, 8). All known cAMP sensors, with the exception of the channels, exert their effects indirectly through the activation of enzyme activities within the cell. Thus, the regulatory (R) subunit of PKA releases an active serine and threonine kinase upon binding to cAMP, and cAMP-bound Epacs promote the exchange of guanosine triphosphate (GTP) for guanosine diphosphate (GDP) on the small guanosine triphosphatases (GTPases) Rap1 and Rap2 (5, 6). Extracellular signal–regulated kinase (ERK) is a member of the mitogen-activated protein kinase (MAPK) family, with protean functional roles in cell physiology (9). In the nervous system, ERK mediates cAMP-dependent intracellular effects, including synaptic plasticity underlying memory formation and neurodegeneration-dependent receptor supersensitivity (10–12). G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors (GPCRs) are the best candidates for stimulating an increase in cAMP concentration in cells of the nervous system that could, in turn, control ERK activation. However, it has long been assumed that GPCR-initiated increases in cAMP concentration and ERK activation are separate and well-insulated signaling pathways (13, 14). In any event, no cAMP sensor leading directly to ERK activation has ever been found.
In 2002, we identified a cAMP-dependent pathway in adreno-medullary chromaffin cells with properties suggestive of a previously un-characterized noncanonical (that is, non-PKA) cAMP sensor that mediates enhanced gene transcription through MAPK signaling (15). This pathway is stimulated by the neuropeptide pituitary adenylate cyclase–activating polypeptide (PACAP) upon binding to its Gs-coupled receptor PAC1 or by other agents that mimic cAMP or increase its abundance, such as the diterpene activator of adenylate cyclase (AC), forskolin. We subsequently identified cAMP sensor cellular functionality not associated with PKA in PC12 and Neuroscreen-1 (NS-1) cells that mediates GPCR-initiated, cAMP-and ERK-dependent signaling for neuritogenesis, the extension and growth of the processes (neurites) that mediate intercellular communication between fully differentiated neurons (16–19). Here, we identified Rapgef2 as a cAMP-binding protein in endocrine cells. Increased intracellular concentrations of cAMP enhanced the Rapgef2-dependent activation of Rap1, which in turn associated with B-Raf to enable the activation of ERK and subsequent neuronal- and endocrine-specific cellular outcomes, such as induction of neuroendocrine-specific genes and extension of neuritic processes (neuritogenesis).
RESULTS
cAMP activates ERK in chromaffin cells and neurons in a PKA-independent manner
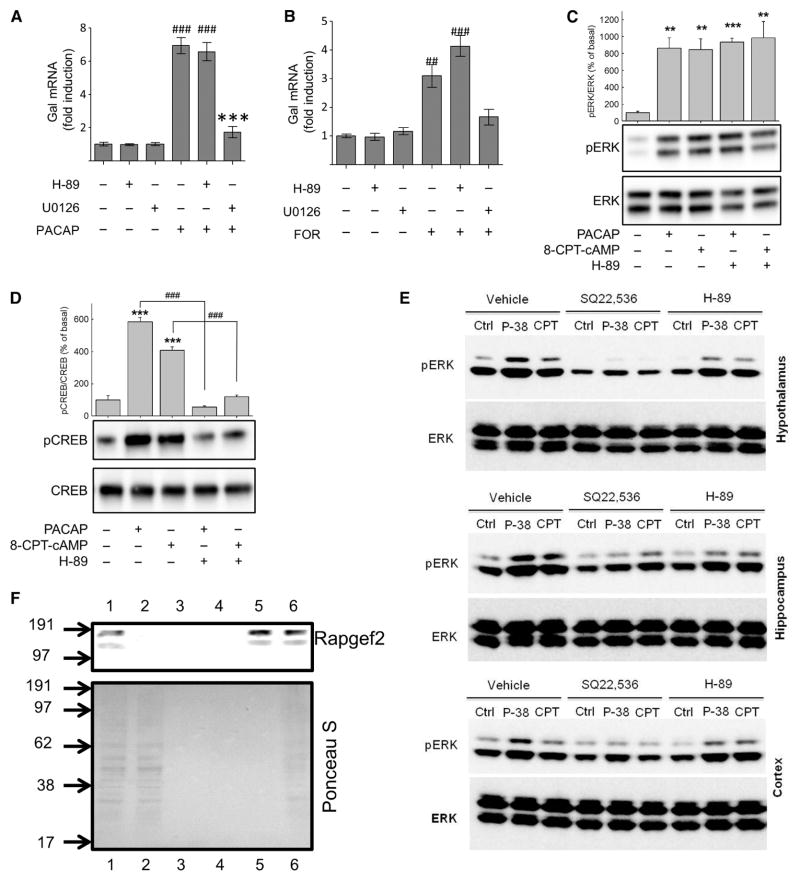
We previously established that a cAMP-dependent pathway that was independent of PKA and Epac mediates ERK activation, leading to neurito-genesis in neuroendocrine cell lines (18, 19). Here, we wished to determine whether this pathway was relevant to cell signaling in primary neuronal and endocrine cells. In primary cultures of bovine chromaffin cells, activating the PAC1 receptor with 100 nM PACAP or increasing the intracellular cAMP concentration with 10 μM forskolin caused statistically significant increases in the abundance of the mRNA of the neuropeptide galanin (Fig. 1, A and B), a hallmark of PACAP-dependent activation of chromaffin cells during stress-induced splanchnic nerve firing in vivo (20). This induction was not blocked by the PKA inhibitor H-89 (Fig. 1, A and B). Treatment of the chromaffin cells with 100 nM PACAP or 100 μM 8-(4-chlorophenylthio)-cAMP (8-CPT-cAMP) (a pan-specific cAMP analog) for 10 min enhanced the extent of ERK phosphorylation compared to that in untreated cells (Fig. 1C). H-89 failed to block ERK activation in response to either agent (Fig. 1C), but it did block the phosphorylation of the transcription factor cAMP response element (CRE)–binding protein (CREB) at Ser133 in response to either PACAP or 8-CPT-cAMP (Fig. 1D). ERK phosphorylation in response to either PACAP- or forskolin-induced increases in cAMP concentration was statistically significantly increased for at least 1 hour (fig. S1). Induction of the expression of the gene encoding galanin by PACAP or forskolin was blocked by the mitogen-activated or extracellular signal–regulated protein kinase kinase (MEK) inhibitor U0126 (Fig. 1, A and B), which inhibits the activation of ERK. These data suggest that cAMP-induced ERK activation and the subsequent increase in progalanin mRNA abundance in chromaffin cells occur through a noncanonical (PKA-independent) signaling pathway.
Fig. 1. PACAP and forskolin stimulate galanin expression and ERK activation.
(A and B) Fold induction in Gal mRNA abundance in bovine chromaffin cells (BCCs). Primary cultures of BCCs (n = 3 wells per condition) were treated with (A) 100 nM PACAP-38 or (B) 25 μM forskolin (FOR) for 6 hours. Where indicated, cells were pre-treated with 10 μM H-89 or U0126 for 30 min. RNA was harvested and subjected to quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis. Data are expressed as mean fold induction ± SEM in Gal mRNA abundance compared to that of untreated controls and were analyzed by one-way analysis of variance (ANOVA) with Bonferroni-corrected t tests. ##P < 0.01, ###P < 0.001, when comparing PACAP-38– or forskolin-treated cells with untreated cells; ***P < 0.001, when comparingtreatmentalone and treatment with inhibitors. Experiments represent quadruplicate wells from a single experiment, and experiments were repeated three or more times with similar results. (C and D) Analysis of ERK and CREB phosphorylation. Western blotting analysis of (C) ERK phosphorylation and (D) CREB phosphorylation at Ser133 in BCCs treated with either 100 nM PACAP-38 or 100 μM 8-CPT-cAMP after 10-min pretreatment with 30 μM H-89 or vehicle [0.02% dimethyl sulfoxide (DMSO)]. Data for each experiment (n = 4) were analyzed by one-way ANOVA with post hoc Bonferroni-corrected t tests. **P < 0.01, ***P < 0.001, compared to untreated controls; ###P < 0.001, when comparing cells with or without H-89. Western blots are from a single experiment and are representative of four independent experiments. (E) Analysis of ERK activation in cultures of primary neurons from rat hypothalamus (upper), hippocampus (middle), and cortex (lower). Neurons were pretreated for 30 min with 10 μM H-89, 1 mM SQ22,536, or vehicle (0.02% DMSO) before being treated for 15 min with 100 nM PACAP-38 or 100 μM 8-CPT-cAMP (CPT) or being left untreated (Ctrl). Samples were analyzed by Western blotting with antibodies specific for phosphorylated ERK (pERK) and total ERK. Data are representative of three independent experiments. (F) Lysates from BCCs were subjected to cAMP-agarose affinity purification. Fractions from affinity chromatography were analyzed by Western blotting for Rapgef2 (upper panel), and membranes were stained with Ponceau S (lower panel). Lane 1: crude lysate (1:5 dilution); lane 2: elution with lysis buffer; lane 3: elution with basic wash buffer; lane 4: pooled elutions with high-salt and no-salt wash buffers; lane 5: elution with 50 mM cAMP; lane 6: heat-treated cAMP-agarose resin (prewashed). Numbers to the left of the images represent molecular mass markers (kD).
The PKA- and Epac-independent pathway also appears to underlie the cAMP-dependent ERK activation stimulated by PACAP in the central nervous system. The hypothalamus is the likely site of action of PACAP in mediating the response of the central nervous system to psychogenic stress (21–24). In primary cultures of hypothalamic neurons, 100 nM PACAP and 100 μM 8-CPT-cAMP stimulated ERK phosphorylation through a signaling pathway that was insensitive to the inhibition of PKA by H-89, but was blocked by the AC inhibitor SQ22,536 (Fig. 1E). Similar results were obtained with primary neuronal cultures of the hippocampus (Fig. 1E), the region in which PACAP affects the balance between long-term potentiation and long-term depression in a cAMP-dependent fashion (25, 26), and in primary neuronal cultures of the cerebral cortex, which we have reported to exhibit both H-89–sensitive and H-89–insensitive gene induction by PACAP and forskolin (27).
Rapgef2 is a cAMP-binding protein present in bovine chromaffin cells
Having established a cellular function for the PKA- and Epac-independent pathway by which cAMP activates ERK signaling, we attempted to identify a cAMP-binding protein in chromaffin cells that might mediate this pathway. We used cAMP-agarose affinity chromatography to perform unbiased selection and enrichment of both known and previously uncharac-terized cAMP-binding proteins (28) from chromaffin cells, which was followed by a combination of mass spectrometric analysis, antibody screening, and expression profiling to identify potential candidates. On the basis of their enrichment and purification by cAMP-agarose, we identified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the PKA regulatory (RIIβ) subunit as cAMP-binding proteins in chromaffin cells, but we eliminated them as candidates for the cAMP-binding protein in the pathway under investigation based on their known signaling pharmacology. We found that Rapgef2 (also known as CNrasGEF, PDZ-GEF1, RA-GEF, and nRapGEP) was enriched upon cAMP-agarose affinity chromatography of proteins in these cells (Fig. 1F). Rapgef2 contains a noncanonical cyclic nucleotide–binding motif (29) and was previously reported as a non–cAMP-binding protein (29–31). However, one group reported that recombinant protein translated from the open reading frame of the cloned human Rapgef2 binds to cAMP and cyclic guanosine monophosphate (cGMP) in vitro, although in that report, the protein was characterized as a cyclic nucleotide–stimulated GEF for Ras rather than Rap (32). Furthermore, Rapgef2 protein is considerably more abundant in the central nervous system than in nonnervous tissue, and genetic ablation of Rapgef2 in mice leads to developmental defects in the central nervous system (33).
cAMP-induced ERK activation and neuritogenesis in NS-1 cells requires Rap1 and B-Raf
If Rapgef2 mediates cAMP-dependent ERK activation in neuroendocrine cell lines, then this pathway would require Rap-dependent activation of a protein kinase, such as Raf, to lead to downstream activation of MEK and ERK. PC12 cells respond to an increase in the intracellular cAMP concentration by assuming a neuronal-like phenotype that includes growth arrest, extension of neurites, and expression of neuronal- and endocrine-specific genes (34, 35). These processes occur by multiple distinct signaling pathways operating in concert (36–38). We previously showed that the small GTPase Rap1 is necessary for PACAP-induced, cAMP-dependent neuritogenesis in PC12 cells (17) and characterized a PKA- and Epac-independent pathway that mediates cAMP-dependent signaling through ERK in NS-1 cells (18, 19).
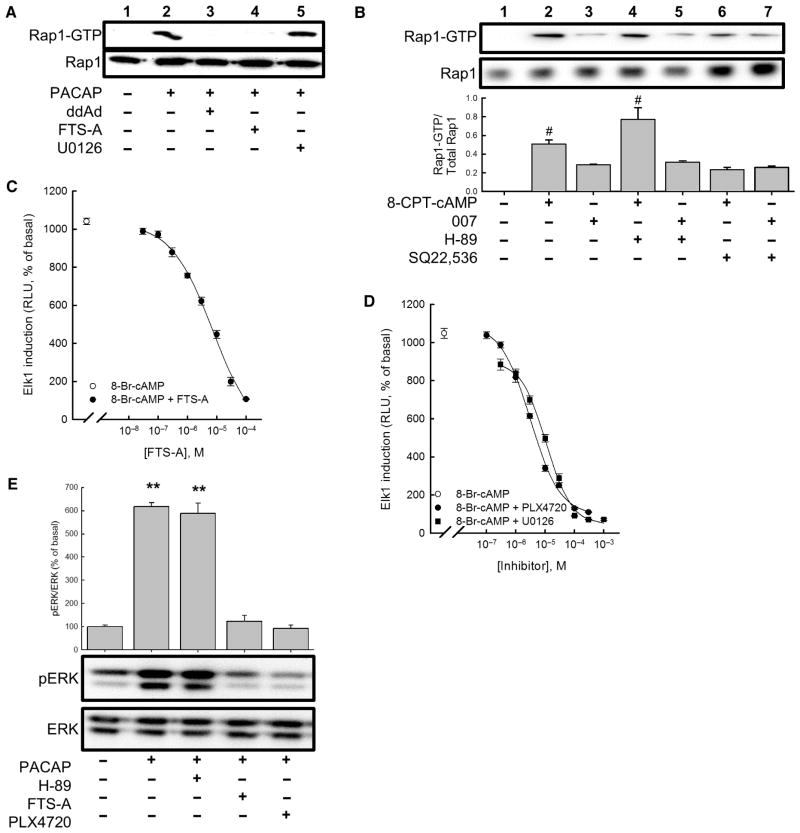
Treatment of NS-1 cells with PACAP resulted in activation of Rap1, as measured by the binding of GTP-loaded Rap1 to agarose-immobilized RalGDS, a recombinant protein that binds Rap1 only when the latter is activated (that is, when it is bound to GTP rather than GDP) (Fig. 2A). PACAP-induced Rap1 activation was blocked by the specific AC inhibitor 2′,5′-dideoxyadenosine (ddAd) (Fig. 2A), indicating that PACAP-induced Rap1 activation was cAMP-dependent. PACAP-induced Rap1 activation was also blocked by trans-farnesylthiosalicylic acid amide (FTS-A) (Fig. 2A), a Rap1 inhibitor that blocks GTP loading of Rap1 and its downstream effector function, that is, the activation by GTP-loaded Rap1 of the kinase activity of B-Raf (39). The pan-specific cAMP analog 8-CPT-cAMP mimicked the effects of PACAP to activate Rap1 in NS-1 cells (Fig. 2B), which was partially blocked by 1 mM SQ22,536 (Fig. 2B). SQ22,536 did not block the effect of the Epac-specific cAMP analog 8-CPT-2′-O-Me-cAMP, which activates only a fraction of the complement of cellular Rap1 that is activated by PACAP or 8-CPT-cAMP (Fig. 2B). We previously reported that SQ22,536 blocks cAMP-dependent ERK activation without affecting Epac-dependent Rap activation or PKA-dependent phosphorylation of CREB in these cells (18). Therefore, the present data suggest that the PKA- and Epac-independent activation of ERK involves the activation of a pool of Rap1 that is distinct from that activated by Epac.
Fig. 2. Rap1 and B-Raf mediate cAMP-dependent activation of ERK.
(A) Measurement of Rap1 activation in NS-1 cells pretreated with vehicle, 100 μM ddAd, 30 μM FTS-A, or 10 μM U0126 before being treated with vehicle or 100 nM PACAP-38 for 5 min. (B) Measurement of Rap1 activation in cells treated with either 100 μM 8-CPT-cAMP or 100 μM 8-CPT-2′-O-Me-cAMP (007) in the presence or absence of 30 μM H-89, 1 mM SQ22,536, or both. Western blotting data are representative of four experiments. Bar graph shows the mean ratios of the abundance of activated Rap1 (Rap1-GTP) to that of total Rap1 protein in each sample and are expressed relative to the amount of activated Rap1 in untreated control cells. #P < 0.05 (Bonferroni). (C) FTS-A inhibited activation of a reporter gene for Elk1 after treatment with 8-Br-cAMP (500 μM). (D) Inhibition of 8-Br-cAMP–dependent Elk1 induction in NS-1 cells by the B-Raf inhibitor PLX4720 and the MEK inhibitor U0126. Points represent means from three experiments, and error bars correspond to SEM. (E) Analysis of ERK activation. NS-1 cells were pretreated with vehicle, 30 μM FTS-A, 10 μM PLX4720, or 30 μM H-89 before being treated with vehicle or 100 nM PACAP-38 for 10 min, and the extent of ERK phosphorylation was determined by Western blotting analysis with antibodies against the indicated proteins. Western blots are representative of four experiments. Bar graph shows the ratio of the abundance of pERK to that of total ERK protein for each sample relative to untreated control cells (set at 100%). **P < 0.01 (Bonferroni), compared to untreated controls.
We wanted to determine whether cAMP-dependent Rap1 activation was necessary for the cAMP-dependent activation of ERK. The transcription factor Elk1 mediates gene transcription in response to ERK activation in mammalian cells (40). We found that 500 μM 8-Br-cAMP, a cell-permeable analog of cAMP, activated Elk1 in NS-1 cells harboring a GAL4-Elk1 fusion protein and a GAL4-luciferase gene, thus acting as an Elk1-dependent gene reporter system. The effect of 8-Br-cAMP was potently inhibited by the Rap1 inhibitor FTS-A (Fig. 2C), further indicating a role for Rap1 in cAMP-dependent ERK activation. To pharmacologically test whether Rap1 was a component of the cAMP-ERK signaling pathway that leads to ERK-dependent neuritogenesis, we treated NS-1 cells with 100 nM PACAP for 48 hours in the absence or presence of varying concentrations of FTS-A, which dose-dependently inhibited PACAP-induced neuritogenesis (fig. S2), suggesting that Rap1 was a component of the cAMP-dependent signaling pathway that leads to neurite extension. PKA-dependent Rap1 activation has been suggested to lead to the differentiation of PC12 cells (41). To examine the contribution of PKA to cAMP-dependent Rap1 activation, we treated NS-1 cells with both H-89 and 8-CPT-cAMP (Fig. 2B). H-89 had no effect on Rap1 activation induced by 8-CPT-cAMP, indicating that PKA was not involved in cAMP-dependent Rap1 signaling, at least in NS-1 cells.
Rap1 activates ERK through B-Raf (41, 42). We therefore measured the potency and efficacy of the B-Raf inhibitor PLX4720 in blocking 8-Br-cAMP–induced Elk1 induction. Inhibition of B-Raf with PLX4720 or of MEK with U0126 effectively inhibited 8-Br-cAMP–dependent Elk1 acti-vation with respective IC50 (median inhibitory concentration) values of 300 nM and 1 μM (Fig. 2D). Inhibition of B-Raf with PLX4720 also blocked PACAP-induced neuritogenesis in NS-1 cells treated for 48 hours (fig. S3). Because FTS-A and PLX4720 inhibited PACAP-induced Elk1 activation and neuritogenesis, we hypothesized that they both would also inhibit PACAP-induced ERK activation. We found that 100 nM PACAP-38 caused a statistically significant increase in ERK phosphorylation (Fig. 2E). Both FTS-A and PLX4720 significantly blocked the effect of PACAP-38 on ERK activation, whereas H-89 had no effect (Fig. 2E), indicating that the targets of these two inhibitors were components of the cAMP-ERK signaling pathway. Although cAMP-dependent Rap1 activation leads to neuritogenesis through ERK, the cAMP-dependent activation of Rap1 that occurs through Epac neither activates ERK nor causes neuritogenesis (19). These results are again consistent with the activation of distinct functional pools of Rap1 in NS-1 cells (43).
Knockdown of Rapgef2 ablates cAMP-dependent ERK activation and neuritogenesis
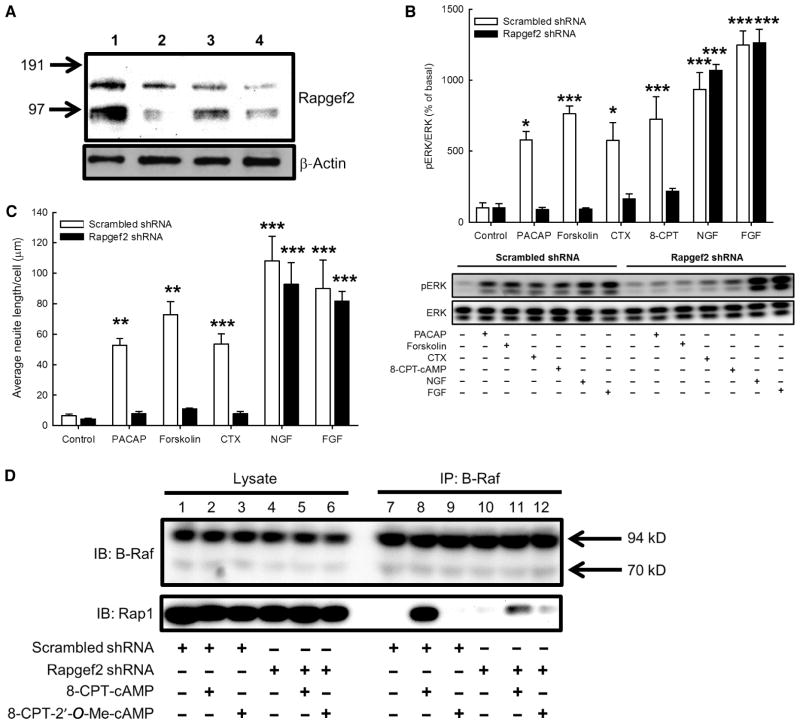
Pharmacological delineation of the complete signaling pathway from GPCR-dependent increases in intracellular cAMP concentration to neuritogenesis enabled us to directly test the hypothesis that the factor mediating PKA-and Epac-independent ERK activation in NS-1 cells was a Rap GEF. There are six members of the Rapgef protein family (fig. S4), four of which have actual or putative cAMP-binding domains (44). We demonstrated that Rapgef2 was purified directly from bovine chromaffin cells through its cAMP-binding property (Fig. 1F), and that PKA- and Epac-independent ERK activation required Rap1 activation (Fig. 2). Accordingly, we performed loss-of-function studies for Rapgef2 in NS-1 cells. We used lenti-virally expressed short hairpin RNA (shRNA) specific for Rapgef2 to generate a stably transduced NS-1 cell line deficient in Rapgef2. In this cell line, Rapgef2 protein abundance was substantially lower than that in wild-type cells or in cells stably transduced with the same lentivirus-based vector encoding scrambled shRNA (Fig. 3A). The generation of cAMP in response to PACAP, cholera toxin, or forskolin was not affected by Rapgef2 knockdown (fig. S5, A and B). Moreover, Epac function, as determined by measuring Rap1 activation upon exposure to the Epac-specific cAMP analog 8-CPT-2′-O-Me-cAMP, was normal in Rapgef2 knockdown cells (fig. S5C).
Fig. 3. Rapgef2 connects cAMP to ERK signaling in NS-1 cells.
(A) Knockdown of Rapgef2 protein in NS-1 cells transduced with lentivirus expressing Rapgef2-specific shRNA. Lane 1: NS-1 cells expressing scrambled shRNA; lanes 2 to 4: stable NS-1 cell lines expressing Rapgef2-specific shRNA constructs. Cells in lane 4 contained <20% of the Rapgef2 protein in cells expressing scrambled shRNA and were propagated for further analyses. (B) Measurement of ERK phosphorylation in NS-1 cells expressing either scrambled shRNA or Rapgef2-specific shRNA. Cells were treated for 10 min with 100 nM PACAP-38, 25 μM forskolin, cholera toxin (CTX; 50 μg/ml), 100 μM 8-CPT-cAMP (8-CPT), NGF (100 ng/ml), or FGF (100 ng/ml). In cells expressing scrambled shRNA, all agents tested caused a statistically significant increase in ERK phosphorylation compared to that in untreated cells (Bonferroni, *P < 0.05, ***P < 0.001). In cells expressing Rapgef2-specific shRNA, only NGF and FGF caused statistically significant ERK phosphorylation (***P < 0.001). Lower panel shows a Western blot that is representative of four independent experiments. (C) Quantification of neurite outgrowth. NS-1 cells expressing either scrambled shRNA or Rapgef2-specific shRNA were treated for 48 hours with 100 nM PACAP-38, 25 μM forskolin, CTX (50 μg/ml), NGF (100 ng/ml), or FGF (100 ng/ml). In cells expressing scrambled shRNA, all agents tested caused significant neurite outgrowth relative to that of untreated control cells (Bonferroni, **P < 0.01, ***P < 0.001). Only NGF and FGF caused statistically significant neurite extension in cells stably expressing Rapgef2-specific shRNA. Bars represent means ± SEM from four fields per condition in three independent experiments. Representative photomicrographs are shown in fig. S6. (D) The association between Rap1 and B-Raf is Rapgef2-dependent. NS-1 cells expressing either scrambled shRNA or Rapgef2-specific shRNA were treated with 100 μM 8-CPT-cAMP or 100 μM 8-CPT-2′-O-Me-cAMP for 10 min. Protein content in cell lysates was normalized, and lysates were precleared and resolved by SDS-PAGE (polyacrylamide gel electrophoresis). Gels were blotted onto nitrocellulose and incubated with antibodies raised against B-Raf and Rap1. Lysates were also subjected to coimmunoprecipitation with a B-Raf–specific antibody coupled to agarose resins. The association between Rap1 and B-Raf in response to 8-CPT-cAMP was greater in extent in cells expressing scrambled shRNA than in cells expressing Rapgef2-specific shRNA. Data are representative of three experiments.
We found that 100 nM PACAP, 25 μM forskolin, cholera toxin (50 μg/ml), or 100 μM 8-CPT-cAMP induced ERK activation in NS-1 cells expressing scrambled shRNA, whereas these agents failed to stimulate ERK activation in cells expressing Rapgef2-specific shRNA (Fig. 3B). In contrast, nerve growth factor (NGF)– and fibroblast growth factor (FGF)–dependent ERK activation was of similar magnitude in cells expressing scrambled shRNA or Rapgef2-specific shRNA. All of the agents tested caused significant neurite elongation in cells expressing scrambled shRNA, whereas in Rapgef2-knockdown cells, only NGF and FGF stimulated neuritogenesis (Fig. 3C and fig. S6). Thus, Rapgef2 was necessary for neuritogenesis dependent on cAMP-ERK signaling, but not for neuritogenesis stimulated by growth factors that activate ERK through cAMP-independent mechanisms.
The cAMP-dependent association between Rap1 and B-Raf requires Rapgef2
Because inhibition of B-Raf blocked cAMP-dependent activation of ERK and Elk1 (Fig. 2, D and E, and fig. S3), we tested the hypothesis that Rapgef2 was necessary for the activation of B-Raf by Rap1 in the context of increased intracellular cAMP concentration. We treated cells expressing either scrambled shRNA or Rapgef2-specific shRNA with 100 μM 8-CPT-cAMP or 100 μM 8-CPT-2′-O-Me-cAMP and measured the association between Rap1 and B-Raf by affinity-purifying B-Raf by immunoprecipitation and then analyzing the samples by Western blotting to detect Rap1. In cells expressing scrambled shRNA, the association between Rap1 and B-Raf was enhanced by 8-CPT-cAMP, but not 8-CPT-2′-O-Me-cAMP (Fig. 3D), and was not observed in cells expressing Rapgef2-specific shRNA. These data suggest that Rapgef2 is necessary for the cAMP-stimulated association between Rap1 and B-Raf. Furthermore, we conclude that the pool of Rap1 activated by Epac does not associate with B-Raf.
Expression of Rapgef2 in human embryonic kidney 293T cells enables cAMP-Rap1-ERK signaling
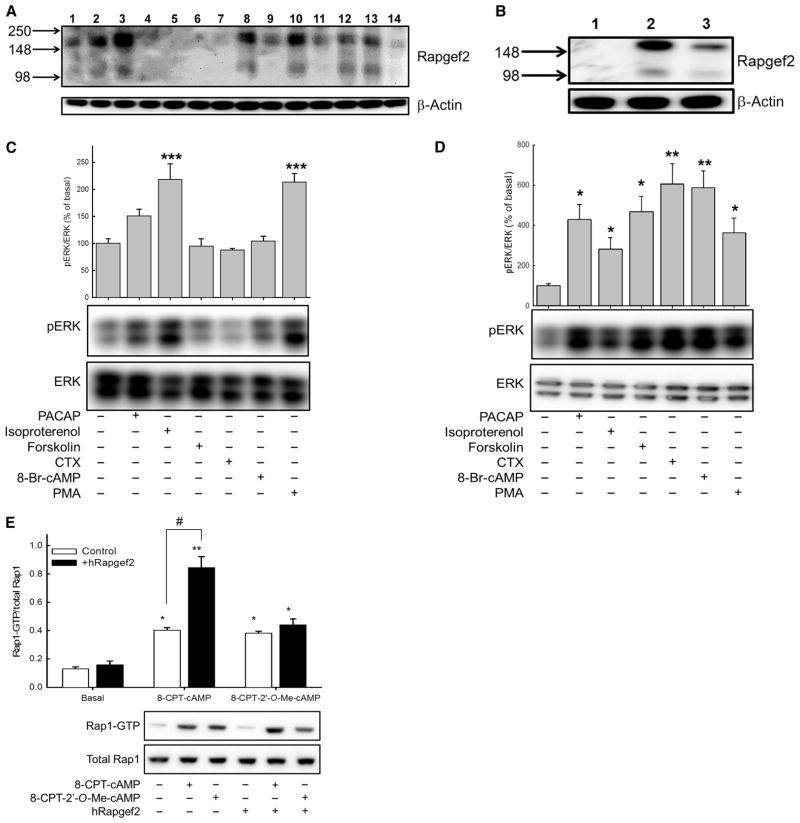
Rapgef2 mRNA and protein are abundant in neural tissues (32, 45). With an antibody against mammalian Rapgef2, we determined that Rapgef2 protein was enriched in cell lines of neuronal and endocrine origin; we detected bands of ~166 kD (major) and ~100 kD (minor) corresponding to Rapgef2 protein that were absent in nonneuronal and nonendocrine cell lines (Fig. 4A). The minor band likely represents a proteolysis fragment of intact Rapgef2 (46). We selected human embryonic kidney (HEK) 293T cells as a system in which to determine whether expression of Rapgef2 could reconstitute cAMP-ERK signaling. Transduction of HEK 293T cells with a retroviral vector expressing human Rapgef2 (hRapgef2) resulted in the detection of the major and minor Rapgef2 antibody–immunoreactive protein bands found endogenously in human neuroblastoma cells (Fig. 4B). We used HEK 293T cells stably expressing exogenous rPAC1 receptors and endogenous β2-adrenergic receptors (βARs) that were or were not transduced with the hRapgef2-expressing retrovirus to test the hypothesis that Rapgef2 was necessary for cAMP-induced ERK phosphorylation initiated through Gαs-coupled GPCR-mediated activation of AC. We measured the extent of ERK phosphorylation after stimulation of cells with the PAC1 ligand PACAP, the βAR agonist isoproterenol, the AC activator forskolin, the Gαs stimulator cholera toxin, the cAMP analog 8-Br-cAMP, or the protein kinase C activator phorbol myristate acetate (PMA) (Fig. 4C). We found that only isoproterenol and PMA significantly increased ERK phosphorylation in HEK 293T cells in the absence of hRapgef2 (Fig. 4C), whereas in HEK 293T cells expressing exogenous hRapgef2, all of the agents tested resulted in substantial increases in ERK phosphorylation (Fig. 4D). These data suggest that expression of Rapgef2 confers upon an epithelial cell line the cAMP-dependent activation of ERK present in Rapgef2-expressing neuroendocrine cells that we have previously characterized (17–19). The expression of Rapgef2 did not result in additional ERK activation in response to isoproterenol, even though isoproterenol and PACAP induced a CRE reporter gene (CRE-luciferase) to a similar extent in the same cell line (table S1). These observations imply that cAMP-dependent activation of ERK is restricted to a subset of Gs-coupled GPCRs (47).
Fig. 4. Rapgef2 protein abundance is enriched in neuroendocrine cells, and its exogenous expression in HEK 293T cells confers cAMP-ERK signaling.
(A) Profile of Rapgef2 protein abundance in human and rodent cell lines. Lane 1: PC12-G; lane 2: NS-1; lane 3: BCC; lane 4: 293T rPAC1hop; lane 5: HEK 293FT; lane 6: HeLa; lane 7: 3T3 Swiss; lane 8: SH-SY5Y; lane 9: NG108-15; lane 10: NBFL; lane 11: FRTL-5; lane 12: B16-F10; lane 13: AtT20; lane 14: Chinese hamster ovary (CHO) K1. Western blotting data are representative of three independent experiments. (B) Western blotting analysis of Rapgef2 abundance in HEK 293T rPAC1hop cells (lane 1), HEK 293T rPAC1hop cells stably expressing hRapgef2 (lane 2), and nontransduced SH-SY5Y cells (lane 3). Western blotting data are representative of three independent experiments. (C and D) Measurement of ERK phosphorylation in (C) 293T rPAC1hop cells and (D) 293T rPAC1hop cells expressing hRapgef2 after treatment for 10 min with 100 nM PACAP-38, 30 μM isoproterenol, 25 μM forskolin, cholera toxin (CTX; 50 μg/ml), 0.5 mM 8-Br-cAMP, or 100 nM PMA. Bar graphs show the ratio of the abundance of pERK to that of total ERK protein for each sample relative to untreated control cells (set at 100%). *P < 0.05, **P < 0.01, ***P < 0.001; n = 4 experiments. (E) Analysis of Rap1 activation in 293T rPAC1hop cells expressing or not expressing the retroviral vector encoding hRapgef2. Cells were treated with vehicle, 100 μM 8-CPT-cAMP, or 100 μM 8-CPT-2′-O-Me-cAMP. *P < 0.05, **P < 0.01, compared to vehicle-treated cells. In cells expressing hRapgef2, 8-CPT-cAMP conferred a significantly greater activation of Rap1 (**P < 0.01) than was observed in nontransduced cells (#P < 0.05, Bonferroni). Data are from four to six independent experiments.
With the same cell lines, we next measured cAMP-induced Rap1 activation. We treated the cells with either 100 μM 8-CPT-cAMP or 100 μM 8-CPT-2′-O-Me-cAMP (Fig. 4E). In cells devoid of Rapgef2, both cAMP analogs caused Rap1 activation to a similar extent (Fig. 4E). That a pan-specific analog of cAMP (8-CPT-cAMP) and a selective Epac activator (8-CPT-2′-O-Me-cAMP) had similar effects on Rap1 activation suggests that cAMP activates only Epac to stimulate Rap1 in cells that do not have Rapgef2. In contrast, 8-CPT-cAMP resulted in greater activation of Rap1 in hRapgef2-expressing cells than did 8-CPT-2′-O-Me-cAMP (Fig. 4E), demonstrating that exogenous Rapgef2 made an additional pool of Rap1 accessible to cAMP.
We showed that Rapgef2 is present in primary endocrine and neuronal cells in which the PKA- and Epac-independent, cAMP-dependent activation of ERK was mediated by an SQ22,536-sensitive mechanism. Knockdown of Rapgef2 in NS-1 cells resulted in loss of cAMP-dependent activation of Rap, B-Raf, ERK, and Elk1 and inhibited neuritogenesis, whereas PKA-and Epac-dependent signaling was completely preserved. Expression of hRapgef2 in HEK 293T cells (which did not previously exhibit ERK activation by PACAP, cholera toxin, or cell-permeant cAMP analogs) conferred ERK activation by all of these agents upon these cells. Together, these results suggest that Rapgef2 mediates, or albeit less likely, is permissive for, GPCR-Gs–initiated, cAMP-regulated activation of ERK. Rapgef2 has also been reported to activate Ras rather than Rap in a cAMP- and cGMP-dependent fashion in vitro; however, we found that activation of Elk1 by cAMP in NS-1 cells was not mimicked by equivalent doses of cGMP (fig. S7). The observed signaling connections leading to ERK activation and neuritogenesis initiated by increased intracellular cAMP are depicted, in the context of parallel signaling through PKA and Epac that leads to other cellular outcomes, in Fig. 5.
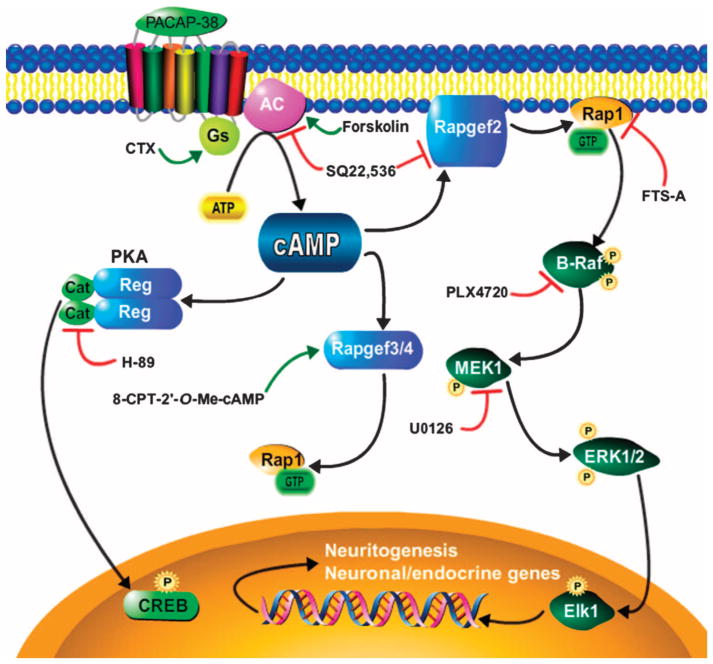
Fig. 5. Proposed Rapgef2-mediated cAMP-ERK signaling pathway in neuronal and endocrine cells.
Demonstrated signaling connections are presented as black arrows, pharmacological activators are connected to their targets with green arrows, and pharmacological inhibitors are connected to their targets by red blunt-ended arrows. SQ22,536 served as a polypharmacological agent: its observed IC50 values at its two targets in NS-1 cells are 10 μM (AC) and 170 μM (Rapgef2). Cat, catalytic subunit of PKA; Reg, regulatory subunit of PKA. The protein product of the Rapgef2 gene has been referred to variously as CNrasGEF, PDZ-GEF1, RA-GEF, nRapGEP, and its activity as the NCS (neuritogenic cAMP sensor) (18, 19). The protein products of the Rapgef3 and Rapgef4 genes are variously referred to as Epac1/2 and cAMP-GEFI/II.
DISCUSSION
Rapgef2 appears to fulfill the criteria to identify it as a mediator of the PKA- and Epac-independent, cAMP-dependent activation of ERK signaling in neuronal and endocrine cells. Rapgef2 is expressed in primary chromaffin cells, in which the cAMP-ERK signaling pathway was first reported, and was enriched from these cells by cAMP-affinity purification. Rapgef2 is expressed in NS-1 cells, where its expression was required for cAMP coupling to a Rap1–B-Raf–ERK pathway that was sufficient for Elk1 activation and neuritogenesis. Expression of Rapgef2 in non-neuroendocrine cells reconstituted the cAMP-Rap1-ERK signaling pathway observed in neuronal and endocrine cells. We propose that Rapgef2 binds and is directly activated by cAMP, leading to GTP loading of Rap1, activation of B-Raf, and the subsequent activation of ERK by MEK, leading to long-term functional effects, including activation of the nuclear transcription factor Elk1, gene transcription in neuronal and endocrine cells, and altered cellular morphology; however, this hypothesis awaits further experimental confirmation, including the demonstration that mutations in Rapgef2 that impair cAMP binding in vitro correspondingly impair Rapgef2-dependent cAMP-initiated activation of ERK in intact cells.
Our finding of a GPCR-dependent signaling pathway connecting increases in cAMP concentration to ERK phosphorylation has implications for neurobiology. First, Rapgef2 in Drosophila and Caenorhabditis elegans is neither neuronal-specific nor dedicated to the activation of ERK (48–50). This suggests that the reported neuronal- and endocrine cell–specific expression of this protein in mammals may reflect a previously unsuspected evolutionary specialization of this pathway. Second, Rap1 is implicated in the cAMP-dependent activation of ERK that is important for hippocampal function underlying learning and memory, albeit the sensor mediating cAMP-dependent Rap1 activation has not yet been identified (11). We suggest that Rapgef2 may be a candidate for this role. Similarly, competition for regulation of ERK activation by cAMP and Ras in tumors of neural crest–derived cells, such as melanoma (51), may involve Rapgef2 or a closely related cAMP sensor. Finally, distinct cellular pools of Rap1 might partition signaling by Rapgef3/4 (Epac) and Rapgef2 (Fig. 5), thus generating two distinct pathways for Rap1 activation by cAMP that depend specifically on their upstream activators.
In the case of Rapgef2, activation appears to be quite specific for at least one member of the family B GPCRs. It is of immediate importance to determine what other GPCR family members might be able to activate the cAMP–Rapgef2–ERK pathway, and which of these might modulate the cAMP-dependent ERK activation that underlies the cellular plasticity required for complex neuronal function, such as memory formation. Our observation of the lack of Rapgef2 activation by β2AR signaling suggests that not all Gαs-coupled GPCRs can activate Rapgef2, either at the level of GPCR-Gαs–AC coupling or of the association between AC and Rapgef2. PDZ-containing signaling proteins can assemble multiple components for signaling, often with a high degree of isoform-specific mutual recognition (52). The PDZ domain of Rapgef2 may function to provide differential linkage to Rap1–B-Raf–MEK–ERK signaling cassettes, as suggested previously (53). Obviously, additional details will need to be filled in to fully define the role of Rapgef2 at the cellular and systems levels in neuronal and endocrine signaling circuits.
MATERIALS AND METHODS
Drugs and reagents
Forskolin, isoproterenol, ddAd, PMA, NGF, and FGF were obtained from Sigma. Cholera toxin (sodium azide–free), H-89, and U0126 were purchased from Calbiochem. SQ22,536 was purchased from Tocris Cookson, and PACAP-38 was purchased from Phoenix Pharmaceuticals. The cAMP analogs 8-bromo-cAMP (8-Br-cAMP), 8-CPT-cAMP, and 8-CPT-2′-O-Me-cAMP (007) were purchased from Biolog Life Science Institute. PLX4720 was purchased from Selleckchem. FTS-A was obtained from Cayman Chemical Company. For all studies with small-molecule inhibitors, cells were pretreated with the compound or vehicle (0.1% DMSO) for 30 min before agonist stimulation for the indicated times.
Primary cultures of bovine chromaffin cells
Chromaffin cells were isolated from steer adrenal glands by collagenase digestion through retrograde perfusion with an adaptation of the Fenwick procedure, as described previously (54). Cells were plated in 5% heat-inactivated fetal bovine serum (HI-FBS) in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with penicillin, streptomycin, fungi-zone, and cytosine arabinofuranoside. Cells were mechanically resuspended in the same medium 24 hours later, collected by centrifugation at 800g for 5 min at room temperature, and replated in T150 flasks in the same medium. This procedure minimized the contamination of chromaffin cells with adherent cells. Cells were again mechanically resuspended 24 to 72 hours after replating, collected by centrifugation, resuspended in phosphate-buffered saline (PBS) at ~2 × 106 cells/ml, and collected by centrifugation. Cells were resuspended in PBS with a blue-tip pipette (800 μl per 110 million cells with a packed volume of about 200 μl) and added dropwise to 5 ml of LyB+ buffer. Lysates were snap-frozen on dry ice and stored at –80°C until needed.
Analysis of ERK activation in primary cultures of neurons
Primary embryonic day 18 (E18) hypothalamic, hippocampal, and cortical neuronal cultures were prepared as previously described from Sprague-Dawley rats (24, 55) with the following modifications. Fetuses were decapitated; brains were removed; meninges were stripped away; and the hypothalamic, hippocampal, and cortical tissues were dissected and collected in ice-cold dissection solution consisting of calcium- and magnesium-free Hanks balanced salt solution (Invitrogen) supplemented with 10 mM Hepes, penicillin (200 U/ml), streptomycin (200 mg/ml), and 10 mM sodium pyruvate before being cut into smaller pieces with a fine pair of scissors. For hypothalamic cultures, the tissue was digested with collagenase type 2 (1.2 mg/ml; Worthington) dissolved in dissection buffer supplemented with 20 mM glucose, bovine serum albumin (BSA; 4 mg/ml), and deoxyribonuclease (DNase; 0.2 mg/ml) for 40 min at 37°C. A final dissociation of the cells was then performed by triturating the suspension up and down with a pipette before centrifugation of the supernatant at 200g for 5 min. The cell pellet was then washed twice in dissection buffer. For hippocampal and cortical cultures, tissues were digested with prewarmed 0.12% trypsin, 0.02% EDTA solution (SAFC Biosciences) and incubated at 37°C for 10 min. The tissue was then washed three times with dissection solution. A final dissociation of the cells was conducted by triturating the suspension up and down with the aid of a pipette. A total of 1 × 106 cells per well were plated onto six-well plates precoated with poly-D-lysine (50 mg/ml; Sigma). Hypothalamic and cortical neurons were plated in medium consisting of DMEM supplemented with penicillin (200 U/ml), streptomycin (200 mg/ml), 0.5 mM L-glutamine, 10 mM sodium pyruvate, and 10% HI-FBS, which was replaced with Neurobasal medium (Invitrogen) supplemented with 2% B-27 (Invitrogen), penicillin (200 U/ml), streptomycin (200 μg/ml), and 0.5 mM L-glutamine 24 hours after plating. The hippocampal neurons were directly plated and maintained in Neurobasal medium as described earlier. Half of the media of all the cultured neurons was changed every 3 to 4 days, and the neurons were used after 10 to 12 days in culture. To analyze pERK and ERK, neurons were pretreated with 0.02% DMSO (vehicle), 1 mM SQ22,536, or 10 μM H-89 for 30 min before stimulation with 100 nM PACAP-38 or 100 μM 8-CPT-cAMP for 15 min, after which time, culture medium was aspirated, and cells were washed once with PBS and lysed in ice-cold radioimmunoprecipitation assay buffer [50 mM tris-HCl, 150 mM NaC1, 0.1 mM EDTA, 10 mM MgCl2, 0.1% SDS, 1% Triton X-100, and 0.5% deoxycholate supplemented with 50 mM NaF, 5 mM Na3VO4, and one tablet of Complete Protease Inhibitor Cocktail (Roche)]. The lysates were briefly sonicated and centrifuged at 8161g for 10 min, and the protein concentration of the supernatant was determined with the Bio-Rad DC Protein Assay. A total of 15 μg of protein from each sample was then resolved on a 4 to 12% NuPAGE bis-tris gel (Invitrogen), transferred onto 0.45-μm Trans-Blot nitrocellulose membranes (Bio-Rad), and first incubated with rabbit anti–phosphor-(Thr202/Tyr204) p44/42 ERK primary antibody (Cell Signaling Technology) followed by horse-radish peroxidase (HRP)–conjugated goat anti-rabbit secondary antibody (Pierce Biotechnology) that was detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce). The membranes were then stripped with Restore Western Blot Stripping Buffer according to the manufacturer’s instructions, and the earlier steps were repeated with rabbit anti-p44/p42 primary antibody (Cell Signaling Technology) to determine total ERK quantities.
Cell lines
Cells were grown in flasks (Techno Plastic Products AG) at 37°C in a humidified environment containing 5% CO2 in media and solutions that were purchased from Invitrogen, unless otherwise noted. NS-1 cells were grown on collagen-coated flasks in RPMI 1640 supplemented with 10% HI-FBS, 5% horse serum, 2 mM L-glutamine, and penicillin and streptomycin. HEK 293T Cre-luc rPAC1 cells were grown in DMEM supplemented with 10% FBS, 2 mM L-glutamine, and hygromycin B (50 μg/ml; Mediatech). PC12-G cells were grown in DMEM supplemented with 7% HI-FBS, 7% horse serum, 25 mM Hepes, 2 mM L-glutamate, and penicillin and streptomycin in an incubator supplying 10% CO2. HeLa, 3T3 Swiss, B16-F10, HEK 293FT, AtT20, and NBFL cells were grown in DMEM supplemented with 10% FBS, 2 mM L-glutamine, and penicillin and streptomycin. NG108-15 cells were grown in DMEM with 10% FBS, 0.1 mM HAT (sodium hypoxanthine, aminopterin, and thymidine) supplement, 2 mM L-glutamine, and penicillin and streptomycin. SH-SY5Y and CHO-K1 cells were grown in a 1:1 mixture of DMEM and Ham’s F-12 medium supplemented with 10% FBS, 2 mM L-glutamine, and penicillin and streptomycin. FRTL-5 cells were grown in Ham’s F-12 medium supplemented with 5% FBS, 2 mM L-glutamine, penicillin and streptomycin, and the following hormones (all from Sigma): 10 nM hydrocortisone, thyrotropin (10 mU/ml), insulin (0.01 mg/ml), transferrin (5 μg/ml), somatostatin (10 ng/ml), and glycyl-L-histidyl-L-lysine acetate (10 ng/ml).
Quantitative RT-PCR
To analyze gene expression in bovine chromaffin cells, total RNA was extracted with a commercial kit (RNAqueous, Ambion) according to the manufacturer’s instructions. Genomic DNA was removed by digestion with ribonuclease-free DNase I (Invitrogen), and an aliquot of each sample, corresponding to 0.5 μg of RNA, was reverse-transcribed with random hexamer primers and SuperScript II Reverse Transcriptase (50 U; Invitrogen). The final complementary DNA (cDNA) synthesis reaction volume of 22 μl was diluted 1:1 in diethyl pyrocarbonate–treated H2O, and 2 μl of the resulting sample was used per well for PCR. Quantitative PCRs were prepared by adding iQ SYBR Green Supermix (Bio-Rad) and a final concentration of 200 nM gene-specific primers [Gal: forward GACAGCCACAGGTCATTTCAAG, reverse GCCGGGCTTCGTCTTCA, amplicon 76 base pairs (bp); GAPDH: forward GCATCGTGGAGG-GACTTATGA, reverse CAGCGCCAGTAGAAGCAGG, amplicon 135 bp] with each cDNA sample run in duplicate. The following quantitative PCR conditions were used on an iCycler iQ Real-Time PCR System (Bio-Rad): initial heating to 95°C for 3 min, 40 cycles of 95°C for 10 s and 55°C for 45 s, followed by 95°C for 60 s and 55°C for 60 s. Fold changes in gene expression relative to untreated controls were determined after quantitative PCR with the ΔΔCt method (56).
cAMP assays
Intracellular cAMP was measured with the cAMP Biotrak enzyme immunoassay kit (Amersham Biosciences). One day after being seeded in 96-well plates, NS-1 cells were pretreated for 20 min with culture medium containing 0.5 mM 3-isobutyl-1-methylxanthine (IBMX) to inhibit phosphodiesterases. After IBMX loading, cells were treated with agonists for 20 min at 37°C. Cells were then lysed, and cAMP accumulation was measured by the nonacetylation protocol as provided by the manufacturer. Data were fit to standard curves with four-parameter logistic regressions.
Elk-luciferase assays
For measurements of Elk1 activation, we generated a stable NS-1 cell line as described previously (18). Briefly, NS-1 cells were sequentially transduced with the pLNCFA2-Elk and pBABE-Luc retroviral vectors, which encode a Gal4-Elk1 fusion protein and a luciferase reporter gene, respectively. NS-1 Elk-luc cells were dispensed into collagen I–coated 96-well plates and incubated overnight. The next day, cells were pretreated with inhibitors for 30 min and then treated with the indicated agonists for 5 hours. Luciferase activity was measured after the addition of Bright-Glo Luciferase Assay Reagent (Promega; 100 μl per well). Luminescence was measured in a Victor3 microtiter plate reader (Perkin Elmer) after 2 min of vigorous shaking.
Neurite outgrowth assays
NS-1 cells were seeded into six-well plates, incubated overnight, and treated for 48 hours, as indicated in the figure legends. Images of cells were then randomly acquired at ×20 magnification with a computer-assisted microscope. Images were randomized, and a blinded observer counted the numbers of cells, numbers of neurites, and lengths of the neurites in each field with NIS-Elements BR (Nikon). Data from neurite outgrowth assays are expressed as the average neurite length per cell.
shRNA-mediated knockdown of Rapgef2
Rapgef2 was knocked down in NS-1 cells with a specific shRNA construct (target sequence: GGAAGTCATTAACCAGGAA) encoded by the psi-Lv-H1 lentiviral vector (GeneCopoeia). To generate an NS-1 cell line with stable Rapgef2 knockdown, NS-1 cells, at 50% confluence, were exposed to the lentiviral vectors overnight. The next day, the medium was changed, and after three more days, transduced cells were selected from the culture by adding puromycin (1 μg/ml) to the medium. In parallel cultures, NS-1 cells were exposed to scrambled shRNA expressed by the psi-Lv-H1 vector. Cells stably expressing scrambled shRNA served as controls for all experiments using cells in which Rapgef2 was stably knocked down.
Human Rapgef2 γ-retroviral construct
The pCR4-TOPO plasmid containing the cDNA encoding hRapgef2 (Thermo Scientific) was cleaved with the restriction enzymes Pme I and Not I. The 4.5-kb Rafgef2 cDNA fragment was then subcloned into the pPRIChpaHA retroviral expression plasmid (57) after cleaving this plas-mid with Bst BI, blunting the overhang, and then cleaving with Not I, thereby linearizing the pPRIChpaHA plasmid in the multiple cloning site. The pPRIChpaHA is a γ-retroviral expression plasmid wherein the cytomegalovirus enhancer and promoter direct the expression of the introduced cDNA (that is, the sequence encoding Rapgef2) and an encephalo-myocarditis virus–derived internal ribosomal entry site directs the translation of the mCherry fluorescent protein. Retroviral vectors were generated by transfecting HEK 293 cells grown in 10-cm dishes to a density of 1.5 × 106 to 2 × 106 cells per plate with this recombinant retroviral genome expression plasmid together with a murine retroviral gag-pol plasmid and a vesicular stomatitis virus G plasmid, as previously described (18).
Western blotting
Cells were seeded in 12-well plates, incubated overnight, and treated as indicated in the figure legends. After treatment, cells were collected in lysis buffer (150 mM NaCl, 50 mM tris-HCl, 1% NP-40, 1 mM EDTA) including the Halt protease and phosphatase inhibitor cocktails (Pierce). The amount of protein for each sample was determined with the Bio-Rad Protein Assay. Equal amounts of protein (20 μg) were added to a solution containing 1× NuPAGE LDS sample buffer and 1× NuPAGE reducing reagent (Invitrogen), which were heated to 95°C for 5 min. Samples were subjected to SDS-PAGE with bis-tris gels containing 4 to 12% polyacrylamide (for analysis of Rapgef2) or 10% polyacrylamide (for analysis of pERK). Gels were blotted onto nitrocellulose membranes (Invitrogen), which were incubated in blocking buffer containing 5% nonfat milk in tris-buffered saline containing 0.05% Tween 20 (TBST) for 2 hours at room temperature. The membranes were then incubated overnight at 4°C with a 1:2000 dilution of antibodies specific for pERK (Thr202/Tyr204), pCREB (Ser133) (both from Cell Signaling Technology), or Rapgef2 (NBP1-06549, Novus Biologicals). After five washes in TBST, membranes were incubated with HRP-conjugated secondary antibodies (1:5000 in blocking buffer). After five washes in TBST, membranes were incubated with SuperSignal West Pico Chemiluminescent Substrate (Pierce) and visualized by a cooled charge-coupled device camera. Antibodies bound to pERK, pCREB, or Rapgef2 were removed with Restore Western Blot Stripping Buffer (Pierce) before the blots were incubated overnight with antibodies specific for total ERK, total CREB, or β-actin (all antibodies from Cell Signaling Technology). Images were quantified with ImageJ [National Institutes of Health (NIH), http://imagej.nih.gov/ij/].
Rap1 assays
The GTP loading of Rap1 was measured with the Active Rap1 Pull-Down and Detection Kit (Pierce) according to the manufacturer’s instructions. Cells were seeded in six-well plates and grown overnight. The next day, cells were treated for 10 min as indicated in the figure legends and then lysed. Protein (500 μg) from each sample was incubated in a solution of 20 μg of glutathione S-transferase–RalGDS–RBD in a glutathione resin slurry for 1 hour. Samples were then centrifuged through spin cups, washed three times with lysis buffer, dissolved in reducing sample buffer, vortexed, and heated to 95°C for 5 min. All samples were then resolved by 12% PAGE, gels were blotted onto nitrocellulose, and membranes were incubated with an antibody specific for Rap1 at a dilution of 1:1000 (Upstate Biotechnology). In parallel, 20-μg samples of unpurified protein, corresponding to samples that were affinity-purified, were loaded onto gels that were incubated with the same antibody to account for differences in Rap1 content between samples
Coimmunoprecipitation
An antibody specific for B-Raf (C-16, Santa Cruz Biotechnology) was dialyzed and directly immobilized onto aldehyde-activated agarose beads (25 μg of antibody/50 μl of resin) that were packed into fritted columns with reagents included in coimmunoprecipitation kits (Pierce, catalog #26149) according to the manufacturer’s instructions. NS-1 cells, grown in six-well plates, were treated as indicated in the figure legends for 10 min, after which they were collected in 0.2 ml of lysis/wash buffer. Lysates were centrifuged at 13,793g for 5 min, and the supernatants were retained. Protein concentrations in the supernatants were determined with the Bio-Rad DC Protein Assay. Samples containing 1 mg of protein were precleared by application to slurries of uncoupled agarose resin (80 μl), which were packed onto fritted columns. Samples were incubated with uncoupled resins for 30 min at 4°C with end-over-end rotation. Columns were centrifuged at 1000g for 1 min, and precleared lysates were reserved and added to antibody-coupled resins, which were incubated overnight at 4°C with end-to-end rotation. Columns were then centrifuged at 1000g for 5 min, and each resin was washed three times with 0.2 ml of lysis/wash buffer. Captured proteins were eluted by adding 50 μl of elution buffer, incubated for 5 min at room temperature, to the column, followed by centrifugation at 1000g for 5 min. Elutants were dissolved in 5× SDS denaturing sample buffer, heated to 95°C for 5 min, and vortexed. Samples were analyzed by Western blotting (as described earlier) with antibodies raised against B-Raf (Santa Cruz Biotechnology) and Rap1 (Upstate).
cAMP-agarose affinity purification of chromaffin cell cAMP-binding proteins
cAMP-binding proteins were purified from bovine chromaffin cells by the method adapted from the affinity purification of regulatory subunits of PKA (58). Briefly, lyophilized cAMP-agarose (Sigma A0144) was swelled in 3.33 ml of LyB [20 mM Hepes (pH 7.4), 200 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.5% Igepal CA-630 (pH 7.4), with BSA (1.5 mg/ml)] for 4 hours at room temperature. The gel was washed with 4 × 5 ml of LyB, suspended in 1 ml/ml-equivalent of original lyophilized cAMP-agarose in LyB+ buffer [LyB containing 1 mM dithiothreitol (DTT) and one tablet of 50 ml of Roche Protease Inhibitor Cocktail]. Gels were stored at 4°C if they were to be used within 1 to 2 weeks; otherwise, they were washed with 3 × 5 ml (per ml-equivalent of original lyophilized cAMP-agarose) in 1 M NaCl and stored in 5 ml (per original ml-equivalent) with 0.5% NaN3 at 4°C. Other buffers used were basic wash buffer (BWB; 10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.1% Igepal CA-630), high-salt wash buffer (HSWB; 25 ml of basic wash buffer, 20 ml of water, 5 ml of 5 M NaCl, 50 μl of 1 M DTT, and one tablet of Roche Protease Inhibitor Cocktail), and no-salt wash buffer (NSWB; 25 ml of basic wash buffer, 25 ml of water, 50 μl of 1 M DTT, and one tablet of Roche Protease Inhibitor Cocktail). Lyophilized cAMP-agarose was initially incubated with LyB containing BSA, washed with LyB, and then packed in columns in a 1-ml packed volume. BCC lysate (0.4 ml; ~1.5 mg of protein) was added to 0.4 ml of cAMP-agarose prepared as above, all in LyB+ buffer, and was incubated overnight with rocking at 4°C. Slurries were then added to Bio-Rad 1-ml columns, and one column volume of the slurry was eluted (1.1). The packed agarose was then rinsed with two successive column volumes of LyB+ (1.2, 1.3) and two column volumes each of BWB (2.1, 2.2), HSWB (3.1, 3.2), and NSWB (4.1, 4.2). Columns were equilibrated with LyB+ buffer, and one column volume of 50 mM cAMP (Biolog) in LyB+ buffer was added. Slurries were incubated with rocking at 4°C overnight directly in Bio-Rad column, after which the first column volume was collected (5.1), and then successive column volumes in LyB+ buffer (for example, 5.2, 5.3, and 5.4) were collected. An aliquot of cAMP-agarose combined with lysate that was not subjected to affinity purification was placed in several volumes of denaturing PAGE buffer, heated to 70°C for 10 min, and clarified by centrifugation (“gel”).
Calculations and statistics
Calculations and statistics were conducted in SigmaPlot 11.0 (Systat). Testing for statistical significance was performed with either one-factor or two-factor ANOVA, as appropriate, followed by Bonferroni-corrected t tests for multiple comparisons. Statistical significance was deemed at P < 0.05. In dose-response experiments, IC50 values were obtained from curves that were fit to data with a four-parameter logistic regression.
Supplementary Material
Acknowledgments
We thank C.-M. Hsu, D. Huddleston, W. Xu, and J. Russ for technical assistance in conducting the experiments, and M. Gerdin, D. Ait-Ali, N. Stroth, and C. Hamelink for performing experiments in chromaffin cells early in the course of this project that were critical to its eventual completion. We are grateful to M. Brownstein, C. Gerfen, S. Bunn, A. Morozov, T. Balla, and J. Marsh for their insightful comments and suggestions about the manuscript. We also thank J. Nagle and D. Kauffman at the National Institute of Neurological Disorders and Stroke (NINDS) DNA sequencing facility. Finally, we thank H. Jaffe, NINDS Protein/Peptide Sequencing Facility, for the identification of chromaffin cell cAMP-binding proteins by mass spectrometry.
Funding: This work was funded by the Intramural Research Program of the NIH Projects 1ZIAMH002386 (to L.E.E.) and 1ZIAMH002592 (to M.V.E.).
Footnotes
www.sciencesignaling.org/cgi/content/full/6/281/ra51/DC1
Fig. S1. Time course of cAMP-dependent ERK activation in bovine chromaffin cells.
Fig. S2. PACAP-dependent neuritogenesis is sensitive to inhibition by FTS-A.
Fig. S3. PACAP-dependent neuritogenesis is PLX4720-sensitive.
Fig. S4. Phylogenetic relationship of members of the human Rapgef protein family.
Fig. S5. Silencing of Rapgef2 does not affect cAMP accumulation or Epac activity in NS-1 cells.
Fig. S6. Rapgef2 is required for cAMP-dependent neuritogenesis in NS-1 cells.
Fig. S7. MAPK-dependent Rapgef2 signaling is initiated by cAMP, but not cGMP.
Table S1. Efficacy of GPCR agonists and the AC activator forskolin in stimulating a CRE reporter gene.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Rat FRTL-5 thyroid cells were obtained from the American Type Culture Collection through a materials transfer agreement (MTA). The pPRIChpaHA retroviral expression vector was obtained by an MTA from R. Y. Tsien (University of California, San Diego).
Author contributions: A.C.E. participated in project design, performed experiments with cell lines, interpreted the data, and drafted and revised the manuscript; M.V.E. generated retroviral expression vectors, contributed to the writing of the manuscript, and provided funding support; T.M. prepared primary cultures of neurons, performed Western blotting, interpreted data, and contributed to drafting the manuscript; L.E.E. performed cAMP-binding assays; was responsible for project supervision, data interpretation, and manuscript writing; and provided funding support.
REFERENCES AND NOTES
- 1.Robison GA, Butcher RW, Sutherland EW. Cyclic AMP. Annu Rev Biochem. 1968;37:149–174. doi: 10.1146/annurev.bi.37.070168.001053. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 3.Hardman JG, Robison GA, Sutherland EW. Cyclic nucleotides. Annu Rev Physiol. 1971;33:311–336. doi: 10.1146/annurev.ph.33.030171.001523. [DOI] [PubMed] [Google Scholar]
- 4.Kuo JF, Greengard P. Cyclic nucleotide-dependent protein kinases. IV. Widespread occurrence of adenosine 3′,5′-monophosphate-dependent protein kinase in various tissues and phyla of the animal kingdom. Proc Natl Acad Sci USA. 1969;64:1349–1355. doi: 10.1073/pnas.64.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawasaki H, Springett GM, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman DE, Graybiel AM. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij J, Zwartkruis FJT, Verheijen MHG, Cool RH, Nijman SMB, Witinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 7.Kaupp UB, Seifert R. Cyclic nucleotide-gated ion channels. Physiol Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 8.Lolicato M, Nardini M, Gazzarrini S, Möller S, Bertinetti D, Herberg FW, Bolognesi M, Martin H, Fasolini M, Bertrand JA, Arrigoni C, Thiel G, Moroni A. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem. 2011;286:44811–44820. doi: 10.1074/jbc.M111.297606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne JK, Zaganjor E, Cobb MH. Signal control through Raf: In sickness and in health. Cell Res. 2012;22:14–22. doi: 10.1038/cr.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerfen CR, Miyachi S, Paletzki R, Brown P. D1 dopamine receptor super-sensitivity in the dopamine-depleted striatum results from a switch in the regulation of ERK1/2/MAP kinase. J Neurosci. 2002;22:5042–5054. doi: 10.1523/JNEUROSCI.22-12-05042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morozov A, Muzzio IA, Bourtchouladze R, Van-Strien N, Lapidus K, Yin D, Winder DG, Adams JP, Sweatt JD, Kandel ER. Rap1 couples cAMP signaling to a distinct pool of p42/44MAPK regulating excitability, synaptic plasticity, learning, and memory. Neuron. 2003;39:309–325. doi: 10.1016/s0896-6273(03)00404-5. [DOI] [PubMed] [Google Scholar]
- 12.Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opin Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Gutkind JS. Regulation of mitogen-activated protein kinase signaling networks by G protein-coupled receptors. Sci STKE. 2000;2000:re1. doi: 10.1126/stke.2000.40.re1. [DOI] [PubMed] [Google Scholar]
- 14.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: Emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 15.Hamelink C, Lee HW, Chen Y, Grimaldi M, Eiden LE. Coincident elevation of cAMP and calcium influx by PACAP-27 synergistically regulates vasoactive intestinal polypeptide gene transcription through a novel PKA-independent signaling pathway. J Neurosci. 2002;22:5310–5320. doi: 10.1523/JNEUROSCI.22-13-05310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaudry D, Chen Y, Ravni A, Hamelink C, Elkahloun AG, Eiden LE. Analysis of the PC12 cell transcriptome after differentiation with pituitary adenylate cyclase-activating polypeptide (PACAP) J Neurochem. 2002;83:1272–1284. doi: 10.1046/j.1471-4159.2002.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravni A, Vaudry D, Gerdin MJ, Eiden MV, Falluel-Morel A, Gonzalez B, Vaudry H, Eiden LE. A cAMP-dependent, PKA-independent signaling pathway mediating neuritogenesis through Egr1 in PC12 cells. Mol Pharmacol. 2008;73:1688–1708. doi: 10.1124/mol.107.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery AC, Eiden MV, Eiden LE. A new site and mechanism of action for the widely used adenylate cyclase inhibitor SQ22,536. Mol Pharmacol. 2013;83:95–105. doi: 10.1124/mol.112.081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emery AC, Eiden LE. Signaling through the neuropeptide GPCR PAC1 induces neuritogenesis via a single linear cAMP- and ERK-dependent pathway using a novel cAMP sensor. FASEB J. 2012;26:3199–3211. doi: 10.1096/fj.11-203042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroth N, Kuri BA, Mustafa T, Chan SA, Smith CB, Eiden LE. PACAP controls adrenomedullary catecholamine secretion and expression of catecholamine bio-synthetic enzymes at high splanchnic nerve firing rates characteristic of stress transduction in male mice. Endocrinology. 2013;154:330–339. doi: 10.1210/en.2012-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Légrádi G, Hannibal J, Lechan RM. Pituitary adenylate cyclase-activating polypeptide-nerve terminals densely innervate corticotropin-releasing hormone-neurons in the hypothalamic paraventricular nucleus of the rat. Neurosci Lett. 1998;246:145–148. doi: 10.1016/s0304-3940(98)00255-9. [DOI] [PubMed] [Google Scholar]
- 22.Grinevich V, Fournier A, Pelletier G. Effects of pituitary adenylate cyclase-activating polypeptide (PACAP) on corticotropin-releasing hormone (CRH) gene expression in the rat hypothalamic paraventricular nucleus. Brain Res. 1997;773:190–196. doi: 10.1016/s0006-8993(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann ML, Mustafa T, Eiden AM, Herkenham M, Eiden LE. PACAP-deficient mice show attenuated corticosterone secretion and fail to develop depressive behavior during chronic social defeat stress. Psychoneuroendocrinology. 2013;38:702–715. doi: 10.1016/j.psyneuen.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroth N, Liu Y, Aguilera G, Eiden LE. Pituitary adenylate cyclase-activating polypeptide controls stimulus-transcription coupling in the hypothalamic-pituitary-adrenal axis to mediate sustained hormone secretion during stress. J Neuroendocrinol. 2011;23:944–955. doi: 10.1111/j.1365-2826.2011.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ster J, De Bock F, Guerineau NC, Janossy A, Barrere-Lemaire S, Bos JL, Bockaert J, Fagni L. Exchange protein activated by cAMP (Epac) mediates cAMP activation of p38 MAPK and modulation of Ca2+-dependent K+ channels in cerebellar neurons. Proc Natl Acad Sci USA. 2007;104:2519–2524. doi: 10.1073/pnas.0611031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang K, Lei G, Jackson MF, Macdonald JF. The involvement of PACAP/VIP system in the synaptic transmission in the hippocampus. J Mol Neurosci. 2010;42:319–326. doi: 10.1007/s12031-010-9372-7. [DOI] [PubMed] [Google Scholar]
- 27.Holighaus Y, Weihe E, Eiden LE. STC1 induction by PACAP, is mediated through cAMP and ERK1/2 but not PKA in cultured cortical neurons. J Mol Neurosci. 2012;46:75–87. doi: 10.1007/s12031-011-9653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerschmidt A, Chatterji B, Zeiser J, Schroder A, Genieser HG, Pich A, Kaever V, Schwede F, Wolter S, Seifert R. Binding of regulatory subunits of cyclic AMP-dependent protein kinase to cyclic CMP agarose. PLoS One. 2012;7:e39848. doi: 10.1371/journal.pone.0039848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan N, Wu J, Anand GS, Yooseph S, Neuwald AF, Venter JC, Taylor SS. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuiperij HB, de Rooij J, Rehmann H, van Triest M, Wittinghofer A, Bos JL, Zwartkruis FJ. Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2. Biochim Biophys Acta. 2003;1593:141–149. doi: 10.1016/s0167-4889(02)00365-8. [DOI] [PubMed] [Google Scholar]
- 31.Liao Y, Kariya K, Hu CD, Shibatohge M, Goshima M, Okada T, Watari Y, Gao X, Jin TG, Yamawaki-Kataoka Y, Kataoka T. RA-GEF, a novel Rap1A guanine nucleotide exchange factor containing a Ras/Rap1A-associating domain, is conserved between nematode and humans. J Biol Chem. 1999;274:37815–37820. doi: 10.1074/jbc.274.53.37815. [DOI] [PubMed] [Google Scholar]
- 32.Pham N, Cheglakov I, Koch CA, de Hoog CL, Moran MF, Rotin D. The guanine nucleotide exchange factor CNrasGEF activates ras in response to cAMP and cGMP. Curr Biol. 2000;10:555–558. doi: 10.1016/s0960-9822(00)00473-5. [DOI] [PubMed] [Google Scholar]
- 33.Bilasy SE, Satoh T, Terashima T, Kataoka T. RA-GEF-1 (Rapgef2) is essential for proper development of the midline commissures. Neurosci Res. 2011;71:200–209. doi: 10.1016/j.neures.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Greene LA, Tischler AS. Establishment of a noradrenergic clonal cell line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 36.Hughes AL, Messineo-Jones D, Lad SP, Neet KE. Distinction between differentiation, cell cycle, and apoptosis signals in PC12 cells by the nerve growth factor mutant Δ9/13, which is selective for the p75 neurotrophin receptor. J Neurosci Res. 2001;63:10–19. doi: 10.1002/1097-4547(20010101)63:1<10::AID-JNR2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 37.Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: Making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 38.Gerdin MJ, Eiden LE. Regulation of PC12 cell differentiation by cAMP signaling to ERK independent of PKA: Do all the connections add up? Sci STKE. 2007;2007:pe15. doi: 10.1126/stke.3822007pe15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mor A, Haklai R, Ben-Moshe O, Mekori YA, Kloog Y. Inhibition of contact sensitivity by farnesylthiosalicylic acid-amide, a potential Rap1 inhibitor. J Invest Dermatol. 2011;131:2040–2048. doi: 10.1038/jid.2011.152. [DOI] [PubMed] [Google Scholar]
- 40.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vossler MR, Yao H, York RD, Pan MG, Rim CS, Stork PJS. cAMP activates MAP kinase and Elk-1 through a B-Raf and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka T, Shimizu K, Yamamori B, Kuroda S, Takai Y. Activation of brain B-Raf protein kinase by Rap1B small GTP-binding protein. J Biol Chem. 1996;271:1258–1261. doi: 10.1074/jbc.271.3.1258. [DOI] [PubMed] [Google Scholar]
- 43.Enserink JM, Christensen AE, de Rooij J, van Triest M, Schwede F, Genieser HG, Døskeland SO, Blank JL, Bos JL. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- 44.Quilliam LA, Rebhun JF, Castro AF. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog Nucleic Acid Res Mol Biol. 2002;71:391–444. doi: 10.1016/s0079-6603(02)71047-7. [DOI] [PubMed] [Google Scholar]
- 45.Ohtsuka T, Hata Y, Ide N, Yasuda T, Inoue E, Inoue T, Mizoguchi A, Takai Y. nRap GEP: A novel neural GDP/GTP exchange protein for rap1 small G protein that interacts with synaptic scaffolding molecule (S-SCAM) Biochem Biophys Res Commun. 1999;265:38–44. doi: 10.1006/bbrc.1999.1619. [DOI] [PubMed] [Google Scholar]
- 46.Pham N, Rotin D. Nedd4 regulates ubiquitination and stability of the guanine-nucleotide exchange factor CNrasGEF. J Biol Chem. 2001;276:46995–47003. doi: 10.1074/jbc.M108373200. [DOI] [PubMed] [Google Scholar]
- 47.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, Madabushi S, Reiter E, Premont RT, Lichtarge O, Lefkowitz RJ. β-Arrestin-dependent, G protein-independent ERK1/2 activation by the β2 adrenergic receptor. J Biol Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 48.Lee JH, Cho KS, Lee J, Kim D, Lee SB, Yoo J, Cha GH, Chung J. Drosophila PDZ-GEF, a guanine nucleotide exchange factor for Rap1 GTPase, reveals a novel upstream regulatory mechanism in the mitogen-activated protein kinase signaling pathway. Mol Cell Biol. 2002;22:7658–7666. doi: 10.1128/MCB.22.21.7658-7666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huelsmann S, Hepper C, Marchese D, Knöll C, Reuter R. The PDZ-GEF Dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development. 2006;133:2915–2924. doi: 10.1242/dev.02449. [DOI] [PubMed] [Google Scholar]
- 50.Boettner B, Van Aelst L. The Rap GTPase activator Drosophila PDZ-GEF regulates cell shape in epithelial migration and morphogenesis. Mol Cell Biol. 2007;27:7966–7980. doi: 10.1128/MCB.01275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amsen EM, Pham N, Pak Y, Rotin D. The guanine nucleotide exchange factor CNrasGEF regulates melanogenesis and cell survival in melanoma cells. J Biol Chem. 2006;281:121–128. doi: 10.1074/jbc.M507595200. [DOI] [PubMed] [Google Scholar]
- 52.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 53.Pak Y, Pham N, Rotin D. Direct binding of the β1 adrenergic receptor to the cyclic AMP-dependent guanine nucleotide exchange factor CNrasGEF leads to Ras activation. Mol Cell Biol. 2002;22:7942–7952. doi: 10.1128/MCB.22.22.7942-7952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anouar Y, MacArthur L, Cohen J, Iacangelo AL, Eiden LE. Identification of a TPA-responsive element mediating preferential transactivation of the galanin gene promoter in chromaffin cells. J Biol Chem. 1994;269:6823–6831. [PubMed] [Google Scholar]
- 55.Martin KR, Corlett A, Dubach D, Mustafa T, Coleman HA, Parkington HC, Merson TD, Bourne JA, Porta S, Arbonés ML, Finkelstein DI, Pritchard MA. Over-expression of RCAN1 causes Down syndrome-like hippocampal deficits that alter learning and memory. Hum Mol Genet. 2012;21:3025–3041. doi: 10.1093/hmg/dds134. [DOI] [PubMed] [Google Scholar]
- 56.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albagli-Curiel O, Lécluse Y, Pognonec P, Boulukos KE, Martin P. A new generation of pPRIG-based retroviral vectors. BMC Biotechnol. 2007;7:85. doi: 10.1186/1472-6750-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaturvedi D, Poppleton HM, Stringfield T, Barbier A, Patel TB. Subcellular localization and biological actions of activated RSK1 are determined by its interactions with subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 2006;26:4586–4600. doi: 10.1128/MCB.01422-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.