Abstract
Chemokines and their receptors are implicated in a wide range of human diseases, including acquired immune deficiency syndrome (AIDS). The entry of human immunodeficiency virus type 1 (HIV-1) into a cell is initiated by the interaction of the virus’s surface envelope proteins with two cell surface components of the target cell, namely CD4 and a chemokine co-receptor, usually CXCR4 or CCR5. Typical anti-HIV-1 agents include protease and reverse transcriptase inhibitors, but the targets of these agents tend to show rapid mutation rates. As such, strategies based on HIV-1 co-receptors have appeal because they target invariant host determinants. Chemokines and their receptors are also of general interest since they play important roles in numerous physiological and pathological processes in addition to AIDS. Therefore, intensive basic and translational research is ongoing for the dissection of their structure – function relationships in an effort to understand the molecular mechanism of chemokine – receptor interactions and signal transductions across cellular membranes. This paper reviews and discusses recent advances and the translation of new knowledge and discoveries into novel interventional strategies for clinical application.
Keywords: HIV, AIDS, CXCR4, CCR5, SDF-1α, vMIP-II, chemokines, chemokine receptors
Introduction
Chemokines are small soluble proteins of approximately 70 amino acid residues with a molecular weight of 8 – 10 kDa.1 They act as potent chemoattractants of a large variety of mononuclear cell types to sites of inflammation or secondary lymphoid organs by interacting with chemokine receptors. Based on the positions of two conserved cysteine residues in their amino (N)-termini, chemokines can be divided into four subfamilies: CC, CXC, CX3C and C.2,3 The two main subfamilies of chemokines are CXC and CC. CXC chemokines are primarily involved in the activation of neutrophils, whereas CC chemokines stimulate other leukocytes such as monocytes, lymphocytes and basophils. The highly conserved three-dimensional structures of all chemokines include a flexible N-terminus, a three-stranded antiparallel β-sheet and a C-terminal α-helix.4 In the typical structure, the first two cysteine residues are situated close together, near the N-terminus, with the third cysteine residue residing in the center of the molecule, and the fourth cysteine residue located close to the C-terminal end.5 An ‘N-loop’ of approximately 10 amino acids follows the first two cysteine residues. Following the N-loop, there is a single-turn ‘310-helix,’ a β-sheet with three β-strands and a C-terminal α-helix, connected by turns called ‘30s,’ ‘40s’ and ‘50s’ loops. The third and fourth cysteine residues are located in the 30s’ and 50s’ loops, respectively. The structures of many chemokines have been determined by nuclear magnetic resonance (NMR) or X-ray crystallography, including those of stromal cell-derived factor (SDF)-1α,6,7 viral macrophage inflammatory protein (vMIP)-II,8,9 macrophage inflammatory protein (MIP)-1β,10 and regulated on activation, normal T-cell expressed and secreted (RANTES).11
Chemokine receptors are a group of transmembrane (TM) proteins that belong to the superfamily of G-protein-coupled receptors (GPCRs).2,12,13 They possess seven TM helices and transmit signals from extracellular ligands to intracellular biological pathways via heterotrimeric G-proteins. Chemokines and their receptors are implicated in a wide range of human acute and chronic inflammatory diseases (i.e. acute respiratory distress syndrome, allergic asthma, psoriasis and arthritis), neurological disorders, and cancer.2–4,14 Chemokine receptors are also involved in the pathogenesis of human immunodeficiency virus type 1 (HIV-1) infection. HIV-1 enters cells through a fusion process in which the HIV-1 envelope glycoprotein gp120 binds to CD4, the main receptor for HIV-1 on the target cell surface.15,16 However, it has long been known that CD4 alone is not sufficient for HIV-1 fusion and entry, and that additional receptors may be needed. In 1996, chemokine receptors CXCR4 and CCR5 were discovered to be the long-sought co-receptors for syncytium-inducing and non-syncytium-inducing HIV-1 strains, respectively.17–20 The viral fusion process involves the initial binding of HIV-1 gp120 to its high-affinity receptor CD4, which results in conformational changes in gp120 and CD4.21–23 The gp120 – CD4 complex then interacts with a chemokine co-receptor, such as CXCR4 or CCR5, to form a heterotrimeric complex of gp120 – CD4 – co-receptor.24–26 During the asymptomatic stage of disease, macrophage (M)-tropic strains of HIV-1 involved in sexual transmission primarily use CCR5 as an entry co-receptor.17–19 However, in 40–50% of HIV-1-infected individuals, T-cell (T)-tropic strains that predominantly use CXCR4 eventually replace M-tropic strains, leading to rapid disease progression.27–29 Among many new directions in AIDS research opened by this discovery, an important area of investigation is the biochemical and biophysical characterization of the interactions of chemokine receptors with HIV-1 and natural as well as synthetic chemokine ligands.
Structure – fuction relationship of CXCR4 and its ligands
CXCR4 is a CXC chemokine receptor 4 which consists of 352 amino acid residues.30 Like other GPCRs, CXCR4 consists of an amino (N)-terminus, three extracellular and intracellular loops, seven TM helices, and a carboxyl (C)-terminus. However, unlike other chemokine receptors which have a number of distinct ligands, CXCR4 has only one endogenous natural ligand known as an SDF-1α.1 However, CXCR4 can also be recognized by an antagonistic ligand, vMIP-II, encoded by the Kaposi’s sarcoma-associated herpes virus.31 CXCR4 – SDF-1α interaction has essential physiological functions in immunomodulation, organogenesis, hematopoiesis and cerebellar neuron migration.32–34 This is further demonstrated by knockout mice of CXCR4 and SDF-1α that die of hematopoietic, cardiac, vascular and cerebellar defects during embryogenesis.32,33,35
vMIP-II displays a broader spectrum of receptor activities than any mammalian chemokine, as it binds with high affinity to a number of both CXC and CC chemokine receptors, including CXCR4 and CCR5, and it inhibits cell entry of HIV-1 mediated by these receptors.36,37 Synthetic peptides derived from the N-terminus of vMIP-II showed that the N-terminus of vMIP-II is the major binding determinant for CXCR438 (Table 1). Only V1 peptide (1–21 residues) from the N-terminus of vMIP-II showed CXCR4 binding, and it selectively prevents CXCR4 signal transduction and co-receptor function in mediating the entry of T- and dual-tropic HIV-1 isolates.38 An all-D-amino acid analog of V1 peptide, designated as DV1 peptide, displayed even higher binding affinity and antiviral activity than V1, demonstrating the remarkable stereochemical flexibility of the CXCR4 – peptide interface.39
Table 1.
List of CXCR4 inhibitors, their chemical structures, sequences and modifications
| Inhibitors | Modification diagrams or chemical structures | Amino acid sequences* | References |
|---|---|---|---|
| ALX40-4C | AcNH-D-Arg-(D-Arg)8-COOH | 55,93,94 | |
| AMD3100 |
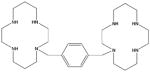
|
82 | |
| CGP64222 |
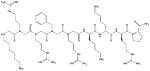
|
97 | |
| R3G |
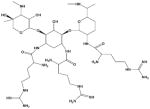
|
97 | |
| T22 |
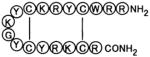
|
RRWCYRKCYKGYCYRKCR | 83 |
| T140 |
|
R-R-Na1-C-Y-R-K-D(K)-P-Y-R-Cit-C-R | 95,96 |
| TC14012 |
|
R-R-Na1-C-Y-Cit-K-D(Cit)-P-Y-R-Cit-C-R | 95 |
| FC131 |
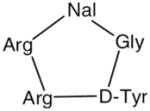
|
96 | |
| SDF-1α |
|
KPVSLSYRCPCRFFESHVARANVKHLKILNTPNCALQIVARLKNNNRQVCIDPKLKWIQEYLEKALNK | 1,6,7,58,84,85,91 |
| vMIP-II |
|
LGASWHRPDKCCLGYQKRPLPQVLLSSWYPTSQLCSKPGVIFLTKRGRQVCADKSKDWVKKLMQQLPVTAR | 8,9,31 |
| V1 |

|
LGASWHRPDKCCLGYQKRPLP | 38 |
| DV1 |

|
LGASWHRPDKCCLGYQKRPLP | 39 |
| RCP168 |
|
LGASWHRPDKCCLGYQKRPLPQVLLSSWYPTSQLCSKPGVIFLTKRGRQVCADKSKDWVKKLMQQLPVTAR | 72 |
D-Amino acids are shown in italic
Prior to the recent publication of high resolution crystal structures of CXCR4 by Wu et al.,30 several groups have endeavored to characterize CXCR4 interactions with HIV-1, natural ligands and de novo designed inhibitors using molecular modeling, chimeras and site-specific mutagenesis. These studies demonstrated that the amino (N)-terminus and the second (ECL2) and third (ECL3) extra-cellular loops (ECLs) of CXCR4 are required for HIV-1 co-receptor activity.40–50 They also indicated a requirement for multiple extracellular and TM domains of CXCR4 in chemokine interactions and receptor signaling.41,42,46,50–55 In addition, a separation of binding and signaling functions was revealed by these chimeric and mutational studies, and it has been exploited to validate the accuracy of a two-site model that was initially developed for the C5a chemoattractant and its receptor. This model has the chemokine core domain being the ‘site one’ docking domain and the chemokine N-terminus being the ‘site two’ signaling trigger.56 According to this model, the motif composed of amino acids 12–17 of the SDF-1α, RFFESH loop, first docks onto the N-terminal domain of CXCR4, and this contact allows the subsequent interaction of the flexible N-terminus of SDF-1α with the receptor groove formed by TM domains and/or ECLs, thereby triggering the receptor function.6,56,57 The N-terminus of SDF-1α, being relatively flexible and unstructured in solution, has been confirmed as essential for CXCR4 recognition and signal transduction.6,7,58
Recently, the long-awaited crystal structure of CXCR4 was published by Wu et al., who reported five independent crystal structures of CXCR4 bound to an antagonist small molecule IT1t and to a cyclic peptide CVX15 at 2.5 to 3.2 Å resolution.30 These structures are now providing new clues about the interactions between CXCR4 and SDF-1α as well as with HIV-1 gp120. All structures have revealed consistent homodimers with an interface, including TM helices V and VI, which may be involved in regulating signaling. Moreover, the peptide and small molecule complexes of CXCR4 have identified the likely ‘site two’ of the chemokine-signaling trigger. The IT1t ligand was shown to occupy part of the binding pocket defined by side chains from helices I, II, III and VII, whereas CVX15 filled most of the binding-pocket volume by inducing major deviations in the base of the receptor N-terminus (residues 29–33), as well as a minor adjustment of extracellular tips of helices VI, VII and V. Compared with previous GPCR structures, the binding pocket of CXCR4 is larger, more open and located closer to the extracellular surface, and it includes acidic Asp187, Glu288 and Asp97 that are important for SDF-1α binding. This suggests that Lys1, the most critical residue in SDF-1α for receptor activation, could reach into the CXCR4 pocket and interact with one of these acidic residues. The importance of Glu288 for SDF-1α signaling was previously demonstrated by our laboratory.50 Similarly, the basic character of gp120 V3 loop, which becomes exposed upon CD4 binding, could potentially penetrate the CXCR4 binding pocket, thereby interacting with one of these acidic residues. Taken together, the crystal structures of CXCR4 provide strong support for the two-site model, and they also suggest the possibility of a three-step interaction between CXCR4 and its ligand. The first step would be the electrostatic interaction of the body of the chemokine with the complementary surface of CXCR4. The second step would be the insertion of the N-terminal of chemokine into the cavity defined by the TM and some extra-cellular domains. The implied third step would be the folding of the N-terminus of CXCR4 across the top of the docked chemokine. As further details are resolved regarding the dynamic changes involved in CXCR4 interactions with its natural ligand and HIV-1 gp120, new opportunities will surely emerge for drug discovery efforts that target specific functional states of the receptor.
Structure – fuction relationship of CCR5 and its ligands
CCR5 is a CC chemokine receptor 5 which consists of 352 amino acid residues. It has multiple natural chemokine ligands, including MIP-1α, MIP-1β, ‘RANTES’ protein and monocyte chemotactic protein (MCP)-2.59 Among these ligands, RANTES and MIP-1α can bind to other CC chemokine receptors, while MIP-1β is known to be most specific for CCR5.59 vMIP-II encoded by the Kaposi’s sarcoma-associated herpes virus displays a broader spectrum of receptor activities, including CCR5.31,36,37 Signal transduction through CCR5 is known to play important roles in both physiological and pathological processes, including inflammation and hematopoiesis.
As the major co-receptor for HIV-1 entry, CCR5 has undergone extensive studies using chimeric receptors,51–54,60,61 site-directed mutagenesis,62–67 NMR and molecular modeling.68 These studies have shown the involvement of all extracellular domains of CCR5 in its HIV-1 co-receptor function, particularly the N-terminal domain and the second ECL (ECL2) of CCR5.51–54,61 The ECL2 of CCR5 is responsible for the binding selectivity of the receptor, and it determines the range of chemokines to which it responds functionally.69 The major interaction sites between MIP-1β and CCR5 include the amino (N)-terminus, N-loop (residues 13–19), 310 turn (Arg18, Lys19 and Arg22), 20s region, 40s loop (Lys45, Arg46 and Lys48), and carboxyl (C)-terminus of MIP-1β,70–73 and the N-terminus and ECL2 of CCR5.51,68,69,74–76 Phe13 of MIP-1β is the most important residue for CCR5 binding.77 In addition to these basic amino acids, Pro21 was found to be important in CCR5 binding, whereas Tyr15 is essential for the proper folding of MIP-1β.70 Thus, the key residues in MIP-1β crucial for CCR5 binding include most of the basic amino acids and two hydrophobic groups, all arranged on one surface of the chemokine.70
As with other chemokines, the N-terminus of MIP-1β is of primary importance for signal transduction and cell activation, but it does not significantly contribute to receptor binding. According to the prevailing two-site model, the core moiety of CC chemokines first binds to the extracellular domains of CCR5 through electrostatic attractions, and the N-terminus subsequently binds to the second binding site spanning the TM helixes to activate the receptor. Several research groups have attempted to locate this second binding site important for receptor activation by alanine scanning mutagenesis of the TM domains.66,78 These studies have revealed that the binding site for small molecule antagonists of CCR5, including TAK-779, AD101 and SCH-C (Table 2), are located near the extracellular surface of the receptor, within a cavity formed between TM helices I, II, III, V and VII. The important residues include Leu33, Tyr37, Asp76, Thr82, Trp86, Tyr108, Phe113, Ile198 and Glu283. Govaerts et al.79 have provided further support by identifying the importance of Thr82 for receptor activation, but not for chemokine binding.
Table 2.
List of CCR5 inhibitors, their chemical structures, sequences and modifications
| Inhibitors | Modification diagrams or chemical structures | Amino acid sequences | References |
|---|---|---|---|
| TAK-779 |
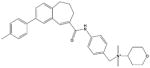
|
66,103,104 | |
| AD101 |
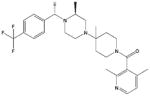
|
78 | |
| SCH-C |
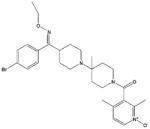
|
78 | |
| MIP-1β |
|
APMGSDPPTACCFSYTARKLPRNFVVDYYETSSLCSQPAVVFGTKRSKQVCADPSESWVQEYVYDLELN | 10,59 |
| RANTES |
|
SPYSSDTTPCCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 11 |
| AOP-RANTES |
|
AOP-PYSSDTTPCCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 101 |
| PSC-RANTES |
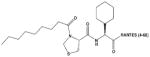
|
PSC-SSDTTPCCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 106 |
| 5P12-RANTES |
|
QGPPLMATQSCCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 80 |
| 5P14-RANTES |
|
QGPPLMSLQVCCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 80 |
| 6P4-RANTES |
|
QGPPGDIVLACCFAYIARPLPRAHIKEYFYTSGKCSNPAVVFVTRKNRQVCANPEKKWVREYINSLEMS | 80 |
Docking of HIV-1 gp120 to CCR5 involves interactions between conserved residues in the V3 stem – C4 region of gp120 and the N-terminus of CCR5, as well as binding of the V3 crown to a second region of CCR5, which a number of studies suggest to be primarily ECL2.51–54,61 Since TAK-779 does not disrupt interactions between gp120 and the N-terminus of CCR5,66 it is certainly possible that the V3 crown interacts with the TM domain residues, although none of the TM domain mutants discussed above were significantly impaired with respect to viral entry. The binding of small molecule antagonists to the TM domains of CCR5 may induce a conformational change in the region of CCR5 (likely to be ECL2) that interact with the V3 crown. Thus, disruption of the binding site on CCR5 for the V3 crown would inhibit the gp120 – CCR5 interaction and viral entry. Interesting, the Hartley group recently identified three analogues of the N-terminally modified chemokine PSC-RANTES, all of which exhibit in vitro potency against HIV-1 comparable to that of PSC-RANTES80 (Table 2). Despite the subtle differences in sequence, volume and mass that distinguish these modified RANTES compounds, all three showed very distinct characteristics with respect to CCR5 binding affinity, G protein-linked signaling and receptor sequestration. The first, 6P4-RANTES, resembles PSC-RANTES in that it is a strong agonist that induces prolonged intracellular sequestration of CCR5. The second, 5P12-RANTES, has no detectable signaling activity and does not bring about receptor sequestration. The third, 5P14-RANTES, induces significant levels of CCR5 internalization without detectable signaling activity. As such, these compounds represent excellent molecular probes for unraveling the details of agonist versus antagonist activity associated with slightly different binding to the second binding site of CCR5. Thus, they can be useful in distinguishing sites important for binding, signaling and internalization.
Chemokine receptor inhibitors
In comparison to anti-HIV-1 agents that are directed against components of the rapidly mutating virus population, co-receptor-based therapeutic strategies have the appeal of targeting relatively invariant host determinants.38,39,55,58,81–91 Furthermore, individuals with CCR5 mutations appear to be both healthy and highly resistant to HIV-1 infection, which demonstrates the feasibility of this approach for inhibiting viral infection.3,92 Although previous studies have demonstrated the potential benefits of chemokine receptor inhibitors for combating AIDS and other diseases, the use of natural, non-specific chemokines in clinical applications is problematic due to the lack of selectivity and its potential side-effects. Consequently, the development of new inhibitors engineered with higher selectivity for targeted receptors and reduced toxicity is clearly desirable for their use in clinical applications and as specific probes in receptor biology, to study the role of particular ligands or receptors.
Several peptides and organic compounds, unrelated to natural chemokines that have antagonistic activity in CXCR4, were discovered much earlier through random screening39,55,87– 90 (Table 1). ALX40-4C (N-a-acetyl-nona-D-arginine amide acetate) and AMD3100 were the first CXCR4 antagonists to enter clinical trials.55,93,94 ALX40-4C is a highly basic peptide designed as an arginine mimic of the HIV Tat protein, and it has been shown to prevent HIV-1 entry via CXCR4.93,94 Moreover, T22,83 an 18-amino acid synthetic polyphemusin, and its downsized analogs, T140 and TC14012 (14 amino acid residues), were shown to inhibit CXCR4-mediated HIV-1 entry in the nanomolar range.95 Subsequent studies show that Arg2, L-3-(2-naphthyl)alanine (NaI)3, Tyr5 and Arg14 constitute the critical pharmacophore of T140.96 Screening of pentapeptide libraries containing these critical residues resulted in the identification of a small cyclic pentapeptide FC131 (cyclo(-NaI-Gly-D-Tyr-Arg-Arg-) that has similar anti-HIV activity to T140. Furthermore, CGP64222, R3G and NeoR were reported as Arg-mimic and cationic CXCR4 antagonists.97 CGP64222, a basic peptide oligomer of nine residues, has been shown to inhibit the replication of a wide range of laboratory HIV strains through selective interaction with CXCR4. However, there is cause for concern regarding undesired side-effects of blocking the normal CXCR4 – SDF-1α function, as knockout mice lacking either CXCR433,35 or SDF-1α32 died during embryogenesis with evidence of hematopoietic, cardiac, vascular and cerebellar defects.98 Even if adverse effects are not observed during clinical trials, any CXCR4 antagonist has the potential to cause harm if it is used chronically as a highly active antiretroviral therapy.
An alternative route of designing specific CXCR4 inhibitors is to use natural chemokine ligands as design templates by mimicking specific regions of a chemokine ligand. This can be used to study the structure – function relationship of the native molecule and to develop novel agonists or antagonists of chemokine receptors.38,39,58,84–87,90,91,99 This approach was first attempted on SDF-1α, the only known natural ligand of CXCR4.58,84,85,91 Peptides derived from the N-terminus of SDF-1α were proven essential for CXCR4 recognition, signal transduction and antiviral activity. However, they were less potent than the native SDF-1α.58,91 The attachment of positively charged residues to the N-terminal peptide sequence was found to enhance its ability to bind to CXCR4 and inhibit CXCR4-mediated T-tropic HIV-1 entry.84 When combined with the observation that peptides and organic compounds, such as AMD3100,82 T22,83 and ALX40-4C,55 have high positive charges and affinity for CXCR4, these studies indicate that electrostatic interaction may play a important role in CXCR4 recognition.84
CXCR4 can also be recognized by vMIP-II, which displays a broad spectrum of receptor activities.36,37 By studying synthetic peptides derived from the N-terminus of vMIP-II, our laboratory demonstrated that the N-terminus of vMIP-II is the major determinant for CXCR4 recognition38 (Table 1). Only V1 peptide (1–21 residues) showed CXCR4 binding, and it selectively prevented CXCR4 signal transduction and the co-receptor function in mediating the entry of T- and dual-tropic HIV-1 isolates. An all-D-amino acid analog of V1 peptide, designated as DV1 peptide, was also synthesized.39 Despite dramatically different conformations of side-chain groups, DV1 displayed higher binding affinity toward CXCR4, and it showed a significant antiviral activity in inhibiting the replication of CXCR4-dependent HIV-1 strains. Unnatural D-peptides can be highly desirable and advantageous over natural L-peptides for therapeutic development, as they are highly stable and resistant to proteolytic degradation.
Understanding the potential adverse effects of the antagonistic inhibitors of CXCR4 in HIV-1 therapy, Sachpatzidis et al. identified RSVM and ASLW as novel allosteric agonists that are insensitive to the CXCR4 antagonists, AMD3100 and T140, or monoclonal antibodies, 12G5 and 44717.111. This was achieved by screening a semi-randomized 17-mer library in a yeast strain that was expressing a functional CXCR4 receptor.90 In chemotaxis assays, RSVM behaves as a partial agonist, while ASLW behaves as a superagonist that displays a chemotactic index greater than the maximum observed in SDF-1α. Allosteric agonists may be therapeutically useful in combination with small molecule antagonists for anti-HIV therapy, since they could maintain essential receptor functions. The data also illustrate that other binding sites may exist for non-physiological agonists. Despite their early stage of development, allosteric agonists and other previously discussed potent CXCR4 peptide antagonists or agonists could serve as leads for the development of new therapeutic agents for HIV-1 infection and other diseases affecting the immune system.
Similarly, several small CCR5 antagonists have been reported to block HIV-1 infection100–102 (Table 2). TAK-779, a non-peptide compound, binds to CCR5 primarily via polar interactions with Glu283 and Tyr37, hydrophobic interactions with Phe10, Phe112, Phe113, Ile198 and Trp248 deeply buried in the TM regions, and a face-to-face aromatic stacking contact with Tyr108.103,104 Moreover, two structurally related small molecules SCH-C (SCH-351125) and AD101 (SCH-350581) inhibit RANTES binding as well as HIV-1 entry and replication, and they have excellent oral bioavailability in rats, dogs, monkeys and humans.78 In addition, Agrawal et al.105 prepared peptide segments from the amino (N)-terminus and ECLs of CCR5 and evaluated their inhibitory effects on HIV-1 entry. In this study, peptides derived from ECL1 and ECL3 inhibited R5-tropic BaL and dual-tropic 89.6 HIV-1 strains, whereas peptides from ECL2 only inhibited the BaL strain. Peptides from the N-terminus of CCR5 showed no effects on HIV-1 inhibition. The inhibitory effects of these CCR5-derived peptides were independent of co-receptors, illustrating that they target HIV envelopes rather than co-receptors.
Aminooxypentane (AOP)-RANTES, a chemically modified analog of RANTES, is a CCR5 antagonist that effectively inhibits CCR5-dependent HIV-1 strains101 (Table 2). Additional efforts to increase the antiviral potency of RANTES derivatives, using unnatural amino acids and systematic SAR, led to the generation of more potent HIV-1 inhibitors. This was exemplified by PSC-RANTES (N-nonanoyl, des-Ser1[L-thioproline2, L-cyclohexylglycine3]-RANTES [2–68]), in which several unnatural, non-coded structures were incorporated into the N-terminal region of RANTES with a 50-fold increase in potency over AOP-RANTES.106 More recently, Gaertner et al.80 used a phage-display strategy to generate fully recombinant chemokine analogs of PSC-RANTES with potent anti-HIV activity, culminating in 5P12-RANTES. 5P12-RANTES is as an effective inhibitor of virus entry via CCR5 as its immediate precursors, but has two important advantages: first, it is a pure antagonist with no demonstrable signaling activity upon CCR5 binding, and second, it is possible to produce 5P12-RANTES by recombinant expression, rather than chemical synthesis, suggesting that production could occur at an ultralow cost.
Although several CCR5 antagonists have been evaluated in clinical trials, only Maraviroc (MCV) has been approved in 2007 by the US Food and Drug Administration (FDA) for treatment of HIV-infected patients experiencing virological failure due to resistance to other classes of antiretroviral drugs.107 Maraviroc was subsequently approved for the treatment of antiretroviral naïve patients as well. Another CCR5 antagonist, Vicriviroc, is currently in advanced clinical development (phase III) but has yet to be approved by the FDA.107 Two other products in development are antibodies to CCR5. PRO 140 is a humanized CCR5 monoclonal antibody that inhibits CCR5-tropic HIV-1 in vitro, and was recently shown to have potent antiviral activity after a single dose in a phase Ib monotherapy, dose-escalation trial.108 HGS004 is a human immunoglobulin G4 monoclonal antibody against CCR5 that was also recently tested in a phase Ib trial that established its safety and in vivo activity against HIV-1.109 Maraviroc and other CCR5 antagonists could potentially be used in a variety of other clinical situations, such as the prevention of HIV transmission, intensification of HIV treatment and prevention of transplant rejection.107 As such, the future roles for CCR5 antagonists in both the prevention and treatment of HIV infection are likely to expand.
Synthetically and modularly modified-chemokines: a new class of unnatural synthetic molecules as leads for therapeutic development
We have been working toward the development of a systematic strategy based on chemokine structures to synthesize a new family of unnatural chemokines that, unlike natural chemokines, have higher receptor binding selectivity. In this regard, we have reported our progress in developing such a strategy by employing SMM (synthetically and modularly modified)-chemokines as a potential general method for the de novo design of novel chemokine receptor-selective ligands.72 Proof of this concept was shown by applying this strategy to transform vMIP-II, a very non-selective chemokine, into new analogs with significantly enhanced selectivity and potency for CXCR4 or CCR5, two principal co-receptors for HIV-1 entry, through modifying only a small N-terminal module of 10 residues. Two representative SMM-chemokines, RCP168 and RCP188, selective for CXCR4 and CCR5, respectively, showed similar or significantly enhanced binding affinities for their corresponding target receptors. However, RCP168 and RCP188 drastically decreased or even completely abolished cross-binding activities for other receptors. Furthermore, RCP168 is more effective in inhibiting HIV-1 infection than SDF-1α, and its anti-HIV activity is comparable to that of T-20.
In addition to high receptor selectivity, another important biological property of such de novo-designed ligands is signaling activity. RCP168, a vMIP-II analog with its N-terminal (1–10) residues replaced with D-amino acids, did not trigger either Ca2+ signaling or receptor internalization,72 which is distinct from the natural ligand of CXCR4, SDF-1α. More interestingly, RCP168 did not interfere with the Ca2+ signaling induced by SDF-1α at its effective CXCR4 binding concentration and only showed its effect at concentrations over 20 times higher than usual. Moreover, RCP168 significantly inhibits HIV-1 entry, in contrast to its much weaker activity in interfering with SDF-1α signaling. Thus, these disparate inhibitory activity profiles of RCP168 in differentiating HIV-1 co-receptor function versus the normal function of CXCR4 may prove to be advantageous in clinical applications, as RCP168 may not induce unwanted Ca2+ signaling or interfere with SDF-1α signaling important for the normal physiological functions at the concentrations used for inhibiting HIV-1 infection. The mechanistic basis for the disparate activities of RCP168 was demonstrated by the mutational mapping analysis of the binding sites of RCP168 and other D-amino acid-containing SMM-chemokines on CXCR4. This analysis revealed that RCP168 binding sites on CXCR4 overlap significantly with HIV-1 but differ from SDF-1α.110
The high-resolution crystal structure of RCP168, when compared with previous reports of vMIP-II,8,9 revealed that the enhanced selectivity of RCP168 was associated with the structural changes at the N-terminus. This was due to the D-amino acid modification and surprisingly the 30s loop as a result of a conformational change propagation from the distal N-terminus through a disulfide bridge.111 Based on this structural insight and with the aim of generating higher CXCR4 selectivity, new analogs containing modifications at the 30s loop were designed by replacing the 30s loop of RCP168 with the corresponding region of SDF-1α. Indeed, this analog, RCP303, exhibited more CXCR4 selective binding profiles than the parent molecule, as the 30s loop substitution led to a substantial reduction in binding to both CCR5 and CCR2. This finding further confirmed the role of the 30s loop in affecting receptor binding selectivity. In light of this finding, one may rationalize a structural basis for the conformational change cascade in chemokine – receptor interactions, which may include: (1) the initial binding of the N-terminus of a chemokine to the receptor; (2) the resulting conformational changes in the N-terminus (including the N-loop) and subsequently the 30s loop as facilitated by the disulfide bridge; and finally (3) the triggered recognition between the 30s loop and the receptor, leading to multipoint (at least including the N-terminus and the 30s loop) contact between the chemokine and its receptor.
To understand the signaling mechanism of CXCR4 or CCR5 in neuronal apoptosis associated with HIV-associated dementia (HAD), we also applied SMM-chemokine analogs as chemical probes of the mechanism(s) whereby these SMM-chemokines prevented or promoted neuronal apoptosis.112 Because of the profound activities of chemokine receptors in HAD, developing selective and potent inhibitors of chemokine receptors, while understanding the physiological or pathological processes of HAD, are crucial when devising novel strategies for clinical interventions. In this study, we demonstrated that unlike natural agonist SDF-1α, which causes neuronal death via a p38 MAPK-dependent pathway,113 antagonistic CXCR4-selective SMM-chemokines can effectively prevent gp120IIIB-induced neuronal apoptosis. Furthermore, compared with natural CCR5 agonist ligands that are known to promote neuronal survival by activating Akt,114 our data on unnatural CCR5 antagonists indicated that inhibition of CCR5 is neurotoxic via the p38 MAPK pathway, demonstrating that distinct CCR5-mediated signaling pathways can be activated by different chemokine ligands, which results in different cell fates. Taken together, using novel synthetic SMM-chemokines as mechanistic probes and/or potential inhibitors, we have obtained new insights into the distinct signaling pathways of neuronal apoptosis associated with HAD, which are activated by different chemokine receptor ligands that are either agonists or antagonists. This study also demonstrated a strategy for using chemically engineered inhibitors of chemokine receptors to study the signaling mechanism and intervention strategy of neuronal cell death and survival. A similar strategy may find its application in the study of other neurodegenerative diseases, as chemokine pathways to neuronal protection or damage may, at least in part, be common to other central nervous system (CNS) disorders including stroke, spinal cord injury and Alzheimer’s disease.
More recently, we tested the functionality of novel ‘dual-moiety’ CXCR4 chemokine agonists in human neural stem cells (hNSCs) that express CXCR4 (unpublished data). Recent advances in regenerative medicine unveiled the importance of chemotaxis during the engagement of neural stem cells toward the areas of neurodegeneration. Exposure of hNSCs to SDF-1α and subsequent induction of CXCR4-mediated signaling triggers a series of intracellular processes, which lead to survival, proliferation and, most importantly, migration of neural stem cells for injury repair. A panel of ‘dual-moiety’ chemokine analogs were designed by linking a highly potent and selective CXCR4 binding moiety, DV1 (D-[1–21]-vMIP-II), to a critical CXCR4-activating moiety, SD1 ([1–8]-SDF-1α), with various spacer sequences in between. Binding assays showed that ‘dual-moiety’ chemokine analogs possess potent CXCR4-selective binding affinity. More importantly, most of these ‘dual-moiety’ chemokine analogs significantly enhanced the calcium influx in CXCR4-expressing cells compared with the effect of SD1. Moreover, the prototypic analog, SDV1a, also chemoattracted CXCR4-expressing cells. Further investigations of SDV1a using hNSCs demonstrated its high capacity to activate CXCR4-downstream signaling events in vitro as well as cause extensive migration inside the transplanted healthy mouse brain. The effectiveness of mobilizing hNSCs with de novo designed agonists may lead to new translational therapeutics for the clinical repair of CNS injuries and other neurodegenerative conditions.
Acknowledgments
This work has been supported by grants from the National Institutes of Health, the Carol M Baldwin Breast Cancer Research Fund, the Connolly Endowment/Hendricks Fund and the LUNGevity Foundation.
Footnotes
Author contributions: W-TC and JA wrote the manuscript.
References
- 1.Premack BA, Schall TJ. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–8. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 2.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 3.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Proudfoot A. Chemokine receptors: multifaceted therapeutic targets. Nat Rev/Immunol. 2002;2:106–15. doi: 10.1038/nri722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–99. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 6.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dealwis C, Fernandez EJ, Thompson DA, Simon RJ, Siani MA, Lolis E. Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 ‘fusin’ coreceptor. Proc Natl Acad Sci USA. 1998;95:6941–6. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez EJ, Wilken J, Thompson DA, Peiper SC, Lolis E. Comparison of the structure of vMIP-II with eotaxin-1, RANTES, and MCP-3 suggests a unique mechanism for CCR3 activation. Biochemistry. 2000;39:12837–44. doi: 10.1021/bi001166f. [DOI] [PubMed] [Google Scholar]
- 9.Liwang AC, Wang ZX, Sun Y, Peiper SC, Liwang PJ. The solution structure of the anti-HIV chemokine vMIP-II. Protein Sci. 1999;8:2270–80. doi: 10.1110/ps.8.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodi PJ, Garrett DS, Kuszewski J, Tsang ML, Weatherbee JA, Leonard WJ, Gronenborn AM, Clore GM. High-resolution solution structure of the beta chemokine hMIP-1 beta by multidimensional NMR. Science. 1994;263:1762–7. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 11.Chung CW, Cooke RM, Proudfoot AE, Wells TN. The three-dimensional solution structure of RANTES. Biochemistry. 1995;34:9307–14. doi: 10.1021/bi00029a005. [DOI] [PubMed] [Google Scholar]
- 12.Kobilka B. Adrenergic receptors as models for G protein-coupled receptors. Annu Rev Neurosci. 1992;15:87–114. doi: 10.1146/annurev.ne.15.030192.000511. [DOI] [PubMed] [Google Scholar]
- 13.Strader CD, Fong TM, Tota MR, Underwood D. Structure and function of G protein-coupled receptors. Annu Rev Biochem. 1994;63:101–32. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 14.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 15.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–48. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 16.Clapham PR, Blanc D, Weiss RA. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by Simian immunodeficiency virus. Virology. 1991;181:703–15. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–8. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio PD, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–73. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–7. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Clements GJ, Proce-Jones MJ, Stephens PE, Sutton C, Schulz TF, Clapham PR, McKeating JA, McClure MO, Thomson S, Marsh M, Kay J, Weiss RA, Moore HP. The V3 loops of the HIV-1 and HIV-2 surface glycoproteins contain proteolytic cleavage sites: a possible function in viral fusion? AIDS Res Hum Retroviruses. 1991;7:3–16. doi: 10.1089/aid.1991.7.3. [DOI] [PubMed] [Google Scholar]
- 22.Gershoni JM, Denisova G, Raviv D, Smorodinsky NI, Buyaner D. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J. 1993;7:1185–7. doi: 10.1096/fasebj.7.12.7690724. [DOI] [PubMed] [Google Scholar]
- 23.Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–93. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapham CK, Ouyang J, Chandrasekhar B, Nguyen NY, Dimitrov DS, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120. Science. 1996;274:602–5. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 25.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–7. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;6605:179–83. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 27.Cheng-Mayer C, Seto D, Tateno M, Levy JA. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–2. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 28.Schellekens PT, Tersmette M, Roos MT, Keet RP, de Wolf F, Coutinho RA, Miedema F. Biphasic rate of CD4+ cell count decline during progression to AIDS correlates with HIV-1 phenotype. AIDS. 1992;6:665–9. doi: 10.1097/00002030-199207000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Tersmette M, Lange JM, de Goede RE, de Wolf F, Eeftink-Schattenkerk JK, Schellekens PT, Coutinho RA, Huisman JG, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–5. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 30.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–44. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 32.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–8. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y, Kottmann A, Kuroda M, Taniuchi I, Littman D. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–9. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronsoni RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–53. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–4. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 36.Boshoff C, Endo Y, Collins PD, Takeuchi Y, Reeves JD, Schweickart VL, Siani MA, Sasaki T, Williams TJ, Gray PW, Moore PS, Chang Y, Weiss RA. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–4. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 37.Kledal TN, Rosenkilde MM, Coulin F, Simmons G, Johnsen AH, Alouani S, Power CA, Luttichau HR, Gerstoft J, Clapham PR, Clark-Lewis I, Wells TNC, Schwartz TW. A broad-spectrum chemokine antagonist encoded by Kaposi’s sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 38.Luo Z, Fan X, Zhou N, Hiraoka M, Luo J, Kaji H, Huang Z. Structure–function study and anti-HIV activity of synthetic peptide analogues derived from viral chemokine vMIP-II. Biochemistry. 2000;39:13545–50. doi: 10.1021/bi000633q. [DOI] [PubMed] [Google Scholar]
- 39.Zhou N, Luo Z, Luo J, Fan X, Cayabyab M, Hiraoka M, Liu D, Han X, Pesavento J, Dong CZ, Wang Y, An J, Kaji H, Sodroski JG, Huang Z. Exploring the stereochemistry of CXCR4-peptide recognition and inhibiting HIV-1 entry with D-peptides derived from chemokines. J Biol Chem. 2002;277:17476–85. doi: 10.1074/jbc.M202063200. [DOI] [PubMed] [Google Scholar]
- 40.Picard L, Wilkinson DA, McKnight A, Gray PW, Hoxie JA, Clapham PR, Weiss RA. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–11. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 41.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–51. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Z, Berson JF, Chen Y, Turner JD, Zhang T, Sharron M, Jenks MH, Wang Z, Kim J, Rucker J, Hoxie JA, Peiper SC, Doms RW. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–31. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang ZX, Berson JF, Zhang TY, Cen YH, Sun Y, Sharron M, Lu ZH, Peiper SC. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 44.Chabot DJ, Zhang PF, Quinnan GV, Broder CC. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J Virol. 1999;73:6598–609. doi: 10.1128/jvi.73.8.6598-6609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doranz B, Orsini M, Turner J, Hoffman T, Berson J, Hoxie J, Peiper S, Brass L, Doms R. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–61. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chabot DJ, Broder CC. Substitutions in a homologous region of extracellular loop 2 of CXCR4 and CCR5 alter coreceptor activities for HIV-1 membrane fusion and virus entry. J Biol Chem. 2000;275:23774–782. doi: 10.1074/jbc.M003438200. [DOI] [PubMed] [Google Scholar]
- 47.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–44. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 48.Kajumo F, Thompson DA, Guo Y, Dragic T. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology. 2000;271:240–7. doi: 10.1006/viro.2000.0308. [DOI] [PubMed] [Google Scholar]
- 49.Zhou N, Luo Z, Luo J, Liu D, Hall JW, Pomerantz RJ, Huang Z. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J Biol Chem. 2001;276:42826–33. doi: 10.1074/jbc.M106582200. [DOI] [PubMed] [Google Scholar]
- 50.Tian S, Choi WT, Liu D, Pesavento J, Wang Y, An J, Sodroski JG, Huang Z. Distinct functional sites for human immunodeficiency virus type 1 and stromal cell-derived factor 1a on CXCR4 transmembrane helical domains. J Virol. 2005;79:12667–73. doi: 10.1128/JVI.79.20.12667-12673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rucker J, Samson M, Doranz BJ, Libert F, Berson JF, Yi Y, Smyth RJ, Collman RG, Broder CC, Vassart G, Doms RW, Parmentier M. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–46. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 52.Atchison RE, Gosling J, Monteclaro FS, Franci C, Digilio L, Charo IF, Goldsmith MA. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–6. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 53.Bieniasz PD, Fridell RA, Aramori I, Ferguson SS, Caron MG, Cullen BR. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alkhatib G, Ahuja SS, Light D, Mummidi S, Berger EA, Ahuja SK. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structure determinants. J Biol Chem. 1997;272:19771–6. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 55.Doranz BJ, Grovit-Ferbas K, Sharron MP, Mao SH, Goetz MB, Daar ES, Doms RW, O’Brien WA. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siciliano SJ, Rollins TE, DeMartino J, Konteatis Z, Malkowitz L, Riper GV, Bondy S, Rosen H, Springer MS. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc Natl Acad Sci USA. 1994;91:1214–8. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta SK, Pillarisetti K, Thomas RA, Aiyar N. Pharmacological evidence for complex and multiple site interaction of CXCR4 with SDF-1alpha: implications for development of selective CXCR4 antagonists. Immunol Lett. 2001;78:29–34. doi: 10.1016/s0165-2478(01)00228-0. [DOI] [PubMed] [Google Scholar]
- 58.Heveker N, Montes M, Germeroth L, Amara A, Trautmann A, Alizon M, Schneider-Mergener J. Dissociation of the signaling and antiviral properties of SDF-1-derived small peptides. Curr Biol. 1998;8:369–76. doi: 10.1016/s0960-9822(98)70155-1. [DOI] [PubMed] [Google Scholar]
- 59.Wells TNC, Power CA, Lusti-Narasimhan M, Hoogewert AJ, Cooke RM, Chung C-w, Peitsch MC, Proudfoot AEI. Selectivity and antagonism of chemokine receptors. J Leuk Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 60.Gosling J, Monteclaro FS, Atchison RE, Arai H, Tsou C-l, Goldsmith MA, Charo IF. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–6. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Picard L, Simmons G, Power CA, Meyer A, Weiss RA, Clapham PR. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–11. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349:41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 63.Saita Y, Kodama E, Orita M, Kondo M, Miyazaki T, Sudo K, Kajiwara K, Matsuoka M, Shimizu Y. Structural basis for the interaction of CCR5 with a small molecule, functionally selective CCR5 agonist. J Immunol. 2006;177:3116–22. doi: 10.4049/jimmunol.177.5.3116. [DOI] [PubMed] [Google Scholar]
- 64.Blanpain C, Doranz BJ, Bondue A, Govaerts C, De Leener A, Vassart G, Doms RW, Proudfoot A, Parmentier M. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J Biol Chem. 2003;278:5179–87. doi: 10.1074/jbc.M205684200. [DOI] [PubMed] [Google Scholar]
- 65.Castonguay LA, Weng Y, Adolfsen W, Di Salvo J, Kilburn R, Caldwell CG, Daugherty BL, Finke PE, Hale JJ, Lynch CL, Mills SG, MacCoss M, Springer MS, DeMartino JA. Binding of 2-aryl-4-(piperidin-1-yl)butanamines and 1,3,4-trisubstituted pyrrolidines to human CCR5: a molecular modeling-guided mutagenesis study of the binding pocket. Biochemistry. 2003;42:1544–50. doi: 10.1021/bi026639s. [DOI] [PubMed] [Google Scholar]
- 66.Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci USA. 2000;97:5639–44. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–13. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 68.Zhou N, Luo Z, Hall JW, Luo J, Han X, Huang Z. Molecular modeling and site-directed mutagenesis of CCR5 reveal residues critical for chemokine binding and signal transduction. Eur J Immunol. 2000;30:164–73. doi: 10.1002/1521-4141(200001)30:1<164::AID-IMMU164>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 69.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–41. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 70.Bondue A, Jao SC, Blanpain C, Parmentier M, LiWang PJ. Characterization of the role of the N-loop of MIP-1 beta in CCR5 binding. Biochemistry. 2002;41:13548–55. doi: 10.1021/bi026087d. [DOI] [PubMed] [Google Scholar]
- 71.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–4. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar S, Choi WT, Dong CZ, Madani N, Tian S, Liu D, Wang Y, Pesavento J, Wang J, Fan X, Yuan J, Fritzsche WR, An J, Sodroski JG, Richman DD, Huang Z. SMM-chemokines: a class of unnatural synthetic molecules as chemical probes of chemical receptor biology and leads for therapeutic development. Chem Biol. 2006;13:69–79. doi: 10.1016/j.chembiol.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 2003;100:1885–90. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, Sakmar TP, Moore JP, Maddon PJ. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–85. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howard OM, Shirakawa AK, Turpin JA, Maynard A, Tobin GJ, Carrington M, Oppenheim JJ, Dean M. Naturally occurring CCR5 extracellular and transmembrane domain variants affect HIV-1 co-receptor and ligand binding function. J Biol Chem. 1999;274:16228–34. doi: 10.1074/jbc.274.23.16228. [DOI] [PubMed] [Google Scholar]
- 76.Rabut GE, Konner JA, Kajumo F, Moore JP, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:3464–8. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laurence JS, Blanpain C, Burgner JW, Parmentier M, LiWang PJ. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry. 2000;39:3401–9. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- 78.Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J Virol. 2003;77:5201–8. doi: 10.1128/JVI.77.9.5201-5208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Govaerts C, Blanpain C, Deupi X, Ballet S, Ballesteros JA, Wodak SJ, Vassart G, Pardo L, Parmentier M. The TXP motif in the second transmembrane helix of CCR5. A structural determinant of chemokine-induced activation. J Biol Chem. 2001;276:13217–25. doi: 10.1074/jbc.M011670200. [DOI] [PubMed] [Google Scholar]
- 80.Gaertner H, Cerini F, Escola JM, Kuenzi G, Melotti A, Offord R, Rossitto-Borlat I, Nedellec R, Salkowitz J, Gorochov G, Mosier D, Hartley O. Highly potent, fully recombinant anti-HIV chemokines: reengineering a low-cost microbicide. Proc Natl Acad Sci USA. 2008;105:17706–11. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amara A, Gall SL, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J-L, Arenzana-Seisdedos F. HIV coreceptor downregulation as antiviral principle: SDF-1a-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–46. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schols D, Struyf S, van Damme J, Este JA, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–8. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–93. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo Z, Zhou N, Luo J, Hall JW, Huang Z. The role of positively charged residues in CXCR4 recognition probed with synthetic peptides. Biochem Biophy Res Comm. 1999;263:691–5. doi: 10.1006/bbrc.1999.1441. [DOI] [PubMed] [Google Scholar]
- 85.Luo J, Luo Z, Zhou N, Hall JW, Huang Z. Attachment of C-terminus of SDF-1 enhances the biological activity of its N-terminal peptide. Biochem Biophys Res Commun. 1999;264:42–7. doi: 10.1006/bbrc.1999.1476. [DOI] [PubMed] [Google Scholar]
- 86.Huang Z. Structural chemistry and therapeutic intervention of protein-protein interaction in immune response, HIV entry and apoptosis. Pharmacol Therapeut. 2000;86:201–15. doi: 10.1016/s0163-7258(00)00052-8. [DOI] [PubMed] [Google Scholar]
- 87.Zhou N, Luo Z, Luo J, Hall JW, Huang Z. A novel peptide antagonist of CXCR4 derived from the N-terminus of the viral chemokine vMIP-II. Biochemistry. 2000;39:3782–7. doi: 10.1021/bi992750v. [DOI] [PubMed] [Google Scholar]
- 88.Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, Moore JP. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–7. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 89.Fenard D, Lambeau G, Maurin T, Lefebvre JC, Doglio A. A peptide derived from bee venom-secreted phospholipase A2 inhibits replication of T-cell tropic HIV-1 strains via interaction with the CXCR4 chemokine receptor. Mol Pharmacol. 2001;60:341–7. doi: 10.1124/mol.60.2.341. [DOI] [PubMed] [Google Scholar]
- 90.Sachpatzidis A, Benton BK, Manfredi JP, Wang H, Hamilton A, Dohlman HG, Lolis E. Identification of allosteric peptide agonists of CXCR4. J Biol Chem. 2003;278:896–907. doi: 10.1074/jbc.M204667200. [DOI] [PubMed] [Google Scholar]
- 91.Loetscher P, Gong JH, Dewald B, Baggiolini M, Clark-Lewis I. N-terminal peptides of stromal cell-derived factor-1 with CXC chemokine receptor 4 agonist and antagonist activities. J Biol Chem. 1998;273:22279–83. doi: 10.1074/jbc.273.35.22279. [DOI] [PubMed] [Google Scholar]
- 92.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 93.O’Brien WA, Sumner-Smith M, Mao SH, Sadeghi S, Zhao JQ, Chen IS. Anti-human immunodeficiency virus type 1 activity of an oligocationic compound mediated via gp120 V3 interactions. J Virol. 1996;70:2825–31. doi: 10.1128/jvi.70.5.2825-2831.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sumner-Smith M, Zheng Y, Zhang YP, Twist EM, Climie SC. Antiherpetic activities of N-alpha-acetyl-nona-D-arginine amide acetate. Drugs Exp Clin Res. 1995;21:1–6. [PubMed] [Google Scholar]
- 95.Tamamura H, Omagari A, Hiramatsu K, Gotoh K, Kanamoto T, Xu Y, Kodama E, Matsuoka M, Hattori T, Yamamoto N, Nakashima H, Otaka A, Fujii N. Development of specific CXCR4 inhibitors possessing high selectivity indexes as well as complete stability in serum based on an anti-HIV peptide T140. Bioorg Med Chem Lett. 2001;11:1897–902. doi: 10.1016/s0960-894x(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 96.Fujii N, Oishi S, Hiramatsu K, Araki T, Ueda S, Tamamura H, Otaka A, Kusano S, Terakubo S, Nakashima H, Broach JA, Trent JO, Wang ZX, Peiper SC. Molecular-size reduction of a potent CXCR4-chemokine antagonist using orthogonal combination of conformation- and sequence-based libraries. Angew Chem Int Ed Engl. 2003;42:3251–3. doi: 10.1002/anie.200351024. [DOI] [PubMed] [Google Scholar]
- 97.Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T. An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 replication. Proc Natl Acad Sci USA. 1997;94:3548–53. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Doranz BJ, Filion LG, Diaz-Mitoma F, Sitar DS, Sahai J, Baribaud F, Orsinis MJ, Benovic JL, Cameron W, Doms RW. Safe use of the CXCR4 inhibitor ALX40-4C in humans. AIDS Res Hum Retroviruses. 2001;17:475–86. doi: 10.1089/08892220151126508. [DOI] [PubMed] [Google Scholar]
- 99.LaBarge MA, Blau HM. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 100.Arenzana-Seisdedos F, Virelizier JL, Rousset D, Clark-Lewis I, Loetscher P, Moser B, Baggiolini M. HIV blocked by chemokine antagonist. Nature. 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 101.Simmons G, Clapham PR, Picard L, Offord RE, Rosenkilde MM, Schwartz T, Buser R, Wells TN, Proudfoot AE. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–9. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 102.Nardese V, Longhi R, Polo S, Sironi F, Arcelloni C, Paroni R, DeSantis C, Sarmientos P, Rizzi M, Bolognesi M, Pavone V, Lusso P. Structural determinants of CCR5 recognition and HIV-1 blockade in RANTES. Nat Struct Biol. 2001;8:611–5. doi: 10.1038/89653. [DOI] [PubMed] [Google Scholar]
- 103.Fano A, Ritchie DW, Carrieri A. Modeling the structural basis of human CCR5 chemokine receptor function: from homology model building and molecular dynamics validation to agonist and antagonist docking. J Chem Inf Model. 2006;46:1223–35. doi: 10.1021/ci050490k. [DOI] [PubMed] [Google Scholar]
- 104.Carrieri A, Pérez-Nueno VI, Fano A, Pistone C, Ritchie DW, Teixidó J. Biological profiling of anti-HIV agents and insight into CCR5 antagonist binding using in silico techniques. Chem Med Chem. 2009;4:1153–63. doi: 10.1002/cmdc.200900101. [DOI] [PubMed] [Google Scholar]
- 105.Agrawal L, VanHorn-Ali Z, Berger EA, Alkhatib G. Specific inhibition of HIV-1 coreceptor activity by synthetic peptides corresponding to the predicted extracellular loops of CCR5. Blood. 2004;103:1211–7. doi: 10.1182/blood-2003-08-2669. [DOI] [PubMed] [Google Scholar]
- 106.Hartley O, Gaertner H, Wilken J, Thompson D, Fish R, Ramos A, Pastore C, Dufour B, Cerini F, Melotti A, Heveker N, Picard L, Alizon M, Mosier D, Kent S, Offord R. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proc Natl Acad Sci USA. 2004;101:16460–5. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gilliam BL, Riedel DJ, Redfield RR. Clinical use of CCR5 inhibitors in HIV and beyond. J Transl Med. 2011;9(Suppl 1):pS9. doi: 10.1186/1479-5876-9-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jacobson JM, Saag MS, Thompson MA, Fischl MA, Liporace R, Reichman RC, Redfield RR, Fichtenbaum CJ, Zingman BS, Patel MC, Murga JD, Pemrick SM, D’Ambrosio P, Michael M, Kroger H, Ly H, Rotshteyn Y, Buice R, Morris SA, Stavola JJ, Maddon PJ, Kremer AB, Olson WC. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. J Infect Dis. 2008;198:1345–52. doi: 10.1086/592169. [DOI] [PubMed] [Google Scholar]
- 109.Lalezari J, Yadavalli GK, Para M, Richmond G, Dejesus E, Brown SJ, Cai W, Chen C, Zhong J, Novello LA, Lederman MM, Subramanian GM. Safety, pharmacokinetics, and antiviral activity of HGS004, a novel fully human IgG4 monoclonal antibody against CCR5, in HIV-1-infected patients. J Infect Dis. 2008;197:721–7. doi: 10.1086/527327. [DOI] [PubMed] [Google Scholar]
- 110.Choi WT, Tian S, Dong CZ, Kumar S, Liu D, Madani N, An J, Sodroski JG, Huang Z. Unique ligand binding sites on CXCR4 probed by a chemical biology approach: implications for the design of selective human immunodeficiency virus type 1 inhibitors. J Virol. 2005;79:15398–404. doi: 10.1128/JVI.79.24.15398-15404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu D, Madani N, Li Y, Cao R, Choi WT, Kumar S, Dong C, Russell JD, Lefebure CR, An J, Wilson S, Gao YG, Pallansch LA, Sodroski JG, Huang Z. Crystal structure and structural mechanism of a novel anti-human immunodeficiency virus and D-amino acid-containing chemokine. J Virol. 2007;81:11489–98. doi: 10.1128/JVI.02845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choi WT, Kaul M, Kumar S, Wang J, Kumar IM, Dong CZ, An J, Lipton SA, Huang Z. Neuronal apoptotic signaling pathways probed and intervened by synthetically and modularly modified (SMM) chemokines. J Biol Chem. 2007;282:7154–63. doi: 10.1074/jbc.M611599200. [DOI] [PubMed] [Google Scholar]
- 113.Kaul M, Lipton SA. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci USA. 1999;96:8212–6. doi: 10.1073/pnas.96.14.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaul M, Medders KE, Desai MK, Lipton SA. Genetic double knockout of CXCR4 and CCR5 chemokine receptors prevents neuronal cell death induced by HIV-1 gp120 upstream of p38 mitogen-activated protein kinase. J Neurovirol. 2004;10:113. [Google Scholar]


