Abstract
MARVEL domain-containing 1 (MARVELD1) is a newly identified nuclear protein; however its function has not been clear until now. Here, we report that mouse MARVELD1 (mMARVELD1), which is highly conserved between mice and humans, exhibits cell cycle-dependent cellular localization. In NIH3T3 cells, MARVELD1 was observed in the nucleus and at the perinuclear region during interphase, but was localized at the mitotic spindle and midbody at metaphase, and a significant fraction of mMARVELD1 translocated to the plasma membrane during anaphase. In addition, treatment of cells with colchicine, a microtubuledepolymerizing agent, resulted in translocation of mMARVELD1 to the plasma membrane, and association of mMARVELD1 and α-tubulin was confirmed by co-immunoprecipitation. Finally, overexpression of mMARVELD1 resulted in a remarkable inhibition of cell proliferation, G1- phase arrest, and reduced cell migration. These findings indicate that mMARVELD1 is a microtubule-associated protein that plays an important role in cell cycle progression and migration.
Keywords: cell migration, cell-cycle progression, G1-phase arrest, MARVELD1, microtubule-associated protein
INTRODUCTION
It is accepted that the cytoskeleton plays a crucial role in a wide variety of cell activities, including cell division, migration and intracellular transport. Multiple proteins associated with the microtubule cytoskeleton are essential for microtubule assembly and its biological function. Among these, microtubule-associated proteins such as NuMA (Lydersen and Pettijohn, 1980; Yang et al., 1992), TPX2 (Wittmann et al., 2000), NuSAP (Raemaekers et al., 2003), INMAP (Shen et al., 2009), and p53 (Giannakakou et al., 2000; Morris et al., 2000) have been found to exhibit cell cycle-dependent localization, residing in the nucleus at interphase but associating with the spindle during mitosis. These proteins shuttle between the nucleus and the microtubule networks to regulate cell cycle progression, cell motility and tumorigenesis (D’Assoro et al., 2002; Lydersen and Pettijohn, 1980; Raemaekers et al., 2003; Shen et al., 2009; Shinmura et al., 2007; Wittmann et al., 2000; Yang et al., 1992). The intracellular translocation of such proteins at different stages of the cell cycle is necessary for their function. This is the case for p53; its multiple critical functions are tightly regulated by its specific subcellular localization in the nucleus and microtubule networks. Thus, cytoplasmic p53 associated with the microtubule cytoskeleton regulates centrosome duplication and cell cycle progression, while upon activation, it is imported into the nucleus to act as a transcription factor (O’Brate and Giannakakou, 2003; Shinmura et al., 2007).
We previously identified MARVELD1 as a nuclear protein that was downregulated in multiple primary tumors (Wang et al., 2009a). Consistent with our findings, mouse MARVELD1 expression was reported to be low in breast cancer and acute myeloid leukemia (Kirstetter et al., 2008; Landis et al., 2005). Interestingly, MARVELD1 was shown to interact with α-tubulin, importin β1 and HSP70 in a pull-down assay using GSTMARVELD1 as bait (Wang et al., 2009b), and both importin β1 and HSP70 moved between the nucleus and microtubules in a cell cycle-dependent manner (Kahana and Cleveland, 2001; Liang and MacRae, 1997; Nachury et al., 2001). In addition, α- tubulin modulates microtubule dynamics and function by interacting with different proteins at different stages of the cell cycle (Downing, 2000). However, the biological significance of the association of MARVELD1 with these proteins remains to be clarified.
Here, we report the cloning and characterization of mouse MARVELD1 (mMARVELD1) in NIH3T3 cells. First, mMARVELD1 exhibits cell cycle-dependent localization; it is nuclear and perinuclear at interphase, but associates with the spindle and plasma membrane during mitosis. Second, overexpression of mMARVELD1 in NIH3T3 cells inhibits cell proliferation by inducing G1-phase arrest and a remarkable reduction in chemotactic motility. We therefore hypothesized that mMARVELD1 is a candidate microtubule-associated protein that plays an important role in cell cycle progression and migration.
MATERIALS AND METHODS
Plasmid construction
The nucleotide sequence of mMARVELD1 (NM_183195.2) was obtained from NCBI (http://www.ncbi.nlm.nih.gov). Total RNA was extracted from NIH3T3 cells with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To obtain a cDNA fragment containing the ORF of mMARVELD1, mRNA was reverse-transcribed (ThermoScript™ RT-PCR system, Invitrogen) with random hexamer primers (reaction conditions: 25℃ for 10 min, 37℃ for 120 min, 85℃ for 5 min) followed by PCR using a specific pair of primers (sense primer: 5′-CGC GGA TCC ATG CTC CCG CCG CCC CC-3′, antisense primer: 5′-CCG CTC GAG CAC ACC ACC TCC TCC TTG-3′). The full length ORF (552 bp) was amplified under the following conditions: 95℃ for 5 min; 98℃ for 10 s, 65℃ for 3 min, 26 cycles; 72℃ for 7 min. After purification, the product fragments were subcloned into a pcDNA3.1/V5-His-TOPO vector, pcDNA3.1/ flag vector, pEGFP-N1 vector or pGEX-6P-1 vector (Invitrogen). Sequences were confirmed to be free of mutations.
Cell culture and transfection
NIH3T3 cells (ATCC, USA) were cultured in DMEM medium supplemented with 10% FBS, 2 mM L-glutamine and 100 U/ml penicillin/streptomycin at 37℃ in 5% CO2. For cell growth analysis, NIH3T3 cells were stably transfected with pcDNA3.1/ mMARVELD1-V5 or pEGFP-N1-mMARVELD1 constructs using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. Stably transfected clones were obtained after a 14- day selection in DMEM complete medium supplemented with 600 μg/ml G418 (Invitrogen). For the subcellular localization assays, NIH3T3 cells were transiently transfected with pEGFPN1- mMARVELD1, pcDNA3.1/Flag-mMARVELD1 or pcDNA3.1/ mMARVELD1-V5 constructs.
Treatment
To observe the translocation of mMARVELD1 into the nucleus, NIH3T3 cells expressing mMARVELD1-EGFP or mMARVELD1- V5 were treated with 2.5 μM PMA (phorbol-12-myristate- 13-acetate; Invitrogen) for 1 h, and control cells were treated with an equal amount of DMSO, followed by confocal microscopy analysis. To confirm the translocation of mMARVELD1 to the plasma membrane, NIH3T3 cells expressing mMARVELD1- EGFP were stimulated with 1 mg/ml colchicine for 12 h (or untreated as a control) before plasma membrane separation and analysis by confocal microscopy.
Tissue extraction of mouse muscle and brain
Tissue lysates were extracted from the muscle and brain of mice (strain B6; Beijing Laboratory Animal Research Center) at P0. Briefly, liquid nitrogen-frozen tissues were ground thoroughly, followed by homogenization in RIPA buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 50 mM Tris pH 8.0) with protease inhibitors (0.5 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin).
Co-immunoprecipitation
For co-immunoprecipitation, control cells or NIH3T3 cells expressing mMARVELD1-EGFP or mMARVELD1-V5 were lysed in RIPA buffer with protease inhibitors as described above, and the cleared lysates were incubated with antibodies against GFP or V5 overnight at 4℃, followed by the addition of 20 μl protein A-Sepharose slurry (GE Healthcare) and incubation for another 3 h with gentle agitation. The beads were then washed four times with IP buffer (20 mM HEPES pH 7.4, 0.5 mM EDTA, 150 mM NaCl and 0.1% Triton X-100). The resulting precipitated protein was analyzed by Western blot.
GST pull-down assay
The GST-mMARVELD1 construct was transformed into E. coli BL21(DE3) strain. The GST fusion protein was purified from E. coli following induction with 0.1 mM IPTG as described previously (Wang et al., 2009a). NIH3T3 cells were lysed and solubilized on ice in cold lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM DTT) with protease inhibitors. The cleared lysate was incubated with glutathione- Sepharose 4B beads (GE Health) linked to a GST-mMARVELD1 fusion protein or GST protein as a control. After 2 h incubation at 4℃, the samples were washed with ice-cold lysis buffer. The samples were then boiled in 5× Laemmli buffer, separated by SDS-PAGE and analyzed by Western blot.
Western blot analysis
For Western blot, protein samples were boiled in 2× Laemmli buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.004% bromophenol blue, 125 mM Tris-HCl, pH 6.8), separated by SDS-PAGE, and transferred onto a PVDF membrane. The membranes were incubated with primary antibody for 1 h at room temperature, washed three times with PBS buffer containing 0.1% Tween 20, and incubated with secondary antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology). ECL (Amersham Biosciences) was used to visualize the immunoblot signals.
Plasma membrane separation of NIH3T3/mMARVELD1- EGFP cells was performed according to the method of Bordier (Bordier, 1981). Antibodies were purchased from Invitrogen (anti-α-tubulin, anti-V5, anti-GST antibody), Santa Cruz (anti- GFP, anti-GAPDH antibody), and BD Transduction Laboratories (anti-Flag, anti-Flotillin antibody). Anti-MARVELD1 polyclonal antibodies were developed in our lab (Wang et al., 2009a).
Immunofluorescence
Immunostaining experiments were performed as described previously (Wang et al., 2009a). Briefly, NIH3T3 cells expressing mMARVELD1 fusion proteins were immunostained, followed by observation by confocal microscopy. The mMARVELD1- EGFP fusion protein was visualized by autofluorescence confocal microscopy. For wound-healing before immunostaining, NIH3T3 cells overexpressing mMARVELD1-EGFP were grown on glass coverslips until confluence, and the monolayer was scratched with a white pipette tip followed by a 6 h wound-healing period. DNA was stained with DAPI and F-actin was stained with Phalloidin-Alexa594 (Invitrogen). The secondary antibody used was TRITC-conjugated goat anti-mouse (Santa Cruz Biotechnology). Localization images of mMARVELD1 were processed using a Zeiss LSM 510 META confocal microscope with a 63× oil immersion objective (NA, 1.4).
Cell proliferation and cell cycle analysis
Cell proliferation was assessed by MTT assay. Briefly, NIH3T3 cells expressing mMARVELD1-EGFP or control vector and untransfected cells were seeded into 96-well plates at a concentration of 2.5 × 103 cells per well, each with eight replicas. Every 2 days, MTT solution was added to the wells at a final concentration of 1 mg/ml, followed by incubation at 37℃ for 4 h. The formazan product was then dissolved in DMSO and the absorbance of the solution was measured at 570 nm with a multimode microplate reader (Infinite 200 NanoQuant, TECAN). Cells were treated and measured every 2 days for 7 days.
For cell cycle analysis, 3 × 106 NIH3T3/mMARVELD1-EGFP cells and NIH3T3/mock cells were harvested and fixed in 70% ethanol overnight at -20℃. The fixed cells were stained with 50 μg/ml propidium iodide (PI) for 30 min at room temperature, followed by flow cytometry analysis (FACSCalibur™, BD Biosciences).
Transwell migration assay
Transwell migration was performed using a Millicell culture plate with an 8.0 μm PET insert (Millipore Corporation, USA). In brief, the cells were serum-starved overnight and 5 × 104 cells were plated into the upper chamber in 200 μl DMEM without serum. The chambers were placed into 24-well plates, each well containing 500 μl of DMEM supplemented with 10% fetal calf serum. After incubation for 4 h at 37℃, the cells that had traversed the membrane were fixed and visualized by crystal violet staining. The cells from five random fields in each transwell chamber were counted using a phase contrast microscope.
Statistical analysis
All values were expressed as the mean ± SD, and statistical analyses were carried out using Student’s t-test. Differences were considered to be statistically significant when p < 0.05.
RESULTS
Identification and characterization of mMARVELD1
The mMARVELD1 gene (GenBank: NM_183195.2) is located on mouse chromosome 19C3 and a mRNA of 3060 bp in length is transcribed. Bioinformatics analysis revealed that mMARVELD1 mRNA contains a 522 bp ORF region encoding a 173 amino acid protein with an expected molecular weight of 19.09. Membrane topology analysis with TMpred (http://www.ch.embnet.org/software/) and WoLF PSORT (http://wolfpsort.org/) predicted that mMARVELD1 is comprised of four transmembrane domains, with the N- and C-terminal domains stretching into the cytosol and containing the primary structural components of the MARVEL domain (MAL and related proteins for vesicle traffic and membrane link) (Sánchez-Pulido et al., 2002). Four candidate PKC phosphorylation sites (Thr15, Ser19, Thr51, Thr120) were also predicted using http://cn.expasy.net/ and http://www.cbs.dtu.dk/.
BLAST search revealed that mMARVELD1 homologs exist in the genomes of vertebrates such as zebrafish, frog and various mammalian species, but not in lower eukaryotes such as fly, worm, and yeast, indicating that it is expressed in vertebrates only (Fig. 1A). Phylogenetic analysis suggested that it is highly conserved between human and mouse (88% identity). This raised the possibility that mMARVELD1 was structurally similar to its human counterpart and may mimic its functions.
Fig. 1. Identification and characterization of mMARVELD1. (A) MARVELD1 amino acid sequences in chimpanzee, human, rhesus monkey, mouse, cattle, zebrafish and frog were aligned by ClustalW http://www.ebi.ac.uk/Tools/clustalw2/ with default settings. The conserved amino acid residues are shaded in gray. The extent of conservation is indicated by the brightness of gray and labeled below according to ClustalW definitions: identical residues (*); conservative substitutions (:); semi-conservative substitutions (.). (B) The expression of mMARVELD1 in NIH3T3 cells and extracts of muscle and brain from mouse strain B6 at P0. mMARVELD1 protein was detected by Western blot with anti- MARVELD1 antibody developed by S. Wang et al. GAPDH was used as a loading control. The molecular weight of endogenous MARVELD1 is approximately 19.
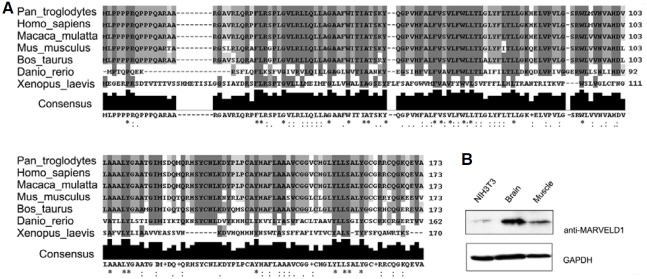
In a previous study, mMARVELD1 mRNA expression was examined in mouse tissues, the results of which will be published elsewhere. Expression of endogenous MARVELD1 protein was confirmed with rabbit polyclonal antibodies raised against a MARVELD1 peptide (Wang et al., 2009a). Exogenous mMARVELD1 protein was detected in neonatal mouse brain and muscle, as predicted by the mRNA expression analysis, but was weakly detected in NIH3T3 cells (Fig. 1B). Therefore, to explore the subcellular localization and functions of mMARVELD1, NIH3T3 cells were transfected with tagged mMARVELD1.
Cell cycle-dependent localization of mMARVELD1
Although human MARVELD1 was thought to localize to the nucleus at interphase, its possible association with α-tubulin suggested that MARVELD1 might have different subcellular localizations at different phases of the cell cycle (Wang et al., 2009a; 2009b). To explore the subcellular distribution of mMARVELD1, three different expression constructs, Flag-mMARVELD1, mMARVELD1-V5 and mMARVELD1-EGFP, were constructed and transiently expressed in NIH3T3 cells. The cellular localization of mMARVELD1 was observed by confocal microscopy 24 h after transfection, and the localization patterns of the three differentially tagged mMARVELD1 constructs are shown in Fig. 2A. At interphase, mMARVELD1 was found to localize to the nucleus and perinuclear region of NIH3T3 cells, which is identical to the subcellular localization of human MARVELD1. Treatment with 2.5 μM PMA, a PKC activator, resulted in further nuclear accumulation of both mMARVELD1-V5 and mMARVELD1-EGFP in NIH3T3 cells (Fig. 2B), suggesting that PKC may regulate the localization of mMARVELD1 through phosphorylation of the predicted PKC target sites in mMARVELD1. However, the predicted phosphorylation sites in mMARVELD1 that are critical for this regulation remain unknown. Interestingly, perinuclear mMARVELD1 showed polarity across the cell body, consistent with the distribution pattern of microtubules (Rosenblatt, 2005) and the microtubule-associated proteins NuSAP (Raemaekers et al., 2003) and NuMA (Lydersen and Pettijohn, 1980) in interphase cells.
Fig. 2. Subcellular localization and dynamic distribution of mMARVELD1 in NIH3T3 cells. (A) mMARVELD1 is localized to the nucleus and perinuclear region in NIH3T3 cells at interphase. NIH3T3 cells were transfected with constructs expressing Flag-mMARVELD1, mMARVELD1- V5 or mMARVELD1-EGFP. The distribution of mMARVELD1 was analyzed by immunofluorescence with anti-Flag or anti-V5 antibody, or by autofluorescence, with confocal microscopy. (B) PMA induces the nuclear translocation of mMARVELD1. Both mMARVELD-V5 and mMARVELD1- EGFP translocated into the nucleus from the perinuclear region upon stimulation with 2.5 μM PMA for 1 h. (C) The localization of mMARVELD1-GFP at different mitotic stages was detected by confocal microscopy. mMARVELD1-EGFP closely colocalizes with the mitotic spindle (white arrows), the cytokinetic midbody (hollow arrow) and the plasma membrane as mitosis progresses.
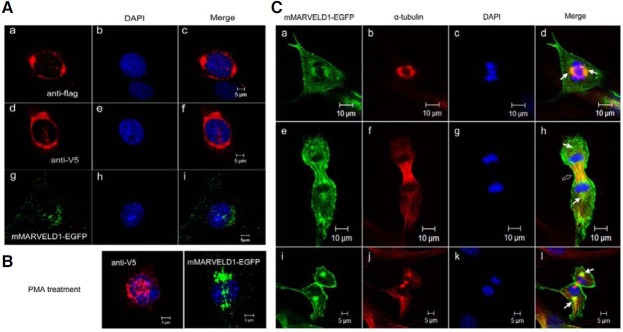
The pattern of mMARVELD1 localization prompted further investigation of a possible interaction between mMARVELD1 and microtubule networks during cell division. NIH3T3 cells stably expressing mMARVELD1-EGFP were immunostained with anti-α-tubulin antibody, and colocalization of microtubules and mMARVELD1-EGFP was observed by confocal microscopy. Although mMARVELD1-EGFP localized in the nucleus and perinuclear region during interphase (Fig. 2A), it undergoes a cell cycle-dependent localization shift. At metaphase, mMARVELD1 colocalized with the spindles (Figs. 2C, a-d), and as the cell cycle progressed, a large fraction of mMARVELD1 associated with microtubules in the midbody bundles at early anaphase (Figs. 2C, e-h). At telophase, with the disassembly of the mitotic spindle, the majority of the fraction of mMARVELD1 that was localized to the microtubules became dissociated, but the fraction of mMARVELD1 at spindle poles remained. By contrast, membrane-bound mMARVELD1 increased significantly (Figs. 2C, i-l). The diverse subcellular localizations of mMARVELD1 suggest that mMARVELD1 may have multiple functions or may exert its function at different sites. As the distribution of mMARVELD1 largely overlapped with the microtubule networks, especially during mitosis, mMARVELD1 may function as a regulator of cell division.
mMARVELD1 interacts directly with α-tubulin
To confirm the interaction between mMARVELD1 and microtubules, NIH3T3 cells expressing recombinant V5-tagged or EGFP-tagged mMARVELD1 were lysed, and whole cell extracts were subjected to immunoprecipitation with anti-V5 or anti-GFP antibody followed by immunoblot analysis. The results showed that both V5- and EGFP-tagged mMARVELD1, but not the mock controls, immunoprecipitated α-tubulin (Fig. 3A). This result suggests that mMARVELD1 associates with microtubules through α-tubulin.
Fig. 3. mMARVELD1 interacts directly with α- tubulin. (A) NIH3T3 cells expressing mMARVELD1- V5 or mMARVELD1-EGFP were lysed, and the cleared cell lysates were immunoprecipitated with monoclonal antibody against V5 or GFP, respectively, and subsequently probed with an antibody against α-tubulin. The results show that α-tubulin co-precipitates with mMARVELD1. (B) Western blot analyses of the GST-pull-down assay. GST and GST-mMARVELD1 fusion proteins immobilized to glutathione-Sepharose 4B beads were incubated with cleared NIH3T3 cell lysate. GST or GST-mMARVELD1 with adsorbed proteins was analyzed by Western blot. Upper panel, detection of α-tubulin; lower panel, detection of GST or GST-mMARVELD1 fusion protein.
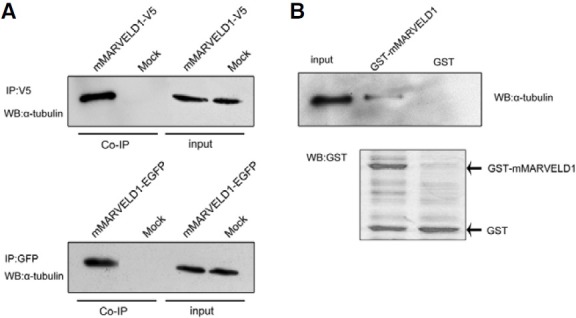
To determine whether the mMARVELD1 and α-tubulin interaction is direct or indirect, an in vitro GST-pull-down assay was performed with purified GST-mMARVELD1 fusion protein. As shown in Fig. 3B, α-tubulin was evident following cell extraction using GST-mMARVELD fusion protein as bait, but not with GST protein control, although the signal was not as strong as from the input control. This result provides strong evidence that mMARVELD1 associates with microtubules by direct interaction with α-tubulin.
Microtubule depolymerization results in translocation of mMARVELD1 to the plasma membrane
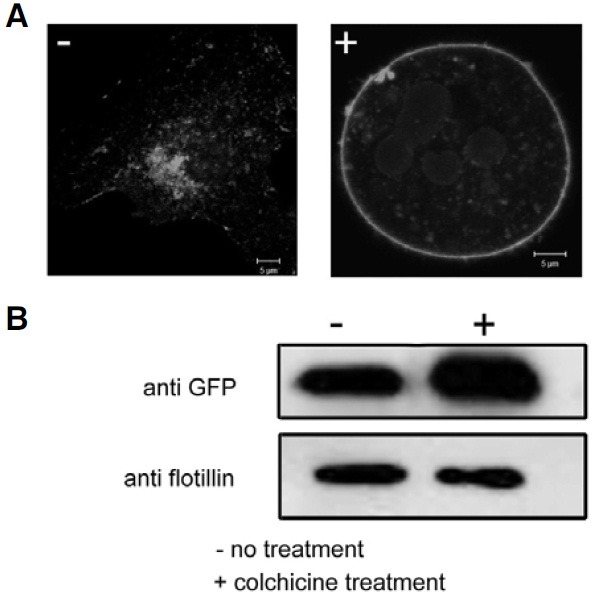
To understand the role of microtubule networks in the distribution of mMARVELD1 during mitosis, NIH3T3 cells stably expressing mMARVELD1-EGFP were treated with colchicine, a microtubule-depolymerizing agent. Surprisingly, treatment with colchicine resulted in the loss of perinuclear and microtubule localization of mMARVELD1 (Fig. 4). Instead, the majority of mMARVELD1-EGFP appeared to have translocated to the plasma membrane. To confirm this, the plasma membrane was isolated from colchicines-treated or control cells, and membrane- bound mMARVELD1 was analyzed by Western blot. As seen in Fig. 4, colchicine treatment increased the amount of mMARVELD1-EGFP in the membrane fraction. These results indicate that microtubule depolymerization prevents the association between microtubules and mMARVELD1, inducing translocation of mMARVELD1 to the plasma membrane. Furthermore it confirms that mMARVELD1 not only colocalizes with, but also directly associates with microtubules in cells, suggesting that it is a microtubule-associated protein. However, the functional role of microtubule-associated mMARVELD1 needs to be clarified.
Fig. 4. Translocation of mMARVELD1 to the plasma membrane after treatment with colchicine. (A) NIH3T3 cells expressing mMARVELD1- EGFP were treated with 1 mg/ml colchicine (right panel). 12 h after treatment, the cells were observed by confocal microscopy. (B) The plasma membrane fraction from colchicine-treated or control NIH3T3 cells expressing mMARVELD1-EGFP was extracted, and the amount of mMARVELD1-EGFP in the plasma membrane fraction was detected by Western blot using anti-GFP antibody. The membrane protein flotillin was used as plasma membrane marker.
mMARVELD1 inhibits cell proliferation
As MARVELD1 is downregulated in multiple human cancers, it may play a role as a negative regulator of cell proliferation and migration. Because mMARVELD1 colocalized with the midbody, a subpopulation of microtubules present during mitosis (Fig. 2C, h), its effect on cell division and proliferation was investigated. NIH3T3 cells transfected with mMARVELD1-EGFP were cultured and subjected to the MTT proliferation assay. The results showed that the growth of NIH3T3 cells expressing mMARVELD1 was markedly reduced compared to control cells (Fig. 5A). To further characterize the inhibitory effect of mMARVELD1 on cell growth, the expression levels of proliferating cell nuclear antigen Ki-67 and cyclin D1 were analyzed. As shown in Figure 5B, a remarkable decrease in the expression of Ki-67 and cyclin D1 is observed in NIH3T3 cells overexpressing mMARVELD1-EGFP. The same result was observed in cells overexpressing mMARVELD1-V5 (data not shown).
Fig. 5. Overexpression of mMARVELD1 inhibits NIH3T3 cell proliferation. (A) NIH3T3 cells expressing mMARVELD1-EGFP or control vector and untransfected cells were seeded into 96-well plates. Cell proliferation was measured by MTT assay at days 0, 2, 4, and 6. (B) Cell lysates from untransfected NIH3T3 cells, NIH3T3/mock and NIH3T3/ mMARVELD1-EGFP transfected cells were analyzed by Western blot using antibodies as shown. (C) NIH3T3 cells stably transfected with mMARVELD1- EGFP or control vector were grown to subconfluency and stained with PI followed by cell cycle analysis with flow cytometry. The experiment was repeated three times. The values are expressed as the mean ± SD.
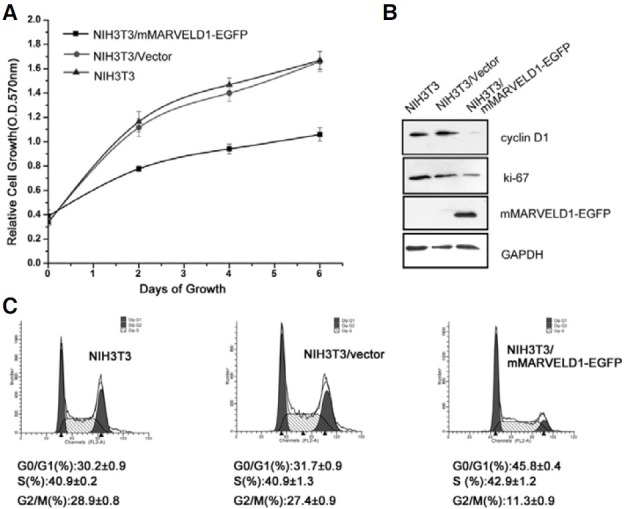
Because cyclin D1, the most important cell cycle progression activator, was downregulated by overexpression of mMARVELD1, it was possible that mMARVELD1 reduced cell growth by inhibiting cell cycle progression. To explore this possibility, flow cytometry was performed to analyze cell cycle characteristics of cells overexpressing mMARVELD1. As shown in Fig. 5C, the expression of mMARVELD1-EGFP in NIH3T3 cells was associated with a significant decrease in the G2/M phase population and a remarkable increase in the G0/G1 population compared to control cells. These results indicate that mMARVELD1 may inhibit cell proliferation by promoting G1-phase arrest.
Overexpressed mMARVELD1 localizes at the leading edge of migrating NIH3T3 cells and inhibits cell migration
When the distribution of mMARVELD1 during cell wound healing was investigated, overexpressed mMARVELD1-EGFP notably localized to the cell junction and the leading edge along the actin cortex at the front lines of directionally migrating NIH3T3 cells (Fig. 6A). This observation is similar to that observed with occludin, which is also a MARVEL domain-containing protein and was found to regulate the directional migration of epithelial cells (Du, 2010).
Fig. 6. Overexpressed mMARVELD1 localizes at the leading edge of migrating NIH3T3 cells and inhibits cell migration. (A) Localization of overexpressed mMARVELD1 in NIH3T3 cells during wound healing. Arrows indicate leading edge localization of mMARVELD1-EGFP, and arrowheads indicate cell junction localization of mMARVELD1-EGFP. (B) NIH3T3 cells expressing mMARVELD1- EGFP or control vector and untransfected cells were used for analysis of chemotactic motility in response to a serum gradient in a Transwell migration assay. Cells that traversed the membrane were visualized by crystal violet staining. Migrated cells in the Millicell plate were counted and statistically analyzed. Results are the average of three independent experiments (P < 0.05).
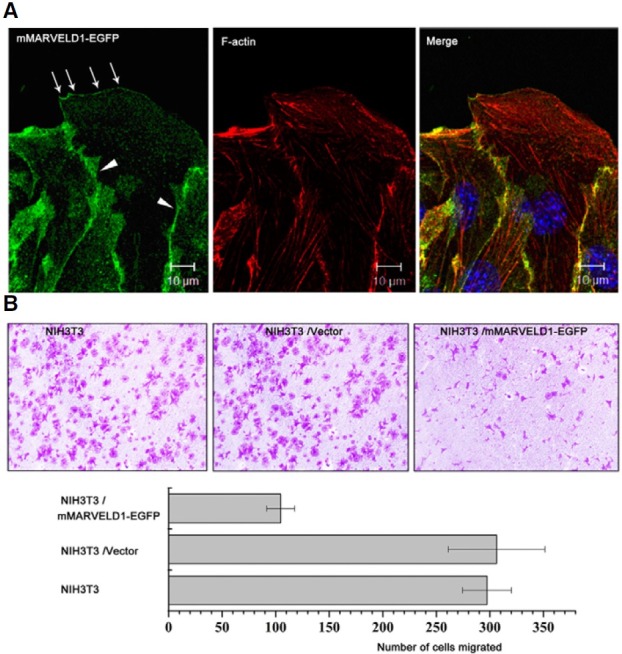
Since mMARVELD1 is associated with α-tubulin and localized to the leading edge of migrating cells, the effect of mMARVELD1 on cell motility was examined using a Transwell migration assay. The results in Fig. 6B show that NIH3T3 cells overexpressing mMARVELD1-EGFP migrated almost 3-fold slower than control cells along a serum gradient, suggesting that mMARVELD1 may negatively regulate cell migration. The negative effects of mMARVELD1 overexpression on cell proliferation and migration support the possibility that mMARVELD1 acts as a tumor-related factor that inhibits cell transformation. The physiological roles of mMARVELD1 in vivo should be clarified in future studies.
DISCUSSION
MARVELD1 was first identified as a tumor-related protein, but its cellular function had not been well characterized (Wang et al., 2009a). Here, we report the characterization of mMARVELD1 as a candidate microtubule-associated protein that exhibits cell cycle-dependent localization and can inhibit cell proliferation and migration. The localization pattern of mMARVELD1 is similar to that of some MAPs, including NuMA (Lydersen and Pettijohn, 1980; Yang et al., 1992), TPX2 (Wittmann et al., 2000), NuSAP (Raemaekers et al., 2003), and INMAP (Shen et al., 2009). Like these MAPs, mMARVELD1 localizes to the nucleus during interphase, but associates closely with the mitotic spindle poles and the midbody during mitosis. In addition, the finding that mMARVELD1 interacts directly with α-tubulin, a component of the mitotic spindle and midbody, further supports this association.
The cell cycle-dependent subcellular localization of mMARVELD1 to the nucleus and spindles is of significant interest, since both the nucleus and the mitotic apparatus are crucial for cell division and other events involved in tumorigenesis. Although the mechanisms involved in the regulation of this localization are still unknown, widely studied tumor suppressors such as p53 and BRCA1 have been found to exhibit a localization pattern similar to mMARVELD1, residing at the centrosomes and the spindle during mitosis, but localized to the nucleus during interphase (Hsu and White, 1998; Shinmura et al., 2007). The translocation of MAPs from the cytoplasm to the nucleus is regulated by the small GTPase Ran and the protein importin (Dasso, 2001; Jason and Don, 2001). As mMARVELD1 was predicted to contain four PKC phosphorylation sites, and treatment of mMARVELD1-expressing cells with the PKC activator PMA could promote its nuclear translocation, we hypothesized that mMARVELD1 is a substrate of PKC and that its nuclear translocation is regulated by PKC phosphorylation. On the other hand, MARVELD1 was also found to associate with importin β1, the nuclear transport protein (Wang et al., 2009a). Therefore, mMARVELD1 may associate with MAPs like NuMA and TPX2a in regulation of localization. However, details regarding the regulation of its cellular distribution await further study.
When the association between microtubules and mMARVELD1 is disrupted, mMARVELD1 translocates to the plasma membrane (Fig. 4). In addition, overexpressed mMARVELD1 localizes at the leading edge of migrating cells (Fig. 6A). These two findings suggest that an essential link may exist between mMARVELD1 and membrane proteins. Like mMARVELD1, several proteins, such as TRPC1 (Bollimuntha et al., 2005) and dysferlin (Azakir et al., 2010), were recently reported to associate with both α-tubulin and the plasma membrane during multiple processes. However, the molecular details of these associations remain unknown. Microtubules, dynamically organized by α-tubulin proteins, are important structures in mitosis, cellular motility, intracellular transport, and cell morphology. In this regard, it is of interest that our study identified mMARVELD1 as a microtubule associated protein, since mMARVELD1 was found to translocate to the plasma membrane when microtubules were destabilized. We propose that mMARVELD1 might interact with signal-transduction pathways connecting the plasma membrane and microtubules.
When mMARVELD1 is overexpressed, it inhibits cell proliferation through G1-phase cell cycle arrest. This effect of mMARVELD1 correlates with the inhibition of cyclin D1 accumulation. Cyclin D1 allows cells to pass through the cell cycle restriction point and enter S phase. Several studies have described the essential function of microtubules, as well as microtubule- associated proteins, in regulation of the G1-S transition (Doxsey, 2001; Khodjakov and Rieder, 2001; Sebastian et al., 2005). Unfortunately, the molecular details of the pathways involved in this regulation remain unclear. A better understanding of this pathway is necessary, as the interaction of MARVELD1 with microtubules may explain why cells over-expressing mMARVELD1 arrest in G1 rather than other phases of mitosis.
Finally, mMARVELD1 inhibits chemotactic cell migration. It is accepted that stabilization of microtubule plus ends towards the membrane is necessary for cell migration during the searchand- capture process (Mimori-Kiyosue and Tsukita, 2003). Further work will be required to elucidate the molecular mechanism by which mMARVELD1 reduces cell mobility.
In conclusion, mMARVELD1 has been characterized as a candidate microtubule-associated protein with cell cycle-dependent subcellular localization. Studies on mMARVELD1, the highly conserved mouse ortholog of human MARVELD1, will provide important insights into the role of human MARVELD1 in regulating proliferation and migration. Further studies are in progress to elucidate the detailed mechanism by which mMARVELD1 shuttles in and out of the nucleus, and regulates cell cycle-progression and cell migration.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant number: 30170516 and 30871271).
References
- 1.Azakir B.A., Di Fulvio S., Therrien C., Sinnreich M. Dysferlin interacts with tubulin and microtubules in mouse skeletal muscle. PLoS One. (2010);5:e10122. doi: 10.1371/journal.pone.0010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollimuntha S., Cornatzer E., Singh B.B. Plasma membrane localization and function of TRPC1 is dependent on its interaction with beta-tubulin in retinal epithelium cells. Vis. Neurosci. (2005);22:163–170. doi: 10.1017/S0952523805222058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. (1981);256:1604–1607. [PubMed] [Google Scholar]
- 4.Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell. (2001);104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 5.D’Assoro A.B., Lingle W.L., Salisbury J.L. Centrosome amplification and the development of cancer. Oncogene. (2002);21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 6.Downing K.H. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu. Rev. Cell Dev. Biol. (2000);16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 7.Doxsey S.J. Centrosomes as command centres for cellular control. Nat. Cell Biol. (2001);3:E105–E108. doi: 10.1038/35074618. [DOI] [PubMed] [Google Scholar]
- 8.Du D., Xu F., Yu L., Zhang C., Lu X., Yuan H., Huang Q., Zhang F., Bao H., Jia L., et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev. Cell. (2010);18:52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Giannakakou P., Sackett D.L., Ward Y., Webster K.R., Blagosklonny M.V., Fojo T. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. (2000);2:709–717. doi: 10.1038/35036335. [DOI] [PubMed] [Google Scholar]
- 10.Hsu L.C., White R.L. BRCA1 is associated with the cen-trosome during mitosis. Proc. Natl. Acad. Sci. USA. (1998);95:12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jason A.K., Don W.C. Cell cycle: Some importin news about spindle assembly. Science. (2001);291:1718–1719. doi: 10.1126/science.1059765. [DOI] [PubMed] [Google Scholar]
- 12.Kahana J.A., Cleveland D.W. Cell cycle: Some importin news about spindle assembly. Science. (2001);291:1718–1719. doi: 10.1126/science.1059765. [DOI] [PubMed] [Google Scholar]
- 13.Khodjakov A., Rieder C.L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. (2001);153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirstetter P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T.A., Pabst T., et al. Modeling of C/EBP alpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. (2008);13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Landis M.D., Seachrist D.D., Montañez-Wiscovich M.E., Danielpour D., Keri R.A. Gene expression profiling of cancer progression reveals intrinsic regulation of transforming growth factor-beta signaling in ErbB2/Neu-induced tumors from transgenic mice. Oncogene. (2005);24:5173–5190. doi: 10.1038/sj.onc.1208712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang P., MacRae T.H. Molecular chaperones and the cytoskeleton. J. Cell Sci. (1997);110:1431–1440. doi: 10.1242/jcs.110.13.1431. [DOI] [PubMed] [Google Scholar]
- 17.Lydersen B.K., Pettijohn D.E. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. (1980);22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 18.Mimori-Kiyosue Y., Tsukita S. “Search-and-capture” of microtubules through plus-end-binding proteins (+TIPs). J. Biochem. (2003);134:321–326. doi: 10.1093/jb/mvg148. [DOI] [PubMed] [Google Scholar]
- 19.Morris V.B., Brammall J., Noble J., Reddel R. p53 localizes to the centrosomes and spindles of mitotic cells in the embryonic chick epiblast, human cell lines, and a human primary culture: an immunofluorescence study. Exp. Cell Res. (2000);256:122–130. doi: 10.1006/excr.2000.4800. [DOI] [PubMed] [Google Scholar]
- 20.Nachury M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. (2001);104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 21.O’Brate A., Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist. Updat. (2003);6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Patzke S., Hauge H., Sioud M., Finne E.F., Sivertsen E.A., Delabie J., Stokke T., Aasheim H.C. Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene. (2005);24:1159–1173. doi: 10.1038/sj.onc.1208267. [DOI] [PubMed] [Google Scholar]
- 23.Raemaekers T., Ribbeck K., Beaudouin J., Annaert W., Van C.M., Stockmans I., Smets N., Bouillon R., Ellenberg J., Car-meliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. (2003);162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenblatt J. Spindle assembly: asters part their separate ways. Nat. Cell Biol. (2005);7:219–222. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- 25.Sánchez-Pulido L., Martín-Belmonte F., Valencia A., Alonso M.A. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. (2002);27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- 26.Shen E., Lei Y., Liu Q., Zheng Y., Song C., Marc J., Wang Y., Sun L., Liang Q. Identification and characterization of INMAP, a novel interphase nucleus and mitotic apparatus protein that is involved in spindle formation and cell cycle progression. Exp. Cell Res. (2009);315:1100–1116. doi: 10.1016/j.yexcr.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Shinmura K., Bennett R.A., Tarapore P., Fukasawa K. Direct evidence for the role of centrosomally localized p53 in the regulation of centrosome duplication. Oncogene. (2007);26:2939–2944. doi: 10.1038/sj.onc.1210085. [DOI] [PubMed] [Google Scholar]
- 28.Wang S., Li Y., Han F., Hu J., Yue L., Yu Y., Zhang Y., He J., Zheng H., Shi S., et al. Identification and characterization of MARVELD1, a novel nuclear protein that is downregulated in multiple cancers and silenced by DNA methylation. Cancer Lett. (2009a);282:77–86. doi: 10.1016/j.canlet.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Wang S., Xiao S., He J., Han F., Li Y. Identification of interaction between MARVELD1 and Importinβ1. Chin. J. Biochem. Mol. Biol. (2009b);25:735–739. (In Chinese) [Google Scholar]
- 30.Wittmann T., Wilm M., Karsenti E., Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. (2000);149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C.H., Lambie E.J., Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J. Cell Biol. (1992);116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]


