Abstract
Tumor suppressor p53 plays a central role in preventing tumor formation. The levels and activity of p53 is under tight regulation to ensure its proper function. Murine double minute 2 (MDM2), a p53 target gene, is an E3 ubiquitin ligase. MDM2 is a key negative regulator of p53 protein, and forms an auto-regulatory feedback loop with p53. MDM2 is an oncogene with both p53-dependent and p53-independent oncogenic activities, and often has increased expression levels in a variety of human cancers. MDM2 is highly regulated; the levels and function of MDM2 are regulated at the transcriptional, translational and post-translational levels. This review provides an overview of the regulation of MDM2. Dysregulation of MDM2 impacts significantly upon the p53 functions, and in turn the tumorigenesis. Considering the key role that MDM2 plays in human cancers, a better understanding of the regulation of MDM2 will help us to develop novel and more effective cancer therapeutic strategies to target MDM2 and activate p53 in cells.
Keywords: MDM2, E3 ubiquitin ligase, gene regulation, p53
Introduction
MDM2 (murine double minute 2) is an oncogene that was originally discovered in a locus amplified on double minute chromosomes in transformed mouse fibroblasts [1]. Over-expression of MDM2 can immortalize rodent primary fibroblasts and induce transformation in cultured cells [2]. MDM2 contains several conserved functional domains, which provide the structural basis for MDM2's oncogenic properties. The N-terminal p53-binding domain plays an essential role in binding to tumor suppressor p53 protein and inhibiting the transcriptional activity of p53. The central region contains the nuclear localization sequence (NLS) and the nuclear export signal (NES), which are responsible for the nucleo-cytoplasmic shuttling of MDM2 protein [3], and the acidic domain and an adjacent zinc finger which mediate the interaction of MDM2 with many ribosomal proteins (RPs) [4,5]. The C-terminus contains the RING finger domain [6–8]. The RING finger domain is responsible for the E3 ubiquitin ligase activity of MDM2, which recruits an ubiquitin-conjugating E2 enzyme to promote the ubiquitination and degradation of the target proteins [9]. MDM2 protein has several substrates. Among them, tumor suppressor p53 is a major substrate of MDM2.
p53 is considered as ‘the cellular gatekeeper’ or ‘the guardian of the genome’. p53 protein can be activated in response to a wide variety of stress signals. As a transcription factor, activated p53 transcriptionally regulates a group of p53 target genes which lead to various cellular responses, including apoptosis, cell cycle arrest, DNA repair, senescence, etc., to maintain the integrity of genome and prevent the tumor initiation and progression [10,11]. MDM2 negatively regulates p53 through multiple mechanisms. As an E3 ubiquitin ligase, MDM2 binds to p53 and ubiquitinates p53 for proteasomal degradation. MDM2 can mono- or poly-ubiquitinate p53 depending upon the levels of MDM2 activity. High levels of MDM2 activity promote poly-ubiquitination and degradation of p53, while low levels of MDM2 activity induce mono-ubiquitination and nuclear exportation of p53 [12]. MDM2 can promote p53 translocation from nucleus to cytoplasm via its NES sequence, which inhibits p53 function in the nucleus by removing it from its nuclear site and decreases the p53 protein levels by p53 degradation in cytoplasm [3]. MDM2 can also directly bind to the N-terminal transactivation domain of p53, which prevents the interaction of p53 with the basal transcriptional machinery, and in turn inhibits the transcriptional activity of p53 [13–15]. In addition, MDM2 negatively regulates p53 translation. Ribosomal protein RPL26 increases p53 translation through binding to the 5′-untranslated region (UTR) of p53 [16]. MDM2 can bind to and ubiquitinate RPL26 for proteasomal degradation, which in turn prevents RPL26-promoted p53 translation [17]. Studies of MDM2-deficient mice have clearly demonstrated the negative regulation of p53 by MDM2, which is indispensable at all stages of life [18]. MDM2 deficiency in mice results in lethality at the blastocyst stage due to excessive p53-dependent apoptosis. Importantly, this phenotype can be completely rescued by the deletion of p53 in mice [18].
MDM2 is classified as an oncogene due to its malignant behaviors in tumors. The over-expression of MDM2 has been observed in a wide variety of human tumors, including sarcoma, leukemia, breast carcinoma, melanoma, and glioblastoma [19]. As a critical negative regulator of p53, high expression levels of MDM2 decrease p53 protein levels and function, which lead to increased cancer risk and/or accelerated tumor formation and progression. In human tumors, the over-expression of MDM2 and p53 mutation are often mutually exclusive [19]. This observation strongly supports the notion that inactivation of p53 by MDM2 contributes to the effect of MDM2 on tumor development. Studies in mice and cultured cells have shown that MDM2 also has p53-independent oncogenic functions, which control proliferation, apoptosis, and tumor invasion and metastasis. For example, MDM2 transgenic mice with increased MDM2 expression levels in p53−/− background developed an altered tumor spectrum with an increased incidence of sarcomas [20]. In cells, MDM2 can interact with and inhibit the function of tumor suppressor retinoblastoma protein (RB) in regulation of cell cycle and proliferation [21]. Foxo3A, which regulates cell cycle, is a target of MDM2 for ubiquitination and proteasomal degradation [22,23]. E-cadherin, which plays a crucial role in cancer metastasis, is also a target of MDM2 [24]. MDM2 is a positive regulator of E2F-1, which plays an important role in cell cycle [25], and XIAP, an anti-apoptotic protein [26]. Therefore, MDM2 is a key player in human cancers and an important cancer therapeutic target.
The increased expression of MDM2 in human tumors is mainly caused by gene amplification. Increased transcription and/or enhanced translation of MDM2 also contribute to MDM2 over-expression in human tumors [19]. In addition, SNP309, a naturally occurring single nucleotide polymorphism (SNP) in the MDM2 gene (a G to T change in the regulatory region in intron 1), increases MDM2 mRNA expression levels, which correlates with increased risk for several human cancers [27]. In tumors, there is also over-expression of cancer-specific alternatively spliced MDM2 transcripts, which are often associated with advanced tumors and poor prognosis [28,29].
MDM2 is highly regulated at both mRNA and protein levels in cells. MDM2 is a transcriptional target of p53. p53 binds to p53 consensus DNA binding element in the first intron of MDM2 gene to transcriptionally induce the expression of MDM2, and forms an auto-regulatory negative feedback loop with MDM2 [30–32]. MDM2 expression levels are also transcriptionally regulated by various oncogenic and tumor suppressive pathways. In addition, many stress signals, including DNA damage, oncogenic activation, ribosomal biogenesis, and chronic stress, and microRNAs regulate MDM2 protein levels, activity, and cellular localization. Here, we will review the regulation of MDM2 by different stress signals and various oncogenic and tumor suppressive pathways.
Regulation of MDM2 at the Transcriptional Level
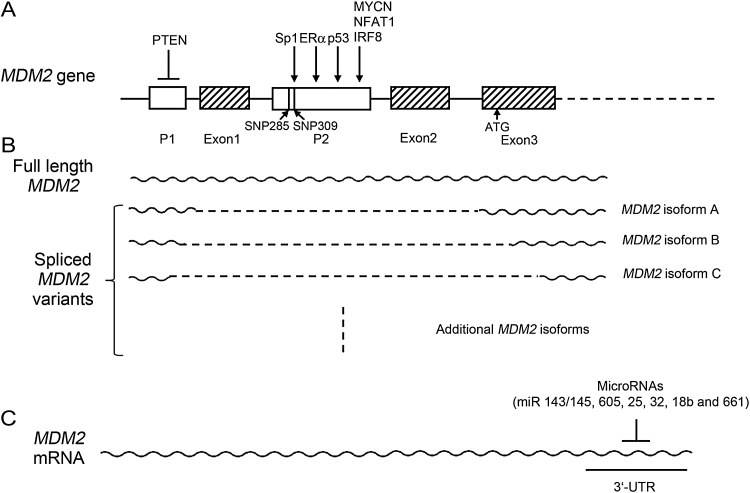
Transcription of the MDM2 gene is regulated by two distinct promoters (P1 and P2). The P1 promoter, which is located at the upstream of the first exon, controls the basal constitutive expression of MDM2 [33]. The P2 promoter, which is located in the first intron, is highly regulated and responsible for the inducible expression of MDM2 (Fig. 1A). Transcripts from both promoters encode identical full-length MDM2; however, they show difference in translation efficiency due to their different 5′-UTR. Transcript from the P1 promoter contains two upstream open reading frames and this mRNA is loaded with ribosomes inefficiently and has lower translation efficiency. In contrast, the 5′-UTR of P2-transcript allows efficient translation with the help of a 5′-UTR-specific RNA-binding protein, the La antigen [34]. Together, the P2 promoter can transcriptionally induce MDM2 expression, and the mRNA has increased translational efficiency. Activated p53, Ras, and estrogen receptor-α all can induce MDM2 transcription from the P2 promoter.
Figure 1.
The regulation of MDM2 at the transcriptional and translational levels (A) The MDM2 gene has two promoters (P1 and P2). The P1 promoter controls the basal constitutive expression of MDM2, and the P2 promoter is highly regulated and responsible for the inducible expression of MDM2. p53 binds to MDM2 P2 promoter to induce the expression of MDM2. In addition to p53, MDM2 can be transcriptionally regulated by several oncogenic and tumor suppressive pathways. (B) Increased expression of alternatively and aberrantly spliced MDM2 variants have been detected in many types of human cancers. Among over 40 MDM2 spliced variants identified in human cancers, MDM2 isoforms A, B, and C are the isoforms that most frequently over-expressed in human cancers. (C) A group of microRNAs bind to the 3′-UTR of the MDM2 mRNA to inhibit its translation.
p53–MDM2 negative feedback loop
p53 is a transcription factor that can be activated by diverse cellular stresses. In response to stress signals, activated p53 can transcriptionally induce MDM2 through binding to two adjacent p53-responsive elements located in the P2 promoter of the MDM2 gene [30,32] (Fig. 1A). The p53-mediated induction of MDM2 can be positively regulated by MDMX, a closely related MDM2 homolog. Similar to MDM2, MDMX binds to p53 transcriptional activation domain and inhibits the transcriptional activity of p53 [35]. Under DNA damage and ribosomal stress conditions, MDMX is required for optimal p53 binding to the MDM2 promoter and selectively increases p53-mediated induction of MDM2, which could be an additional mechanism by which MDMX down-regulates p53 [36,37]. The induction of MDM2 by p53 can also be negatively regulated by orphan receptor TR3, which mainly functions as a transcription factor. TR3 down-regulates p53 transcriptional induction of MDM2 through direct interaction of TR3 with p53 to block its acetylation [38].
As p53 and MDM2 regulate each other, they form an auto-regulatory negative feedback loop. One feature of this p53–MDM2 feedback loop is that MDM2 and p53 levels oscillate, especially in response to stress [39,40]. MDM2 protein levels decrease immediately in response to stress, such as gamma irradiation. The immediate decrease of MDM2 levels after stress results in an increase of p53 protein levels, which can then transcriptionally induce MDM2 expression, and in turn results in the decrease of p53 protein levels mediated by MDM2. The dynamic pattern of p53 and MDM2 varies depending upon the stress signals. For example, p53 shows a series of repeated pulses in response to double-stranded DNA breaks. Different dynamic behaviors are important signals, which lead to very different cellular responses and cell fate. A recent report showed that sustained p53 activation in cells preferentially induces p53 target genes that are involved in senescence, which is very different with the group of p53 target genes induced in cells with repeated p53 pulses [41]. In turn, cells have different fate: cells with repeated p53 pulses recover from DNA damage, while cells with sustained p53 activation often become senescent.
The importance of p53–MDM2 feedback loop has been further characterized in vivo in a recent study employing a genetically engineered mouse with a mutated p53-binding element in the P2 promoter of the MDM2 gene [42]. Mice that are deficient for p53–MDM2 feedback loop are viable. While p53 cannot induce MDM2 expression through the P2 promoter, constitutive MDM2 levels from the P1 promoter are sufficient for maintaining the proper p53 protein levels, including the degradation of p53 protein after stress. However, loss of P2-induced MDM2 expression enhances p53-dependent DNA damage response in these mice. Increased p53 activity, especially p53-dependent apoptosis in response to stress sensitizes hematopoietic stem cells (HSCs), which causes drastically increased lethality upon gamma irradiation in these mice [42]. These observations challenged the prevailing view of the p53–MDM2 feedback loop by providing in vivo evidence showing that constitutive MDM2 levels from the P1 promoter are sufficient for development, homeostasis, and longevity, while p53-induced MDM2 levels from the P2 promoter are essential for attenuation of p53 function, especially in the HSCs after DNA damage.
Transcriptional regulation of MDM2 by oncoproteins and tumor suppressors
In addition to p53, MDM2 can be transcriptionally regulated by several oncogenic and tumor suppressive pathways (Fig. 1A). For example, MDM2 has been shown to be a direct transcriptional target of MYCN oncogene. MYCN binds to a consensus E-box in MDM2 P2 promoter and transcriptionally induces MDM2 expression levels. In neuroblastoma cells with MYCN amplification, inhibition of MYCN decreases MDM2 levels, and in turn leads to the increase of p53 levels and function [43]. MDM2 can also be induced by the transcription factor of the human nuclear factor of activated T cells (NFAT) family, NFAT1 [44]. Dysregulation of the NFAT signaling promotes malignant transformation, and the over-expression of NFAT has been observed in human tumors. The MDM2 P2 promoter contains a consensus NFAT1-binding element. NFAT1 directly binds to this binding element and transcriptionally induces the expression levels of MDM2, which in turn reduces p53 function in response to stress signals. Both NFAT1 and MDM2 are over-expressed in human liver cancers, and there is a positive correlation between the expression levels of NFAT1 and MDM2 in human tumors [44]. This positive regulation of MDM2 by NFAT1 could contribute to the role of NFAT1 in tumor development and progression. IFN regulatory factor 8 (IRF8), a transcription factor, has been reported to induce the expression of MDM2 in germinal center (GC) B-cells. IRF8 binds to MDM2 P2 promoter to transcriptionally induce the MDM2 expression levels. In GC B-cells of IRF8-deficient mice, the MDM2 expression levels are greatly down-regulated [45]. The expression of MDM2 can be negatively regulated by PTEN. PTEN is a tumor suppressor with lipid phosphatase activity. PTEN down-regulates MDM2 P1 promoter activity through its lipid phosphatase activity; MDM2 transcripts from P1 promoter are up-regulated in cell lines deficient for PTEN [46]. Clearly, regulation of the mRNA expression levels of MDM2 could be an important mechanism by which oncogenes promote tumorigenesis while tumor suppressors prevent tumor development.
Polymorphisms in the MDM2 gene
The expression levels of MDM2 can be transcriptionally regulated by naturally occurring polymorphisms in the MDM2 gene (Fig. 1A). The G to T change in SNP309, a common SNP in the first intron of the MDM2 gene, enhances the binding affinity of transcriptional activator Sp1, and leads to the increased MDM2 transcriptional levels (increased by 2- to 4 fold) in humans, which in turn attenuate p53 function and activity [27]. The transcription of MDM2 from SNP309 G allele by Sp1 can be regulated by estrogen; estrogen preferentially enhances the Sp1 binding towards the MDM2 SNP309 G allele. The MDM2 expression levels in estrogen-responsive cells with the SNP309 G allele are significantly higher than that in cells with the SNP309 T allele [47]. SNP309 G allele has been shown to correlate with increased risk for several cancers and/or early onset age of cancers, often in a gender-specific manner; SNP309 G allele in females is often associated with the highest increased risk for cancers, including breast cancer [48,49]. MDM2 SNP309 is a common SNP with the frequencies differing among different ethnic backgrounds. The G allele is about 40% in Caucasian population and about 10% in African Americans. A recent study revealed a second rare functional SNP in the MDM2 promoter, SNP285 with a G to C change. SNP285 resides on the SNP309 G allele and only exists in Caucasian population with ∼8% allele frequency. The SNP285C/309G haplotype decreases the binding affinity of Sp1 towards MDM2 promoter, and is associated with reduced risk for cancers, including breast and ovarian cancers in Caucasian population [50]. Therefore, understanding the collection of functional SNPs in the MDM2 gene, which modulates the MDM2 expression levels and in turn affects the p53 function and the cancer risk, may help in cancer prevention as well as selection of a better treatment for cancer.
Alternative splicing variants of MDM2
In human tumors, in addition to the over-expression of full-length MDM2, there is often increased expression of alternatively and aberrantly spliced MDM2 variants. MDM2 spliced variants have been detected in high percentage of many types of cancers, including invasive breast cancer (∼30%), pediatric rhabdomyosarcoma (∼80%), and soft tissue sarcoma (∼50%) [51–53]. So far, over 40 MDM2 spliced variants have been identified in human tumors. Among these variants, MDM2 isoform A, isoform B, and isoform C, are frequently over-expressed in many types of human tumors (Fig. 1B). The mechanisms for the over-expression of MDM2 isoforms in tumors are not well understood. It has been reported that ultraviolet (UV) irradiation can induce the alternative splicing of the MDM2 gene, which results in the increased MDM2 isoform B levels in cultured human cells [54]. The over-expression of MDM2 spliced variants are often associated with advanced tumors and poor prognosis [29]. The over-expression of some MDM2 isoforms in transgenic mouse models promotes tumor formation [55]. However, the function of MDM2 isoforms is not well understood. An interesting feature of many MDM2 isoforms is that they lose the p53-binding domain, the NLS and NES, but retain the C-terminus. Therefore, most MDM2 isoforms cannot directly bind to p53 to regulate its levels and activity, while they still have the ability to interact with full-length MDM2. As full-length MDM2 negatively regulates p53 protein levels and activity, MDM2 isoforms can indirectly regulate p53. It has been reported that MDM2 isoforms can increase wild-type p53 levels in in vitro cultured cells [56,57]. However, p53 activation by MDM2 isoforms in cells cannot explain the promoting effect of MDM2 isoforms on tumorigenesis observed in vivo. p53 is the most frequently mutated gene in human tumors; ∼50% of human tumors contain the mutant p53. Mounting evidence suggests that many tumor-associated mutant p53 proteins gain new functions to promote tumorigenesis in addition to the loss of wild-type p53's function in tumor suppression, defined as gain of function (GOF) [58–60]. The results from our recent study suggest that MDM2 isoforms may regulate the levels and function of mutant p53 to promote tumorigenesis. We found that MDM2 isoform B, the MDM2 isoform most frequently over-expressed in human tumors, interacts with full-length MDM2 to inhibit MDM2-mediated mutant p53 degradation, which in turn promotes mutant p53 accumulation and GOF in tumorigenesis [61]. Furthermore, a MDM2 isoform similar to human MDM2 isoform in structure is over-expressed in majority of tumors of mice with knock-in of R172H mutant p53 (equivalent to human R175H), and correlated with mutant p53 accumulation in tumors. Consistently, this MDM2 isoform promotes mutant p53 accumulation and tumorigenesis [61]. In addition, MDM2 isoforms have p53-indpendent function in tumorigenesis since the promoting effect of MDM2 isoforms on tumorigenesis has also been observed in p53-deficient mice [54,55,62]. Future studies on the function of MDM2 isoforms and the mechanisms for their over-expression in tumors will further increase our understanding of their role in tumorigenesis.
Regulation of MDM2 at the Translational Levels
In addition to the transcriptional regulation of MDM2, enhanced translation of the MDM2 transcripts also increases MDM2 protein levels, which has been suggested to be an important mechanism for the over-expression of MDM2 in human tumors. As described in the above section, MDM2 transcripts from P1 or P2 promoter have different translation efficiency due to their differences in the 5′-UTR. Another important group of factors that regulate MDM2 translation is microRNAs which target for MDM2 (Fig. 1C). MicroRNAs are a group of small non-coding RNAs, which regulate the translation of gene products through binding to its partially complementary sites in the 3′-UTRs of target mRNAs, leading to translational repression of their target genes. Several microRNAs that target MDM2 have been reported, including miR-143/145, miR-605, miR-25, miR-32, miR-18b, and miR-661 [63–67]. Some of these microRNAs, including miR-143/145, miR-605, and miR-32 can be regulated by p53 at transcriptional or post-transcriptional levels. These microRNAs inhibit MDM2 translation, which in turn increases p53 protein levels and function. Many of these microRNAs show tumor suppressive properties, and often have decreased expression levels in many human tumors. For example, the decreased expression levels of miR-143/145 have been observed in many different cancers, including breast cancer, colon cancer, prostate cancer, bladder cancer, etc. [68]. The expression levels of miR-25 in human colon cancer tissues have been shown to be decreased compared with adjacent normal tissues [69]. The decreased expression levels of miR-18b have been observed in a panel of melanoma cell lines and primary melanoma cancers, mainly due to hyper-methylation [66]. Another recent study showed that miR-661 targets both MDM2 and MDMX to activate p53 in a cell-type-dependent manner [67]. The role of miR-661, either pro- or anti-tumorigenic, depends upon p53 status. While low expression levels of miR-661 correlate with poor survival probability in breast cancers with wild-type p53, high miR-661 expression levels correlate with aggressive malignant behaviors in various cancers harboring mutant p53 [67]. Therefore, these microRNAs join the MDM2-p53 feedback loop to down-regulate MDM2 levels and increase p53 protein levels and function.
Regulation of MDM2 Protein
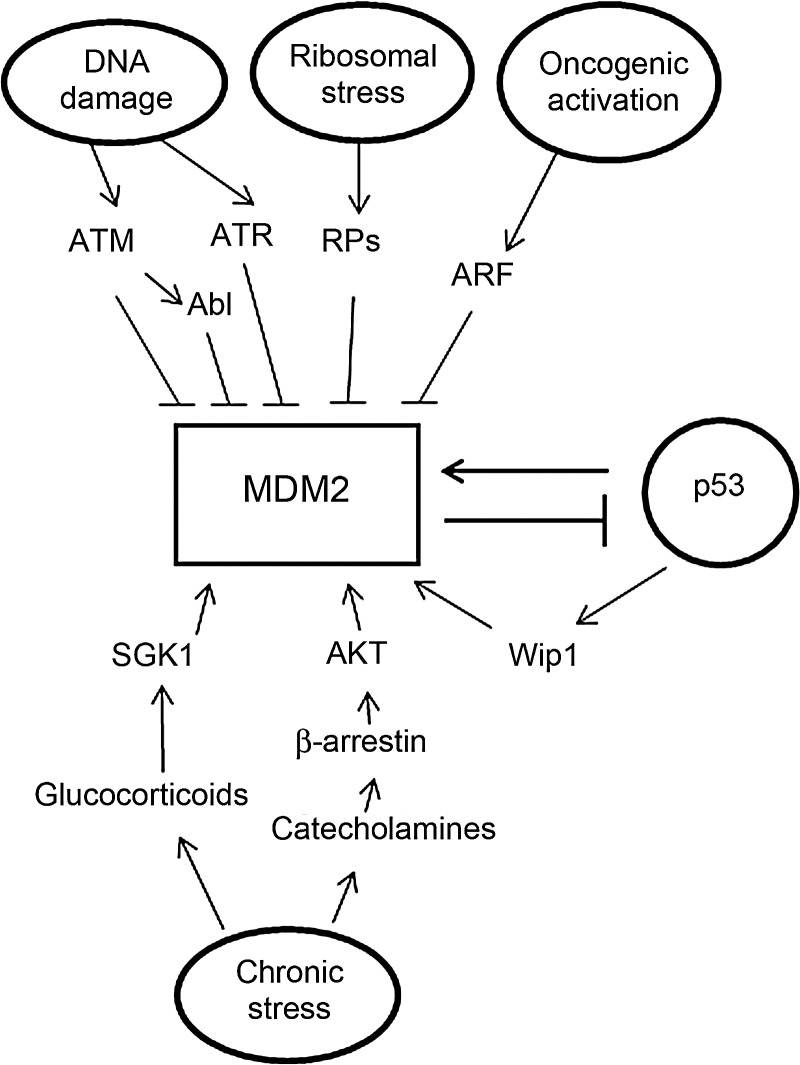
MDM2 protein levels and activity are highly regulated by many extracellular and intracellular stress signals, including genotoxic stress signals, oncogenic activation, ribosomal stress, and psychological stress signals. These stress signals function through distinct signaling pathways to regulate the ability of MDM2 to interact with p53, MDM2 E3 ubiquitin ligase activity, and the cellular localization of MDM2 (Fig. 2).
Figure 2.
The regulation of MDM2 protein levels and activity MDM2 protein levels and activity are highly regulated by many stress signals and factors. These stress signals regulate the ability of MDM2 to bind to p53, MDM2 E3 ubiquitin ligase activity, cellular localization of MDM2, and the stability of MDM2 protein. Genotoxic stress signals, such as IR and UV, negatively regulate MDM2 protein levels and activity mainly through post-translational modification (phosphorylation) of MDM2. Oncogenic activation negatively regulates MDM2 E3 ligase activity, and promotes MDM2 localization in nucleoli through ARF. Ribosomal stress signals negatively regulate MDM2 E3 ubiquitin ligase activity through interaction of RPs with MDM2. Some oncogenes, including AKT and Wip1, directly increase MDM2 activity. Chronic psychological stress signals increase the levels of neurohormones, including glucocorticoids and catecholamines, which can increase MDM2 activity through distinct pathways.
Genotoxic stress signals
Genotoxic stress signals such as ionizing radiation (IR) and UV irradiation could cause DNA damage that activates protein kinases (ATM for IR and ATR for UV). ATM and ATR phosphorylate MDM2 at Ser-395 and Ser-407, respectively [70–72]. The phosphorylation of MDM2 at these two positions blocks the ability of MDM2 to export p53 to cytoplasm [70]. MDM2 Ser-395 phosphorylation also reduces the E3 ligase activity of MDM2 [73], and promotes the interaction of MDM2 with p53 mRNA to enhance p53 protein synthesis [74,75]. ATM also regulates MDM2 indirectly through following mechanisms: first, ATM activates c-Abl, another kinase to phosphorylate MDM2 at multiple sites which in turn reduces the function of MDM2 to negatively regulate p53 [76]; secondly, ATM reduces the levels of herpesvirus-associated ubiquitin-specific protease (HAUSP), a specific deubiquitinase for MDM2 that prevents MDM2 from auto-ubiquitination. The decrease of HAUSP results in the increased MDM2 degradation through auto-ubiquitination [77]. All these regulations on MDM2 result in a rise of p53 protein levels and activity.
Oncogenic activation
Aberrant activation of a list of oncogenes, including E2F-1, beta-catenin, Myc, and Ras, has been shown to increase the levels of ARF, a tumor suppressor, to negatively regulate MDM2 function [78–80]. The down-regulation of MDM2 function in turn activates p53, which is essential for tumor suppression. ARF negatively regulates MDM2 through binding to the central domain of the MDM2 protein to promote MDM2 accumulation in the nucleoli. This interaction also down-regulates the E3 ubiquitin ligase activity of MDM2. In turn, MDM2 is segregated from p53 and the activity of MDM2 to negatively regulate p53 is decreased [81,82]. Deletion and silencing of ARF are the events often observed in cancers that disrupt the ARF-MDM2-p53 signaling for tumor surveillance [83]. In addition to ARF-mediated negative regulation of MDM2, results from a recent report show that E2F1 can directly inhibit MDM2 expression by suppressing its promoter activity in a p53-dependent manner [84].
There are oncogenes, including AKT, Wip1, that directly regulate the activity of MDM2. The IGF-1/AKT oncogenic pathway plays critical roles in the regulation of cell proliferation and survival. MDM2 is a substrate of AKT kinase; AKT directly phosphorylates MDM2 at Ser-166/186 [85]. MDM2 Ser-166/186 phosphorylation promotes MDM2 to localize in the nucleus, increases the interaction of MDM2 with p300, which promotes MDM2-mediated p53 degradation, and inhibits the interaction of MDM2 with ARF [86,87]. Altogether, the phosphorylation of MDM2 at Ser-166/186 by AKT increases MDM2 activity to down-regulate p53 protein levels and function. Wip1, a serine/threonine phosphatase with oncogenic activity, is often over-expressed in several human cancers. Wip1 can be transcriptionally regulated by p53 and is often induced by p53 at a later time point after genotoxic stress [88,89]. Wip1 can dephosphorylate MDM2 at Ser-395, the site which is often phosphorylated by ATM, thereby restoring the activity of MDM2 to negatively regulate p53 [89]. Therefore, Wip1 is an important regulator for the p53–MDM2 feedback loop.
Ribosomal stress
Ribosomal biogenesis is a coordinated cellular process that plays an essential role in a number of important cellular activities [90]. Perturbations of ribosomal biogenesis induce ribosomal stress, which can impact upon MDM2 activity and p53 function. Several RPs, including RPL5, RPL11, RPL23, RPL26, RPS3, RPS7, RPS14, and RPS27/L, have been shown to interact with MDM2 [91,92]. RPs mainly bind to the central region of MDM2. The RP–MDM2 interaction inhibits the E3 ubiquitin ligase activity of MDM2, which results in the accumulation and activation of p53 [93]. A possible mechanism by which the RP–MDM2 interaction at the MDM2 central region inhibits MDM2 E3 ubiquitin ligase activity is to reduce the flexibility of MDM2 protein and therefore inhibits its function [4]. While it is currently unclear why so many RPs bind to and regulate MDM2 in response to ribosomal stress, it is clear that the RP-MDM2-p53 pathway is important in monitoring the proper ribosomal biogenesis in cells.
Chronic psychological stress
Ample epidemiological evidence suggests that chronic psychological stress has significant negative impact upon the onset, progression, and mortality of human cancers [94–96]. The results from recent studies show that the neurohormones elevated during chronic stress, including glucocorticoids and catecholamines, can activate MDM2 to down-regulate p53 through distinct pathways [97,98]. Elevation of glucocorticoid hormones under chronic stress conditions activates glucocorticoids receptor pathway, which then transcriptionally induces SGK1 kinase. SGK1 phosphorylates MDM2 at Ser-166/186, the same sites phosphorylated by AKT, to activate MDM2 [97,99], which in turn down-regulates p53 function. Catecholamines, including epinephrine and norepinephrine, activate MDM2 through binding to one type of adrenergic receptor (AR), β2-AR, to activate PKA-β-arrestin pathway [98]. The activation of β-arrestin-1 promotes AKT-mediated activation of MDM2, as well as the interaction of MDM2 with p53. The activation of MDM2 by neurohormones to negatively regulate p53 under chronic stress could be an important mechanism by which chronic stress promotes tumorigenesis.
MDMX
In addition to the above stresses, the E3 ubiquitin ligase activity of MDM2 can be regulated by MDMX, a close homolog of MDM2. MDMX is another important negative regulator of p53. The importance of negative regulation of p53 by MDMX is demonstrated by the early embryonic lethality of MDMX-null mice which can be rescued by the loss of p53 [100]. Unlike MDM2, MDMX is not under transcriptional regulation of p53. Furthermore, MDMX has no intrinsic E3 ubiquitin ligase activity despite the high similarity of the RING domain between MDM2 and MDMX [101]. It has been shown that MDMX can bind to the transactivation domain of p53 and block the transcriptional activity of p53 [102]. Importantly, MDMX can modulate the levels and activity of MDM2, which in turn regulates p53 protein levels and function [103,104]. While both MDM proteins can form homodimers through their RING domains, heterodimers of MDMX and MDM2 are more stable than homodimers of either protein [105]. Hetero-dimers of MDMX and MDM2 preferentially reduce auto-ubiquitination of MDM2 and increase MDM2-mediated ubiquitination and degradation of p53 [103,106,107]. Consistently, the MDM2-binding defective mutant MDMX knock-in mice are embryonic lethal due to p53 activation [108], demonstrating the regulation of MDM2 by MDMX as an important mechanism for p53 regulation.
Conclusion
Oncoprotein MDM2 is a key negative regulator of p53; the main function of MDM2 is to degrade p53 protein and inhibit p53 activity. Many human tumors have high MDM2 protein levels, which could in turn inactivate p53 function. Therefore, targeting MDM2 is a valuable therapeutic strategy for cancer treatment. Several agents have been developed to target the protein–protein interaction of p53–MDM2 or the E3 ubiquitin ligase activity of MDM2 to increase p53 protein levels and activity. Small molecules, including Nutlins and MI-219, bind to MDM2 in its p53-binding pocket to disrupt p53–MDM2 interaction [109,110]. Another small molecule RITA binds to the p53 N-terminus to block p53–MDM2 interaction [111]. A recently identified small molecule RO-5963 is a dual inhibitor of p53–MDM2 and p53–MDMX interaction, which in turn leads to the activation of p53 [112]. Stapled peptides with improved stability have also been utilized to target p53–MDM2; stapled peptides SAH-p53s bind to MDM2 and block p53–MDM2 interaction [113]. Another group of agents targets MDM2 E3 ubiquitin ligase activity. For example, small molecule HLI98 binds to MDM2 C-terminus to compromise its E3 ubiquitin ligase activity [114]. Other small molecules that belong to this category include MPD compounds, MEL23 and MEL24 [115,116]. More studies in the future are needed to develop MDM2 inhibitors that can be used safely and effectively for cancer treatment in clinic. It is worth noting that MDM2 also retains the ability to interact with and regulate mutant p53 protein. For example, in transgenic mice with knock-in of tumor-associated hotspot p53 mutations (R172H or R270H, which equivalent to human R175H and R273H, respectively), mutant p53 protein levels are kept low in normal tissues and only accumulate in tumor tissues [117–119]. Loss of MDM2 in these mice clearly increases mutant p53 protein levels in normal tissues, and further promotes tumor development of these mice [118]. Since some tumor-associated mutant p53 proteins gain new activities in promoting tumorigenesis, targeting MDM2, especially its function in negative regulation of p53, in patients carrying mutant p53 in their tumors may cause unwanted oncogenic effects. It is therefore very important to determine the status of p53 of cancer patients before giving MDM2 inhibitors. While most studies on MDM2 focus on its regulation of p53, increasing evidence has shown that MDM2 also has p53-independent functions. It is unclear whether MDM2 inhibitors currently developed have any impact upon p53-independent functions of MDM2. Studies to further understand p53-independent functions of MDM2 are needed with the goal to target MDM2, especially its oncogenic function, in cancers.
Funding
This work was supported by the grants from National Institutes of Health (NIH) Grant (1R01CA160558-01), DOD (W81XWH-10-1-0435), CINJ new investigator award, and the Ellison Foundation.
References
- 1.Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. doi:10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 2.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. doi:10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. doi:10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. doi:10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddy MN, Freemont PS, Borden KL. The p53-associated protein MDM2 contains a newly characterized zinc-binding domain called the RING finger. Trends Biochem Sci. 1994;19:198–199. doi: 10.1016/0968-0004(94)90020-5. doi:10.1016/0968-0004(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 7.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. doi:10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 8.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. doi:10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. doi:10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. doi:10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 11.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. doi:10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. doi:10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. doi:10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. doi:10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 16.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. doi:10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 17.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. doi:10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. doi:10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 19.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. doi:10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci USA. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. doi:10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. doi:10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 22.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. doi:10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, Nawaz Z, et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284:13987–14000. doi: 10.1074/jbc.M901758200. doi:10.1074/jbc.M901758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, Broglio K, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26:7269–7282. doi: 10.1128/MCB.00172-06. doi:10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huart AS, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem. 2009;284:32384–32394. doi: 10.1074/jbc.M109.052647. doi:10.1074/jbc.M109.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. doi: 10.1016/j.ccr.2009.03.002. doi:10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. doi:10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. doi:10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H. Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res. 1998;58:609–613. [PubMed] [Google Scholar]
- 30.Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci USA. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. doi:10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. doi:10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 32.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendrysa SM, Perry ME. The p53 tumor suppressor protein does not regulate expression of its own inhibitor, MDM2, except under conditions of stress. Mol Cell Biol. 2000;20:2023–2030. doi: 10.1128/mcb.20.6.2023-2030.2000. doi:10.1128/MCB.20.6.2023-2030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. doi:10.1016/S1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 35.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. doi:10.1128/MCB.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biderman L, Manley JL, Prives C. Mdm2 and MdmX as regulators of gene expression. Genes Cancer. 2012;3:264–273. doi: 10.1177/1947601912455331. doi:10.1177/1947601912455331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biderman L, Poyurovsky MV, Assia Y, Manley JL, Prives C. MdmX is required for p53 interaction with and full induction of the Mdm2 promoter after cellular stress. Mol Cell Biol. 2012;32:1214–1225. doi: 10.1128/MCB.06150-11. doi:10.1128/MCB.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao BX, Chen HZ, Lei NZ, Li GD, Zhao WX, Zhan YY, Liu B, et al. p53 mediates the negative regulation of MDM2 by orphan receptor TR3. EMBO J. 2006;25:5703–5715. doi: 10.1038/sj.emboj.7601435. doi:10.1038/sj.emboj.7601435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. doi:10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. doi:10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 41.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. doi:10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pant V, Xiong S, Jackson JG, Post SM, Abbas HA, Quintas-Cardama A, Hamir AN, et al. The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev. 2013;27:1857–1867. doi: 10.1101/gad.227249.113. doi:10.1101/gad.227249.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA. 2005;102:731–736. doi: 10.1073/pnas.0405495102. doi:10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Zhang Z, Cheng J, Li M, Wang W, Xu W, Wang H, et al. Transcription factor NFAT1 activates the mdm2 oncogene independent of p53. J Biol Chem. 2012;287:30468–30476. doi: 10.1074/jbc.M112.373738. doi:10.1074/jbc.M112.373738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, Morse HC., III IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188–3194. doi: 10.4049/jimmunol.0803693. doi:10.4049/jimmunol.0803693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CJ, Freeman DJ, Wu H. PTEN regulates Mdm2 expression through the P1 promoter. J Biol Chem. 2004;279:29841–29848. doi: 10.1074/jbc.M401488200. doi:10.1074/jbc.M401488200. [DOI] [PubMed] [Google Scholar]
- 47.Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci USA. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. doi:10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. doi:10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt MK, Tommiska J, Broeks A, van Leeuwen FE, Van't Veer LJ, Pharoah PD, Easton DF, et al. Combined effects of single nucleotide polymorphisms TP53 R72P and MDM2 SNP309, and p53 expression on survival of breast cancer patients. Breast Cancer Res. 2009;11:R89. doi: 10.1186/bcr2460. doi:10.1186/bcr2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knappskog S, Bjornslett M, Myklebust LM, Huijts PE, Vreeswijk MP, Edvardsen H, Guo Y, et al. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell. 2011;19:273–282. doi: 10.1016/j.ccr.2010.12.019. doi:10.1016/j.ccr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 51.Lukas J, Gao DQ, Keshmeshian M, Wen WH, Tsao-Wei D, Rosenberg S, Press MF. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001;61:3212–3219. [PubMed] [Google Scholar]
- 52.Bartel F, Taylor AC, Taubert H, Harris LC. Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol Res. 2001;12:451–457. doi: 10.3727/096504001108747459. [DOI] [PubMed] [Google Scholar]
- 53.Harris LC. MDM2 splice variants and their therapeutic implications. Curr Cancer Drug Targets. 2005;5:21–26. doi: 10.2174/1568009053332654. doi:10.2174/1568009053332654. [DOI] [PubMed] [Google Scholar]
- 54.Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502–9508. doi: 10.1158/0008-5472.CAN-05-4271. doi:10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 55.Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 2003;63:5703–5706. [PubMed] [Google Scholar]
- 56.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20:4041–4049. doi: 10.1038/sj.onc.1204533. doi:10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- 57.Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222–1230. [PubMed] [Google Scholar]
- 58.Blandino G, Deppert W, Hainaut P, Levine A, Lozano G, Olivier M, Rotter V, et al. Mutant p53 protein, master regulator of human malignancies: a report on the Fifth Mutant p53 Workshop. Cell Death Differ. 2012;19:180–183. doi: 10.1038/cdd.2011.148. doi:10.1038/cdd.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. doi:10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. doi:10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 61.Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, Yu H, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4:2996. doi: 10.1038/ncomms3996. doi:10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okoro DR, Arva N, Gao C, Polotskaia A, Puente C, Rosso M, Bargonetti J. Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS ONE. 2013;8:e77643. doi: 10.1371/journal.pone.0077643. doi:10.1371/journal.pone.0077643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32:61–69. doi: 10.1038/onc.2012.28. doi:10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 64.Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S, Lee TJ, Kim T, et al. MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci USA. 2012;109:5316–5321. doi: 10.1073/pnas.1202465109. doi:10.1073/pnas.1202465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao J, Lin H, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30:524–532. doi: 10.1038/emboj.2010.347. doi:10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, et al. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst. 2013;105:433–442. doi: 10.1093/jnci/djt003. doi:10.1093/jnci/djt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffman Y, Bublik DR, Pilpel Y, Oren M. miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.146. doi:10.1038/cdd.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–180. [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q, Zou C, Han Z, Xiao H, Wei H, Wang W, Zhang L, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335:168–174. doi: 10.1016/j.canlet.2013.02.029. doi:10.1016/j.canlet.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 70.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. doi:10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. doi:10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene. 2003;22:8870–8880. doi: 10.1038/sj.onc.1207176. doi:10.1038/sj.onc.1207176. [DOI] [PubMed] [Google Scholar]
- 73.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. doi:10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. doi:10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 75.Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V, Fahraeus R. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25–35. doi: 10.1016/j.ccr.2011.11.016. doi:10.1016/j.ccr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 76.Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, Oren M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J. 2002;21:3715–3727. doi: 10.1093/emboj/cdf384. doi:10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell. 2012;45:801–813. doi: 10.1016/j.molcel.2012.01.021. doi:10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu W, Feng Z, Levine AJ. The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer. 2012;3:199–208. doi: 10.1177/1947601912454734. doi:10.1177/1947601912454734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–1589. doi: 10.1101/gad.1941710. doi:10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. doi:10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. doi:10.1016/S1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 82.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. doi:10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 83.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. doi:10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 84.Tian X, Chen Y, Hu W, Wu M. E2F1 inhibits MDM2 expression in a p53-dependent manner. Cell Signal. 2011;23:193–200. doi: 10.1016/j.cellsig.2010.09.003. doi:10.1016/j.cellsig.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 85.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. doi:10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 86.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. doi:10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 87.Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. doi:10.1016/S1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 88.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. doi:10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. doi:10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 90.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. doi:10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 91.Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS ONE. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. doi:10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. doi:10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 93.Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2:234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lillberg K, Verkasalo PK, Kaprio J, Teppo L, Helenius H, Koskenvuo M. Stressful life events and risk of breast cancer in 10,808 women: a cohort study. Am J Epidemiol. 2003;157:415–423. doi: 10.1093/aje/kwg002. doi:10.1093/aje/kwg002. [DOI] [PubMed] [Google Scholar]
- 95.Stommel M, Given BA, Given CW. Depression and functional status as predictors of death among cancer patients. Cancer. 2002;94:2719–2727. doi: 10.1002/cncr.10533. doi:10.1002/cncr.10533. [DOI] [PubMed] [Google Scholar]
- 96.Chida Y, Hamer M, Wardle J, Steptoe A. Do stress-related psychosocial factors contribute to cancer incidence and survival? Nat Clin Pract Oncol. 2008;5:466–475. doi: 10.1038/ncponc1134. doi:10.1038/ncponc1134. [DOI] [PubMed] [Google Scholar]
- 97.Feng Z, Liu L, Zhang C, Zheng T, Wang J, Lin M, Zhao Y, et al. Chronic restraint stress attenuates p53 function and promotes tumorigenesis. Proc Natl Acad Sci USA. 2012;109:7013–7018. doi: 10.1073/pnas.1203930109. doi:10.1073/pnas.1203930109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hara MR, Kovacs JJ, Whalen EJ, Rajagopal S, Strachan RT, Grant W, Towers AJ, et al. A stress response pathway regulates DNA damage through beta2-adrenoreceptors and beta-arrestin-1. Nature. 2011;477:349–353. doi: 10.1038/nature10368. doi:10.1038/nature10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Amato R, D'Antona L, Porciatti G, Agosti V, Menniti M, Rinaldo C, Costa N, et al. Sgk1 activates MDM2-dependent p53 degradation and affects cell proliferation, survival, and differentiation. J Mol Med (Berl) 2009;87:1221–1239. doi: 10.1007/s00109-009-0525-5. doi:10.1007/s00109-009-0525-5. [DOI] [PubMed] [Google Scholar]
- 100.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. doi:10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 101.Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem. 2002;277:49668–49675. doi: 10.1074/jbc.M208593200. doi:10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 102.Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, Wahl GM. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273–285. doi: 10.1016/j.ccr.2006.03.014. doi:10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 103.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. doi:10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. doi:10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 105.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. doi:10.1016/S0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 106.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363:433–450. doi: 10.1016/j.jmb.2006.08.027. doi:10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 107.Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem. 2011;286:23725–23734. doi: 10.1074/jbc.M110.213868. doi:10.1074/jbc.M110.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci USA. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. doi:10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. doi:10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 110.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. doi:10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. doi:10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 112.Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Di Lello P, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. doi:10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. doi:10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. doi:10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 115.Roxburgh P, Hock AK, Dickens MP, Mezna M, Fischer PM, Vousden KH. Small molecules that bind the Mdm2 RING stabilize and activate p53. Carcinogenesis. 2012;33:791–798. doi: 10.1093/carcin/bgs092. doi:10.1093/carcin/bgs092. [DOI] [PubMed] [Google Scholar]
- 116.Herman AG, Hayano M, Poyurovsky MV, Shimada K, Skouta R, Prives C, Stockwell BR. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 2011;1:312–325. doi: 10.1158/2159-8290.CD-11-0104. doi:10.1158/2159-8290.CD-11-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. doi:10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 118.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. doi:10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. doi:10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]