Significance
Light-harvesting antenna are essential for photosynthetic systems, which comprise photosystems I and II (PSI and PSII, respectively). Phycobilisome (PBS) is a dominant and efficient antenna for PSII in cyanobacteria and some algae, whereas the attachment of PBS to PSI is a long-standing open question. We isolated a unique PBS–PSI supercomplex from a nitrogen-fixing cyanobacterium. Biochemical and spectral studies revealed that PBS is functionally connected to the PSI tetramer via a new universal connecting component. A pseudoatomic model explains the configuration of the PSI tetramer and how the PBS is connected to PSI. Such antenna may play an important role for light harvesting in PSI-driven cyclic electron transport to facilitate nitrogen fixation and other reactions.
Abstract
Oxygenic photosynthesis is driven by photosystems I and II (PSI and PSII, respectively). Both have specific antenna complexes and the phycobilisome (PBS) is the major antenna protein complex in cyanobacteria, typically consisting of a core from which several rod-like subcomplexes protrude. PBS preferentially transfers light energy to PSII, whereas a PSI-specific antenna has not been identified. The cyanobacterium Anabaena sp. PCC 7120 has rod–core linker genes (cpcG1-cpcG2-cpcG3-cpcG4). Their products, except CpcG3, have been detected in the conventional PBS. Here we report the isolation of a supercomplex that comprises a PSI tetramer and a second, unique type of a PBS, specific to PSI. This rod-shaped PBS includes phycocyanin (PC) and CpcG3 (hereafter renamed “CpcL”), but no allophycocyanin or CpcGs. Fluorescence excitation showed efficient energy transfer from PBS to PSI. The supercomplex was analyzed by electron microscopy and single-particle averaging. In the supercomplex, one to three rod-shaped CpcL–PBSs associate to a tetrameric PSI complex. They are mostly composed of two hexameric PC units and bind at the periphery of PSI, at the interfaces of two monomers. Structural modeling indicates, based on 2D projection maps, how the PsaI, PsaL, and PsaM subunits link PSI monomers into dimers and into a rhombically shaped tetramer or “pseudotetramer.” The 3D model further shows where PBSs associate with the large subunits PsaA and PsaB of PSI. It is proposed that the alternative form of CpcL–PBS is functional in harvesting energy in a wide number of cyanobacteria, partially to facilitate the involvement of PSI in nitrogen fixation.
In photosynthesis, light-harvesting antennas are essential to efficiently collect solar energy. Photosynthetic organisms have diverse antenna protein-pigment complexes, which are specifically associated with photosystems I or II (PSI or PSII, respectively) (1). Light-harvesting chlorophyll a/b-binding proteins form the peripheral antenna of PSI or PSII in green plants (2) and light-harvesting chlorophyll a/c-binding proteins are present in brown algae and related organisms (3, 4). In cyanobacteria the phycobilisome (PBS) serves as a major antenna for PSII. No specific antenna has been isolated for PSI in cyanobacteria, although PBS transfers light energy to PSI under conditions of state transition (5), a temporal energy redistribution mechanism between PSII and PSI (6, 7).
Generally, the PBS is a supercomplex of rod and core subcomplexes, which consist of various phycobilin-binding proteins connected by several classes of colorless linker proteins (8). Whereas phycocyanin (PC) is the major phycobiliprotein of the rod, allophycocyanin (APC) is the major phycobiliprotein of the core cylinders. The rod–core linker cyanobacterial phycocyanin protein G (CpcG), which connects the rod to the core, plays a key role in the assembly of the PBS (9). The chromosome of the filamentous, N2-fixing cyanobacterium Anabaena sp. PCC 7120 (hereafter “Anabaena”) bears tandem repeats of rod–core linker genes (cpcG1-cpcG2-cpcG3-cpcG4). Except for CpcG3, the products of these genes have been detected in the PBS (10–13). Only cpcG3 encodes a hydrophobic region at the C terminus, although all four copies share the sequence for the highly conserved N-terminal linker domain (Fig. S1). Given the distinct role of the hydrophobic CpcG variant protein in docking to PSI as shown in this article, the hydrophobic CpcG is hereafter renamed “CpcL.”
In this study, a supercomplex comprising PSI and its specific antenna was isolated from Anabaena and biochemically and structurally characterized. We found that within the supercomplex, PSI is organized into tetramers, which were previously suggested from biochemical data (14). Single-particle electron microscopy (EM) shows that in fact they are structurally organized as a dimer of dimers. In addition, we describe a specific association of CpcL–PBS rods with the PSI pseudotetramers and discuss the role of these unique supercomplexes.
Results
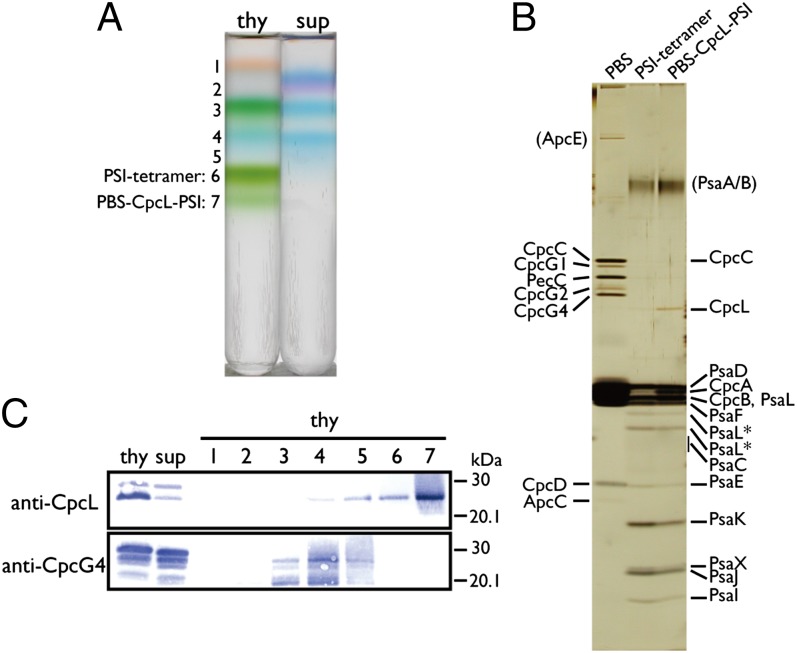
The thylakoid membrane from Anabaena was solubilized with n-dodecyl-β-d-maltoside (DM) and fractionated using a linear sucrose density gradient centrifugation with 0.005–0.05% DM (Fig. S2A). Three green bands (PSI/PSII monomer, PSI/PSII dimer, and PSI tetramer) were resolved at 0.03–0.05% DM, in agreement with our previous results (14). The solubilized complexes were aggregated into precipitation at 0.005% DM. At 0.01–0.02% DM, PSI was mostly recovered in the tetramer band, and bluish-green band 7 was newly detected below the PSI tetramer (band 6) (Fig. 1A and Fig. S2A). Blue native PAGE revealed that both fractions 6 and 7 contained the PSI tetramer (Fig. S2B). Subsequent SDS/PAGE and N-terminal sequencing showed that fraction 7 contained CpcL, CpcC, CpcB, and CpcA in addition to the PSI subunits (PsaA/B, PsaC, etc.) (Fig. 1B and Table S1). Neither the other rod–core linker proteins (CpcG1, CpcG2, and CpcG4) nor any of the PBS core components (such as ApcA or ApcB) were detected in this fraction. Immunoblot analysis showed that CpcL was predominantly associated with thylakoids (Fig. 1C). After solubilization with DM, CpcL was predominantly recovered in fraction 7, whereas CpcG4 was detected in fractions 3–5, which included PBS subcomplexes. Thus, CpcL specifically interacts with PSI to form a unique PBS–CpcL–PSI supercomplex in the bluish-green fraction 7. The band of CpcL was appreciably less intense than the band of the PSI subunit PsaD in fraction 7, suggesting the substoichiometric abundance of CpcL in the PSI tetramer (Fig. 1B). This suggests that only one or two PBS rods are attached to the tetramer.
Fig. 1.
Isolation of PBS–CpcL–PSI supercomplex by centrifugation through a linear sucrose gradient containing low-salt buffer and 0.01% DM. (A) Fractionation profile of thylakoid (thy) and supernatant (sup) after centrifugation at 130,000 × g for 6 h at 4 °C. (B) Silver staining following SDS/PAGE of the conventional PBS, PSI tetramer (see 6 in A) and supercomplex (see 7 in A). N-terminal sequences of the bands are listed in Table S1. Asterisks indicate degradation products of PsaL. (C) Western blotting of CpcL and CpcG4. Lane numbers correspond to the numbered bands in A.
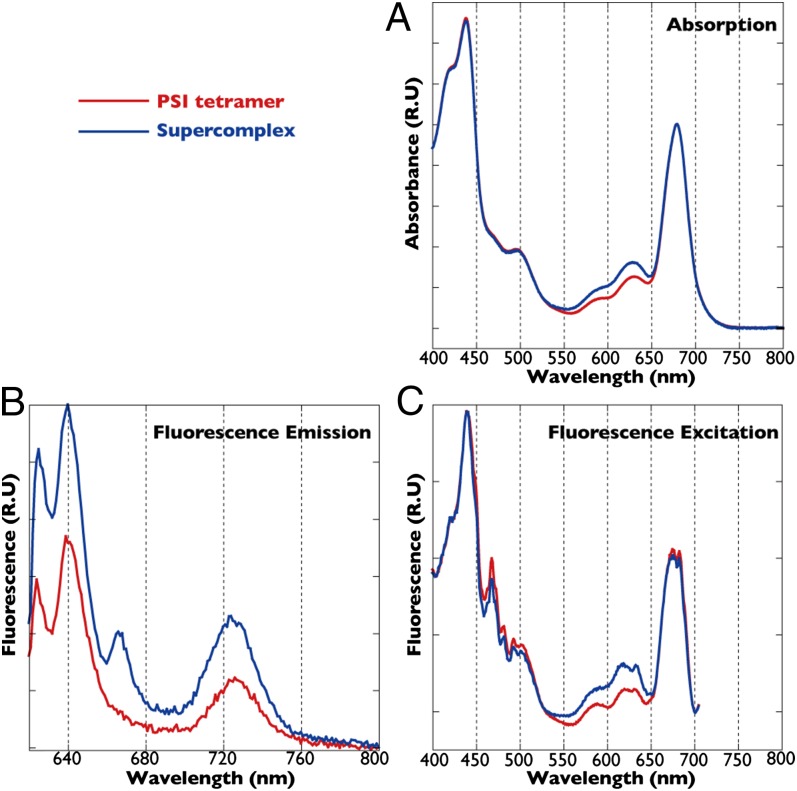
The absorption spectra revealed that the bluish-green fraction 7 contained PC (Fig. 2A). The absence of the APC shoulder around 650 nm, together with the absence of the core polypeptides, strongly supports the idea that PC and CpcL directly interact with the PSI tetramer to form a supercomplex. Energy transfer from PC to the PSI tetramer was estimated through analysis of 77-Kelvin (K) fluorescence spectra (Fig. 2B). Following excitation at 600 nm, which excites PC efficiently and chlorophyll less efficiently, emission at 725 nm was higher from PSI in the PBS–CpcL–PSI supercomplex than from the PSI tetramer (Fig. 2B). The higher excitation peak for fluorescence emission from PSI corresponded well to the higher absorption peak of PC in the supercomplex than the PSI tetramer, confirming the energy transfer from PC to PSI in the supercomplex (Fig. 2C).
Fig. 2.
Absorption spectra and 77-K fluorescence spectra of PSI tetramer (red) and supercomplex (blue) fractions. (A) Absorption spectra were normalized relative to the chlorophyll peak at 678 nm. (B) Emission spectra excited at 600 nm were normalized relative to the chlorophyll concentration. (C) Excitation spectra were normalized relative to the chlorophyll peak at 435 nm. PSI fluorescence was measured at 725 nm.
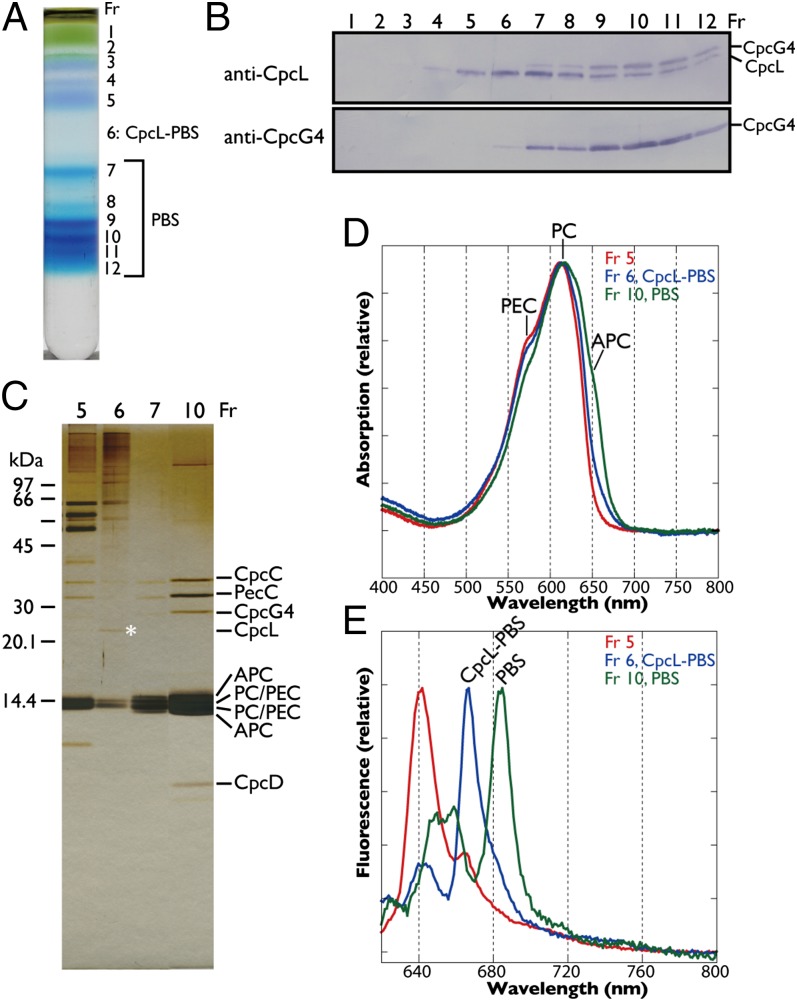
CpcL–PBS was isolated from vegetative cells of Anabaena with high phosphate linear sucrose gradient centrifugation with modification (Fig. 3A). CpcL and CpcG4 were detected in fractions 5–12 and 7–12, respectively (Fig. 3B). Because the antigenic peptide for CpcL was partially shared with CpcG4 (Fig. S1A), the upper band in the anti-CpcL Western blot was derived from the weak cross-reaction with CpcG4, which corresponded to the band in the anti-CpcG4 blot.
Fig. 3.
Fractionation of CpcL–PBS by linear sucrose gradient centrifugation in the presence of high concentrations of phosphate. (A) Fractionation profile. Fr, fraction number. (B) Western blotting. Note that anti-CpcL also weakly detected CpcG4. (C) Silver-stained SDS/PAGE gel. The asterisk indicates CpcL in fraction 6. (D) Absorption spectra, normalized to PC. Absorption of PEC, PC, and APC are indicated. (E) Seventy-seven–kelvin fluorescence spectra with 600 nm excitation, normalized at each peak.
The conventional PBS that contained CpcG4 was resolved into at least five discrete bands with a linear sucrose gradient (Fig. 3A), but only one condensed band with a typical stepwise gradient (13). Because the multiple bands showed nearly identical polypeptide compositions, and band 7 corresponded to the intact PBS band of Synechocystis, the multiple bands may have been derived from the oligomerization of the conventional rod–core PBS complex. Although Western analysis showed that fractions 7–12 also contained some CpcL, silver staining of SDS gels and protein sequencing detected CpcC, PecC, and CpcG4, but not CpcL, probably due to its very low abundance (Fig. 3C).
Fraction 6 that mainly contained CpcL in Western analysis was a pale blue region between the two blue bands in the gradient (Fig. 3A). The absorption spectra revealed that the pale blue fraction 6 contained PC and phycoerythrocyanin (PEC) but not APC (Fig. 3D). After concentration, the bands of CpcL, PC, and PEC, but not CpcG4 or APC, were visible in the silver-stained SDS gel, indicative of CpcL–PBS complex (Fig. 3C), which is consistent with Western analysis. The CpcL–PBS emitted fluorescence at 665 nm at 77 K, even though it did not contain APC (Fig. 3E). This is different from the conventional CpcG–PC rod that emits at 652 nm (15). Notably, the same fluorescence at 665 nm was also detected in the PBS–CpcL–PSI supercomplex (Fig. 2B). Red-shifted fluorescence of PC due to CpcL may help energy transfer from PBS to PSI without APC.
We further investigated the PBS–CpcL–PSI supercomplex during N2 fixation. The N2-fixing heterocyst cells were isolated by mild disruption of the N2-fixing filaments. Immunoblot analysis indicated that heterocysts contained high levels of CpcL but very little CpcG4 (Fig. 4). Some of CpcL was recovered in the heterocyst supernatant. When expressed in terms of chlorophyll content, total CpcL was approximately four times more abundant in heterocysts than in vegetative cells.
Fig. 4.
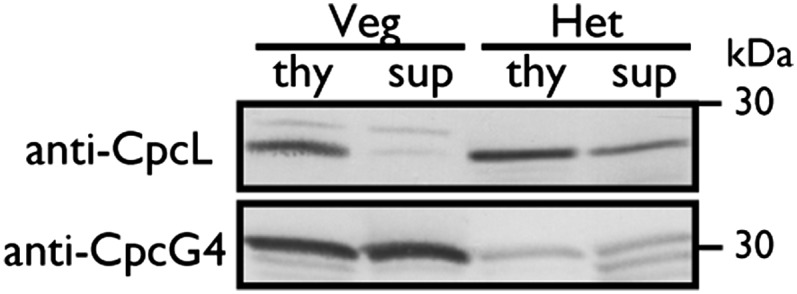
Western blotting of CpcL and CpcG4 in isolated heterocysts. Protein of thylakoid and supernatant fractions (0.25 mg protein per lane) from isolated heterocysts (Het) (0.4 µg Chl per lane) and BG11-grown vegetative cells (Veg) (1.6 µg Chl per lane) were subjected to SDS/PAGE.
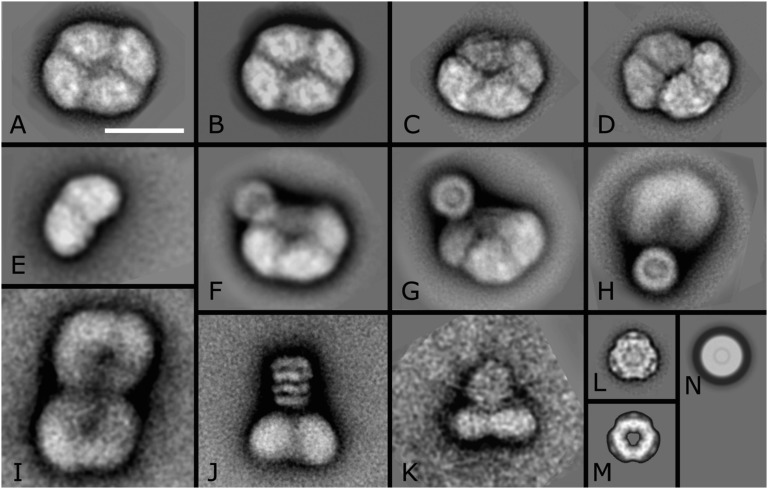
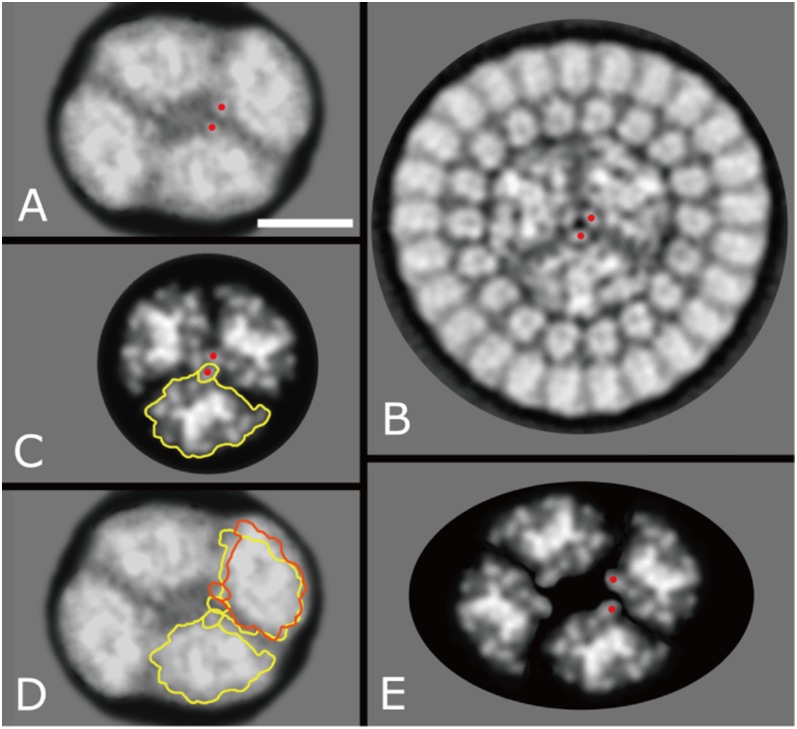
The purified PBS–CpcL–PSI supercomplex was studied by single-particle EM. About 20,000 tetrameric PSI particles were present in an almost nontilted view parallel to the membrane plane (Fig. 5 A and B) or differently tilted views (Fig. 5 C and D). Small numbers of dimeric complexes (4% of the tetramers) were also present (Fig. 5E). About 4,000 tetramers had an attached CpcL–PBS in one specific position, with the tetramers seen in a more slightly tilted position (Fig. 5 F and G). The CpcL–PBS was bound at the periphery of the PSI tetramer, at the interface of monomers, as present in the dimer map of Fig. 5E. In a less-abundant second position, the averaged projection map looks more blurred (Fig. 5H), but nevertheless it can be assigned to the interface between two dimers, if compared with a map showing two tetramers attached, in which the upper monomer is seen in a similar way (Fig. 5I). From an average made of 1,000 CpcL–PBS supercomplexes in a side view position, it appears that most particles are composed of two hexameric PC units, each consisting of two layers. Some other side views show the hexamers in a face-on position (Fig. 5K), indicating that the association between PSI and CpcL–PBS molecules is rather loose. This is also apparent by observing and analyzing smaller particles in the electron micrographs, which showed multiple single PC hexamers that became disconnected from the PSI tetramers likely due to an artifact such as freezing. An average of 2,000 PC hexamers (Fig. 5L) is very compatible with the X-ray structure of the PC hexamer (Fig. 5M) except that the density in the center, which can now be assigned to the CpcL linker protein, is lacking in the X-ray structure. The fact that the CpcL–PBS molecules attached to the PSI tetramers appear smaller than the single PC hexamers is a matter of a variation in rotational position, as can be seen in comparison with a rotationally averaged view of the PC hexamer (Fig. 5M). In addition to the observation that the binding between PSI and CpcL–PBS molecules is possible at two positions, the total number of CpcL–PBS rods can vary between one and three per tetramer. In addition to the two PC hexamers, the rods occasionally accommodate one to two additional hexamers (Fig. S3), likely containing PEC.
Fig. 5.
Single-particle EM analysis of projections of Anabaena PSI fractions. (A) Averaged projection map showing the PSI tetrameric complex in an almost nontilted view parallel to the membrane plane. (B) Twofold rotationally symmetrized projection map of A. (C and D) Projection maps of tilted tetramers. (E) Projection map of the dimer, in an equivalent position to the left half of the tetramer. (F) Projection map of tilted tetramer with a CpcL–PBS rod attached. (G and H) Projection maps of a stronger tilted tetramer with a CpcL–PBS rod attached. (I) Map of two associated tetramers, both in a tilted position. (J) PSI complex in side-view position with an attached CpcL–PBS rod composed of two hexameric units seen in side view. (K) PSI complex in side-view position with a CpcL–PBS rod attached in the top-view position. (L) Projection of a PC hexamer in the top-view position. (M) Map of PC hexamer, derived from the X-ray structure of P. yezoensis. (N) Projection map of L, rotationally averaged. (Scale bar = 200 Å.)
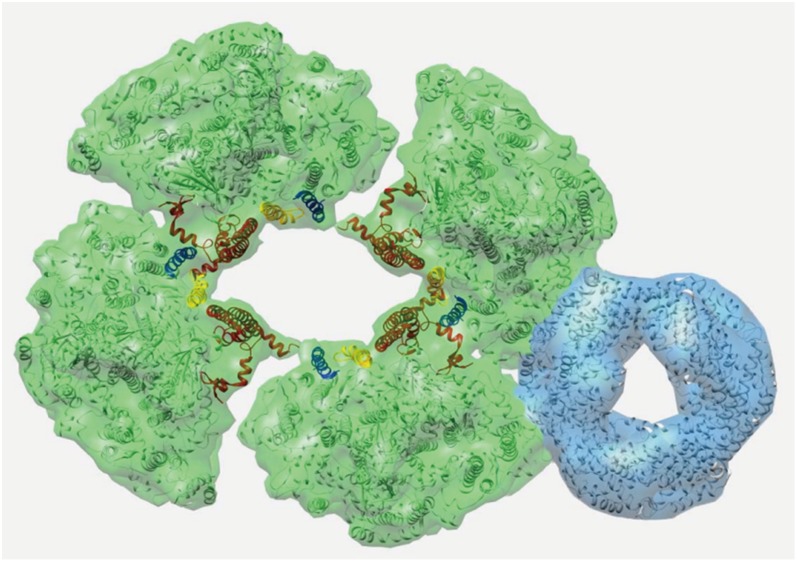
The map of the Anabaena PSI tetramer (Fig. 6A) with a resolution of about 17 Å can be compared with the high-resolution X-ray structure of the PSI trimer (16) to see which subunits are involved in linking the monomers and to assign the handedness of the tetramer (Fig. 6C). Close comparison with the map of a PSI–IsiA supercomplex from Thermosynechococcus elongatus (17) at 15 Å (Fig. 6B) tells us that the tetramer as presented is seen from its stromal side. By an overlay of the circumference of the monomer from the trimer (yellow contour line in Fig. 6C) on the tetramer we can see the difference in association of the monomers in trimers and tetramers, which is mostly a matter of different angles. In the pseudotetramer, the angles between the monomers are 2 × 73° and 2 × 107°, instead of 3 × 120° in the trimer and 4 × 90° for a symmetric tetramer. There is an additional shift of about 5 Å to optimize the fit (orange contour line in Fig. 6D). Based on the EM maps, we constructed a pseudoatomic model of the typical PSI supercomplex with one associated CpcL–PBS molecule on the most frequently occupied interface position (Fig. 7). The model shows the tetramer (green) with an associated PC hexamer (blue) at the interface of two monomers. The PsaL (red), PsaI (yellow), and PsaM (blue) subunits are involved in linking the monomers into dimers and into the pseudotetramer. The model clearly shows how these three small subunits are positioned in a different way in the two types of connections of the monomers.
Fig. 6.
Interpretation of the Anabaena PSI tetrameric projection map. (A) Projection map of the tetramer. Red dots mark the densities of PsaL subunits linking monomers into dimers. (B) Projection map of the PSI–IsiA supercomplex from T. elongatus (from ref. 17). (C) Projection map of the high-resolution trimer structure (16), truncated to 15 Å for comparison, seen from the stromal side of the membrane. The circumference of one monomer is indicated (in yellow). (D) Map of the tetramer, with two monomers, as positioned in the trimer, overlaid (in yellow). The trimer in C has been shifted and rotated clockwise by 13° to optimize the fit in the tetramer (orange contour). (E) Model of the tetramer, composed of the truncated X-ray monomers of the projection map of C. (Scale bar = 100 Å.)
Fig. 7.
Model of the most abundant PBS–CpcL–PSI supercomplex, with a CpcL–PBS rod (blue) associated at tetrameric PSI (green) at the interface of two monomers. The PsaL (red), PsaI (yellow), and PsaM (blue) subunits, linking the monomers into dimers and dimers into the pseudotetramer, have been highlighted. The model shows the supercomplex from the stromal side of the membrane. Pigment molecules have been left out.
Discussion
The precise interaction between PSI and PSII with any type of PBS is unknown because it was never possible to purify a photosystem with a PBS component attached. In this study we show the unique structural data of a specific association of CpcL–PBSs to PSI complexes in Anabaena cells by EM. This was achieved by purifying a PBS–CpcL–PSI supercomplex, which efficiently transfers energy from PBS to PSI. We also show that CpcL–PBS is specific for cyanobacterial PSI. This CpcL–PBS rod is distinctly different from the conventional hemidiscoidal PBS in which APC, PC, PEC, and the linkers CpcG1/CpcG2/CpcG4 are involved. The PSI complex is also special because it has an unusual tetrameric configuration, although the same PsaI, PsaL and PsaM subunits are involved in linking the monomers as in trimeric PSI (16, 18, 19). However, it should be noted that the PsaI and PsaL subunits that can potentially make tetramers have a special amino acid sequence (14).
The CpcL–PBS rods bind at the periphery of the PSI tetramer in one to two or possibly three copies (Fig. 5 and Fig. S3). Because CpcL appears to be located in the center of the PC hexamer (Fig. 5L), it could be in direct contact with the membrane. This makes sense if we assume that the C-terminal hydrophobic tail of CpcL anchors the PBS rods at the membrane close to or in direct contact with PSI via hydrophobic interactions. Sequence comparison of related CpcL species reveals a conserved region that harbors a hydrophobic segment of 23-aa residues (Fig. S4). Most of the interaction of the CpcL–PBS can be direct, because the docking sites are outside the large hydrophilic ridge of the stromally located PSI subunits PsaC, -D, and -E (16).
We previously reported that a CpcG variant (CpcG2), which possesses a hydrophobic segment in the C-terminal region, binds a rod subcomplex (CpcG2–PBS) without APC core subunits in the unicellular cyanobacterium Synechocystis sp. PCC 6803 (20). Although a specific association of CpcG2–PBS with PSI was not shown (21), selective energy transfer from PBS to PSI is implicated in mutants of the conventional CpcG1 but not CpcG2 (22). Hence, CpcG2 in Synechocystis is functionally similar to the Anabaena CpcL. Synechocystis CpcG2 and related sequences, which harbor the C-terminal hydrophobic segment, are widely distributed among cyanobacteria (Fig. S5). Although not close relatives of Anabaena CpcL, we call CpcG2 “CpcL” and propose that the supercomplex configuration, as characterized for Anabaena, will be present in other cyanobacterial species as well. A functional role for the PBS–CpcL–PSI supercomplex may be in N2 fixation, because the level of supercomplexes is appreciably higher under the N2-fixing condition than the nonfixing condition in Anabaena (Fig. 4).
Clustering analysis illustrated the unique evolutionary features of the CpcG/CpcL superfamily (Fig. S5), which is a member of the clan of linker proteins (23). First, clades of hydrophilic rod–core linker CpcG and hydrophobic rod–PSI linker CpcL coexist in the phylogeny. Second, their phylogeny can be separated into three sets of CpcG and CpcL, depending on the grouping of organisms: (i) group I, which consists of thermophilic species and some heterocyst-forming species that typically carry multiple copies that are encoded by a gene cluster [cpcG1-cpcG2-cpcG3(cpcL)-cpcG4]; (ii) group II, which consists of major cyanobacterial species and eukaryotic algae that each carry one copy of CpcG and CpcL, although algae and some cyanobacteria only have the hydrophilic CpcG; and (iii) group III, which consists of all of the marine Synechococcus and related species with one copy of CpcG and one or two copies of CpcL. Third, the CpcL clade is closely related to its cognate CpcG clade in each group, suggesting convergent evolution of the hydrophobic CpcL from the hydrophilic CpcG. And fourth, there were some intermediary species between groups I and II. The heterocyst-forming species Nostoc punctiforme possesses single copies of CpcG and CpcL that belong to group II (24), whereas the closely related Cylindrospermopsis raciborskii CS-505 possesses the multicopy-type group I cpcG1-cpcG2-cpcG4 and group II cpcL (25). These results suggest that the heterocyst-forming cyanobacteria of group I acquired the multiple cpcG copies by lateral gene transfer from the thermophilic cyanobacteria and later created cpcL from them.
Cyclic electron transport around PSI provides the ATP needed for N2 fixation in heterocyst cells of Anabaena. In heterocysts, efficient energy transfer has been suggested to occur from PC to PSI (26), although the transfer mechanism and structural details were unknown. With the discovery of CpcL–PBS as a specific antenna for PSI in which CpcL is a unique and indispensable component of the PBS–CpcL–PSI supercomplex, it is relevant to mention that levels of CpcL were approximately fourfold higher in heterocysts than in vegetative cells (Fig. 4). It should also be mentioned that CpcL of groups I or II is present in many species that fix nitrogen in the daytime (heterocystous species and Trichodesmium) but mostly absent in the species that fix in the dark (Crocosphaera and Cyanothece American Type Culture Collection 51142; Fig. S5). This suggests that increasing levels of PBS–CpcL–PSI supercomplexes could be efficient in N2 fixation, especially under light-limited conditions. Under normal light conditions, it has been thought that the core antenna chlorophylls of PSI and state transition using PSII-associated PBS (5) harvest sufficient light energy for PSI in cyanobacteria. Interestingly, in some cyanobacteria, cpcL expression is induced under PSII-exciting green or orange light in chromatic acclimation (22, 24, 27). Thus, the spectral range of light for PSI chlorophyll may be expanded using CpcL–PBS to improve the overall photosynthetic activity. CpcL–PBS appears to be widely distributed in many cyanobacteria (Fig. S4). Together, these facts suggest that optimal photosynthesis could be achieved by both short-term state transition of PBS and long-term PSI-specific CpcL–PBS in cyanobacteria. This mechanism would be a general concept for efficient photosynthesis in photosynthetic organisms.
Developmental regulation of CpcL is another interesting issue. The expression of the hydrophobic CpcL (CpcG2) is transcriptionally regulated by the cyanobacteriochrome photoreceptor CcaS and its cognate response regulator CcaR in Synechocystis and N. punctiforme (24, 27), whereas a CcaS and CcaR homolog was not found in the Anabaena genome. Activation of a specific promoter for cpcL under nitrogen starvation was suggested by RNA-sequencing analysis, although the regulation mechanism is unclear (28). In cyanobacteria, phycobiliproteins are specifically degraded by the NblA system under nitrogen starvation. In Anabaena, they are transiently degraded before development of nitrogen fixation (29, 30). Thus, heterocyst cells show reduced phycobiliprotein content, whereas cells in the nblA disruptant are highly fluorescent due to the lack of their degradation (29, 31). The reduced content of CpcG4 in heterocysts may be due to protein turnover by the NblA system. It is more likely that specific activation of cpcL and enhanced degradation of the conventional PBS may underlie the selective accumulation of CpcL–PBS in heterocyst cells, although it remains unclear how the NblA system acts on the conventional PBS and CpcL–PBS in heterocyst and vegetative cells.
Materials and Methods
Growth Conditions.
Anabaena cells were grown at 31 °C in liquid BG11 or N-free medium (BG110) supplemented with 20 mM Hepes⋅potassium hydroxide (pH 8.2) (32) and with bubbling of 1% CO2 under continuous illumination with white fluorescent lamps (ca. 20 µmol photons⋅m–2⋅s–1).
Isolation of Heterocyst Cells and Cell Fractionation.
Heterocysts were purified as described (33) with minor modifications. Cells grown in BG110 were harvested and washed with a buffer [8% (wt/vol) sucrose, 50 mM EDTA, and 50 mM Tris⋅HCl (pH 8.0)]. The cells were resuspended in the same buffer supplemented with 5% (vol/vol) Triton X-100 and 1 mg⋅mL−1 lysozyme and vortexed for a few minutes. Cells were then broken with a French pressure cell with two passages at 210 kg⋅cm–2 and 350 kg⋅cm–2. Heterocyst cells were collected by centrifugation at 600 × g for 3 min, washed twice with the buffer, and the purity was confirmed with light microscopy.
Thylakoids and soluble proteins were isolated from the heterocyst and vegetative cells with a low salt buffer. Cells were harvested and resuspended in buffer A containing 50 mM 4-Morpholineethanesulfonic acid⋅sodium hydroxide (pH 6.5), 10 mM MgCl2, 5 mM CaCl2, and 25% (wt/vol) glycerol. The cells were disrupted with zirconia beads with a bead beater (Micro Smash MS-100R; TOMY). Cells were agitated at 4 °C for 15 s and then cooled on ice for 2 min, and this cycle repeated 10 times. After removal of unbroken cells, the resulting supernatant was centrifuged at 300,000 × g for 30 min at 4 °C to separate thylakoid membranes and soluble proteins.
Isolation of Photosystem Complexes.
Thylakoids from vegetative cells [1 mg chlorophyll (Chl)⋅mL–1] were solubilized with 1% DM (Sigma-Aldrich) on ice for 30 min, followed by centrifugation at 300,000 × g for 30 min at 4 °C. The supernatant was diluted with three volumes of buffer A without glycerol, loaded onto a 10–30% linear sucrose density gradient in buffer A containing DM but not glycerol, and centrifuged at 130,000 × g at 4 °C.
Blue-Native PAGE.
Blue-native PAGE was performed as described (34). Thylakoids (1 mg Chl⋅mL–1) were solubilized with 1% DM on ice for 30 min, followed by centrifugation at 30,000 × g for 30 min at 4 °C, and then subjected to blue-native PAGE with a gradient of 3–13% (wt/vol) acrylamide in a separation gel at 4 °C.
SDS/PAGE and Immunoblotting.
Proteins were separated with SDS-urea-PAGE with a 16–22% (wt/vol) linear gradient of polyacrylamide gel containing 7.5 M urea (35), followed by silver staining (36). Immunoblotting was performed as described (22), except that a 16% (wt/vol) polyacrylamide gel containing 7.5 M urea was used. The antipeptide antibodies were produced by Takara Bio. Synthetic peptides (for CpcL, CTTDRPFSFTPRYGAD; for CpcG4, QTDWRFTLDKFYSRKSC; the underlined Cys was added for conjugation) were conjugated to keyhole limpet hemocyanin and injected into rabbits.
N-Terminal Sequencing.
SDS/PAGE gels were blotted to a Immobilon-PSQ membrane (Millipore) and subjected to N-terminal sequencing using the Edman degradation method (PPSQ-21; Shimadzu).
Absorption and 77-K Fluorescence Spectroscopy.
Absorption spectra were measured at room temperature using a spectrophotometer (UV-2400PC; Shimadzu). Fluorescence was measured with a spectrofluorometer (RF-5300PC; Shimadzu). Emission spectra were recorded with excitation at 600 nm (PC). Fluorescence excitation spectra were recorded with an emission at 725 nm (PSI).
Isolation of PBS.
PBS was isolated as described (37) with modifications as noted below. Cells grown in BG11 were harvested, washed twice with 0.9 M potassium phosphate buffer (pH 7.0), and broken with zirconia beads with a bead beater (Micro Smash MS-100R; TOMY). The cell extract was treated with 2% (vol/vol) Triton X-100 in 0.9 M phosphate buffer for 30 min and centrifuged at 20,000 × g for 20 min at 18 °C to separate into the upper green Triton X-100 layer and the lower blue aqueous layer. The blue layer was loaded onto a 10–50% (wt/vol) linear sucrose density gradient with 0.9 M phosphate buffer and centrifuged at 130,000 × g for 16 h at 18 °C.
Clustering Analysis.
Clustering analysis was performed by automatic sequence alignment and classification with the neighbor-joining algorithm using ClustalX (http://www.clustal.org/). Hydropathy plots were obtained with the SOSUI program (http://bp.nuap.nagoya-u.ac.jp/sosui/).
EM and Analysis.
Supercomplexes were negatively stained with 2% uranyl acetate for EM. Imaging was performed on a Philips CM120 equipped with a LaB6 tip operating at 120 kV. The GRACE system for semiautomated specimen selection and data acquisition (38) was used to record 2048 × 2048 pixel images at 130,000 × magnification using a Gatan 4000 SP 4K slow-scan CCD camera with a pixel size of 0.225 nm. Over 3,000 images were recorded and a total of 30,000 particle projections were collected and analyzed by single-particle averaging with Groningen image processing software including multireference and nonreference alignments, multivariate statistical analysis and classification, as in ref. 39. The best class averages were used as references to sharpen the images in the subsequent alignments. For the final class-sums the best ∼30% of the projections were summed. The tetramer structure was generated from the cyanobacterial PSI monomeric structure [Protein Data Bank (PDB) ID code 1JB0]. The structure of Porphyra yezoensis (PDB code 1kn1) was used for modeling the PC hexamer. For final modeling, University of California, San Francisco Chimera 1.8, EMAN 2, Adobe Photoshop CS4, and GIMP software were used.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Education and Science [Grants-in-Aid for Young Scientists (to R.N.), Scientific Research and the Global Center of Excellence (GCOE) program (to M.I.)] and the Core Research of Evolutional Science and Technology program and Precursory Research for Embryonic Science and Technology from the Japan Science and Technology Agency. This work was further supported by the HARVEST Marie Curie Research Training Network (PITN-GA-2009-238017) (to E.J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320599111/-/DCSupplemental.
References
- 1.Neilson JAD, Durnford DG. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth Res. 2010;106(1-2):57–71. doi: 10.1007/s11120-010-9576-2. [DOI] [PubMed] [Google Scholar]
- 2.Minagawa J. State transitions—the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim Biophys Acta. 2011;1807(8):897–905. doi: 10.1016/j.bbabio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda Y, et al. Photosystem I complexes associated with fucoxanthin-chlorophyll-binding proteins from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta. 2008;1777(4):351–361. doi: 10.1016/j.bbabio.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Veith T, Büchel C. The monomeric photosystem I-complex of the diatom Phaeodactylum tricornutum binds specific fucoxanthin chlorophyll proteins (FCPs) as light-harvesting complexes. Biochim Biophys Acta. 2007;1767(12):1428–1435. doi: 10.1016/j.bbabio.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Mullineaux CW. Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res. 2008;95(2-3):175–182. doi: 10.1007/s11120-007-9249-y. [DOI] [PubMed] [Google Scholar]
- 6.Bellafiore S, Barneche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433(7028):892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- 7.Kargul J, Barber J. Photosynthetic acclimation: Structural reorganisation of light harvesting antenna—role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FEBS J. 2008;275(6):1056–1068. doi: 10.1111/j.1742-4658.2008.06262.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu LN, Chen XL, Zhang YZ, Zhou BC. Characterization, structure and function of linker polypeptides in phycobilisomes of cyanobacteria and red algae: An overview. Biochim Biophys Acta. 2005;1708(2):133–142. doi: 10.1016/j.bbabio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Bryant DA. In: Cell Culture and Somatic Cell Genetics of Plants. Bogorad L, Vasil IK, editors. Vol 7B. New York: Academic; 1991. pp. 255–298. [Google Scholar]
- 10.Glauser M, et al. Phycobilisome structure in the cyanobacteria Mastigocladus laminosus and Anabaena sp. PCC 7120. Eur J Biochem. 1992;205(3):907–915. doi: 10.1111/j.1432-1033.1992.tb16857.x. [DOI] [PubMed] [Google Scholar]
- 11.Bryant DA, et al. A small multigene family encodes the rod-core linker polypeptides of Anabaena sp. PCC7120 phycobilisomes. Gene. 1991;107(1):91–99. doi: 10.1016/0378-1119(91)90301-q. [DOI] [PubMed] [Google Scholar]
- 12.Ducret A, Sidler W, Wehrli E, Frank G, Zuber H. Isolation, characterization and electron microscopy analysis of a hemidiscoidal phycobilisome type from the cyanobacterium Anabaena sp. PCC 7120. Eur J Biochem. 1996;236(3):1010–1024. doi: 10.1111/j.1432-1033.1996.01010.x. [DOI] [PubMed] [Google Scholar]
- 13.Cai YA, Schwartz SH, Glazer AN. Transposon insertion in genes coding for the biosynthesis of structural components of the Anabaena sp. phycobilisome. Photosynth Res. 1997;53:109–120. [Google Scholar]
- 14.Watanabe M, Kubota H, Wada H, Narikawa R, Ikeuchi M. Novel supercomplex organization of photosystem I in Anabaena and Cyanophora paradoxa. Plant Cell Physiol. 2011;52(1):162–168. doi: 10.1093/pcp/pcq183. [DOI] [PubMed] [Google Scholar]
- 15.Pizarro SA, Sauer K. Spectroscopic study of the light-harvesting protein C-phycocyanin associated with colorless linker peptides. Photochem Photobiol. 2001;73(5):556–563. doi: 10.1562/0031-8655(2001)073<0556:ssotlh>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Jordan P, et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature. 2001;411(6840):909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 17.Chauhan D, et al. A novel photosynthetic strategy for adaptation to low-iron aquatic environments. Biochemistry. 2011;50(5):686–692. doi: 10.1021/bi1009425. [DOI] [PubMed] [Google Scholar]
- 18.Chitnis VP, Chitnis PR. PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1993;336(2):330–334. doi: 10.1016/0014-5793(93)80831-e. [DOI] [PubMed] [Google Scholar]
- 19.Naithani S, Hou JM, Chitnis PR. Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of Photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res. 2000;63(3):225–236. doi: 10.1023/A:1006463932538. [DOI] [PubMed] [Google Scholar]
- 20.Kondo K, Geng XX, Katayama M, Ikeuchi M. Distinct roles of CpcG1 and CpcG2 in phycobilisome assembly in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res. 2005;84(1-3):269–273. doi: 10.1007/s11120-004-7762-9. [DOI] [PubMed] [Google Scholar]
- 21.Kondo K, Mullineaux CW, Ikeuchi M. Distinct roles of CpcG1-phycobilisome and CpcG2-phycobilisome in state transitions in a cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res. 2009;99(3):217–225. doi: 10.1007/s11120-008-9399-6. [DOI] [PubMed] [Google Scholar]
- 22.Kondo K, Ochiai Y, Katayama M, Ikeuchi M. The membrane-associated CpcG2-phycobilisome in Synechocystis: A new photosystem I antenna. Plant Physiol. 2007;144(2):1200–1210. doi: 10.1104/pp.107.099267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe M, Ikeuchi M. Phycobilisome: Architecture of a light-harvesting supercomplex. Photosynth Res. 2013;116(2-3):265–276. doi: 10.1007/s11120-013-9905-3. [DOI] [PubMed] [Google Scholar]
- 24.Hirose Y, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc Natl Acad Sci USA. 2010;107(19):8854–8859. doi: 10.1073/pnas.1000177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stucken K, et al. The smallest known genomes of multicellular and toxic cyanobacteria: Comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS ONE. 2010;5(2):e9235. doi: 10.1371/journal.pone.0009235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson RB, Dolan E, Calvert HE, Ke B. Energy transfer from phycobiliproteins to photosystem I in vegetative cells and heterocysts of Anabaena variabilis. Biochim Biophys Acta. 1981;634(2):237–248. doi: 10.1016/0005-2728(81)90142-0. [DOI] [PubMed] [Google Scholar]
- 27.Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci USA. 2008;105(28):9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA. 2011;108(50):20130–20135. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collier JL, Grossman AR. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994;13(5):1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baier K, Lehmann H, Stephan DP, Lockau W. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology. 2004;150(Pt 8):2739–2749. doi: 10.1099/mic.0.27153-0. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Sherman LA. Characterization of Synechocystis sp. strain PCC 6803 and deltanbl mutants under nitrogen-deficient conditions. Arch Microbiol. 2002;178(4):256–266. doi: 10.1007/s00203-002-0446-y. [DOI] [PubMed] [Google Scholar]
- 32.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 33.Golden JW, Whorff LL, Wiest DR. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173(22):7098–7105. doi: 10.1128/jb.173.22.7098-7105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe M, Iwai M, Narikawa R, Ikeuchi M. Is the photosystem II complex a monomer or a dimer? Plant Cell Physiol. 2009;50(9):1674–1680. doi: 10.1093/pcp/pcp112. [DOI] [PubMed] [Google Scholar]
- 35.Ikeuchi M, Inoue Y. A new 4.8-kDa polypeptide intrinsic to the PS II reaction center, as revealed by modified SDS-PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 1988;29:1233–1239. [Google Scholar]
- 36.Aro EM, et al. Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J Exp Bot. 2005;56(411):347–356. doi: 10.1093/jxb/eri041. [DOI] [PubMed] [Google Scholar]
- 37.Gray BH, Gantt E. Spectral properties of phycobilisomes and phycobiliproteins from the blue-green alga-nostoc sp. Photochem Photobiol. 1975;21(2):121–128. doi: 10.1111/j.1751-1097.1975.tb06638.x. [DOI] [PubMed] [Google Scholar]
- 38.Oostergetel GT, Keegstra W, Brisson A. Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy. 1998;74:47–59. [Google Scholar]
- 39.Boekema EJ, Van Roon H, Van Breemen JF, Dekker JP. Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur J Biochem. 1999;266(2):444–452. doi: 10.1046/j.1432-1327.1999.00876.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.