Significance
Mediator is a megadalton multisubunit molecular switchboard involved in gene regulation in eukaryotes and is structurally conserved between species. It bridges the general transcription machinery and function-specific DNA binding proteins. It plays a dynamic role in regulating a wide range of processes, involving, for example, thyroid and vitamin D receptors. The role of Mediator appears to be in the fine tuning of the activation and repression of gene expression in many organisms, yet the underlying mechanisms of how its own function is regulated remains to be unraveled. Here we demonstrate how Mediator autoregulates its own function by cross-talk between the tail module and the Cdk8 kinase module in an active process involving priming of the mediator component Med3 for ubiquitin-ligase (Grr1)–mediated degradation by Cdk8 phosphorylation.
Abstract
Mediator, an evolutionary conserved large multisubunit protein complex with a central role in regulating RNA polymerase II–transcribed genes, serves as a molecular switchboard at the interface between DNA binding transcription factors and the general transcription machinery. Mediator subunits include the Cdk8 module, which has both positive and negative effects on activator-dependent transcription through the activity of the cyclin-dependent kinase Cdk8, and the tail module, which is required for positive and negative regulation of transcription, correct preinitiation complex formation in basal and activated transcription, and Mediator recruitment. Currently, the molecular mechanisms governing Mediator function remain largely undefined. Here we demonstrate an autoregulatory mechanism used by Mediator to repress transcription through the activity of distinct components of different modules. We show that the function of the tail module component Med3, which is required for transcription activation, is suppressed by the kinase activity of the Cdk8 module. Med3 interacts with, and is phosphorylated by, Cdk8; site-specific phosphorylation triggers interaction with and degradation by the Grr1 ubiquitin ligase, thereby preventing transcription activation. This active repression mechanism involving Grr1-dependent ubiquitination of Med3 offers a rationale for the substoichiometric levels of the tail module that are found in purified Mediator and the corresponding increase in tail components seen in cdk8 mutants.
Gene expression is regulated through a number of distinct yet integrated processes that can be broadly defined as epigenetic and nonepigenetic. The regulation of transcription per se, where initiation is the first critical step in a highly regulated control process, occurs following the opening or relaxation of chromatin to establish a permissive structure for gene activation. Shutting off transcription before the establishment of a repressive chromatin structure over control regions involves inhibiting recruitment of the transcription machinery and direct inhibition of the RNA polymerase II holoenzyme and Mediator function (1).
Earlier work proposed chromatin-independent repression mechanisms, where coregulators repressed transcription modestly in vitro in the absence of chromatin (2). Genetic and biochemical experiments demonstrated interactions between corepressors and subunits of the Mediator complex (3) and suppression of coactivator function through targeted degradation (4), suggesting active mechanisms for the direct repression of the transcription machinery.
Mediator is a multisubunit protein complex involved in the regulation of activator-dependent transcription and has been identified in most eukaryotes (1, 5). Based on structural, biochemical, and genetic data, Mediator has been divided into four distinct modules; head, middle, tail, and Cdk8. The head and middle modules form core Mediator and are relatively stable, remaining tightly associated during biochemical purification. In contrast the tail module, composed of the mediator components Med15/Gal11, Med2, Med3/Hrs1/Pgd1, and Med16/Sin4, has been isolated in substoichiometric amounts (6), suggesting that it may interact transiently with core Mediator or be subjected to regulated turnover to modulate its function. Furthermore, a triad of tail components, Med15, Med2, and Med3, have been isolated and shown to recruit Mediator to a target gene promoter to activate transcription (7). The Cdk8 module, which contains the protein kinase Cdk8 (Srb10), it’s associated cyclin CycC (Srb11), Med12, and Med13, has a dynamic association with Mediator and is absent from Mediator preparations isolated under specific growth conditions. Cdk8 has been shown to regulate the function of DNA-binding transcriptional activators including Msn2, where it controls nuclear accumulation, and Ste12 and Gcn4, where it regulates protein stability (8, 9).
At least three different activation models have been proposed for Mediator that involve either the Cdk8 or tail modules. These models include interaction between core Mediator and Cdk8 away from the promoter preventing Mediator recruitment (10); association of the tail module or the triad with promoters independently from core Mediator and the subsequent recruitment of core Mediator (7); and transient interactions between Cdk8 and promoter-bound Mediator modulating the functional status of positioned Mediator (11). However, the molecular mechanisms underlying the regulation of Mediator function have yet to be fully elucidated.
Here we show that the kinase activity of Cdk8 regulates the coactivator function of Med3 by site-specific phosphorylation, marking it for targeted degradation. This mechanism requires the E3 ubiquitin ligase Grr1 and appears analogous to phosphorylation-dependent SRC-3 ubiquitination involving SCFFbw7α (4). The active repression mechanism, and the concomitant increase in tail components seen in cdk8 mutants, offers a rationale as to why the tail module can be found in substoichiometric levels in purified Mediator and reveals how Mediator function is autoregulated through interactions between components of distinct modules.
Results
Cdk8 Interacts with Med3 to Modulate Target Gene Transcription.
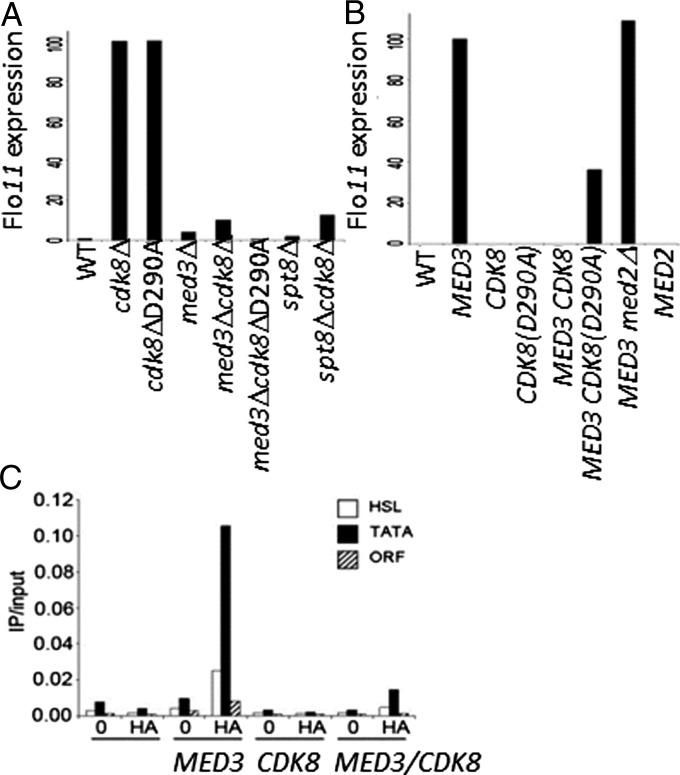
Cdk8 has been shown to play roles in both positive and negative regulation of gene expression (12). The mechanism by which Cdk8 represses transcription through its association with Mediator remains to be fully elucidated. In contrast, Med3 functions as a potent activator of transcription when artificially recruited to a test promoter (3). Interestingly, both Mediator components interact with Ssn6-Tup1, a member of the Groucho/TLE family of general corepressors, suggesting that Cdk8 and Med3 could be positioned close enough to permit molecular interactions. To test this possibility, we examined genetic interactions between CDK8 and MED3 using the shared flocculation protein encoding gene target FLO11 (YIR019C, MUC1) as a model. FLO11 was greatly elevated (∼100-fold) in cdk8Δ cells compared with WT, where it was at background levels and only very slightly elevated in med3Δ (Fig. 1A), demonstrating the requirement for Cdk8 in maintaining FLO11 repression. Conversely, high levels of FLO11 were observed when Med3 was overexpressed (Fig. 1B), confirming its coactivator function. This function was due to direct recruitment as ChIP analysis demonstrated that Med3 was specifically localized to the FLO11 promoter region and absent from the coding region (Fig. 1C). Two promoter regions, the heat shock–like element (−488 to −342 from ATG; Fig. 1C, white bars) (13) and the TATA box (−183 to −52 from ATG; Fig. 1C, black bars) were both occupied by Med3. This observation is consistent with the requirement for Med3 in the establishment of a stable preinitiation complex (14, 15) and its role in the recruitment of core Mediator (7, 11).
Fig. 1.
Genetic regulation of Med3 target gene expression. Total RNA was isolated, and FLO11 expression levels were determined using qRT-PCR. (A) MED3 and SPT8 are required for CDK8-mediated FLO11 derepression. (B) CDK8 suppresses the activation function of MED3. Strains carrying plasmids expressing MED3 or MED2 (3) together with CDK8 or CDK8 (D290A) (29) were maintained in appropriate selective media. Values are given as a percentage relative to either (A) cdk8Δ or (B) MED3 set at 100% and normalized to ACT1. (C) Med3 recruitment to the FLO11 promoter is regulated by CDK8. Chromatin immunoprecipitates were prepared from cells expressing Med3, Cdk8, or Med3 and Cdk8 using anit-HA and then analyzed for the presence of Med3 on the heat shock–like element (−488 to −342 from ATG, white bars) (13), TATA box (−183 to −53 to ATG, black bars), and 3′ ORF (3,628–3,765 to ATG, hashed bars) of FLO11 using qRTPCR. 0, no antibody control.
The above results demonstrated the opposing roles of Cdk8 and Med3 in regulating transcription of the same gene. We therefore tested for genetic interactions between the two Mediator components in cdk8Δmed3Δ cells. In contrast to cdk8Δ, in a cdk8Δmed3Δ double mutant, FLO11 was only very slightly de-repressed, showing that MED3 is essential for FLO11 expression (Fig. 1A). This result established a link between the tail- and Cdk8-modules suggesting that cross-talk between these modules could regulate Mediator function.
To test the specificity of this interaction we examined whether a similar effect was observed with Med2, a second component of the tail module. Med2 was previously shown to be a target for Cdk8 phosphorylation, although this was found to occur only in the presence of Med3. Despite being a potent activator when artificially recruited to a test promoter (3), MED2 overexpression did not result in up-regulation of FLO11 expression (Fig. 1B). Furthermore, when Med3 was expressed in med2Δ cells, Med3-dependent activation was unaffected (Fig. 1B). It appears that Med2 is not involved in the activation of FLO11 and that Med3 functions independently of Med2 in FLO11 regulation.
Cdk8 Kinase Activity Regulates Med3 Activation Potential and Promoter Recruitment.
To determine whether the role of Cdk8 in FLO11 repression was due to its kinase function, a strain harboring a point mutation (D290A) in the catalytic domain of the kinase was assessed. Here high levels of FLO11 expression equal to those in cdk8Δ cells were detected (Fig. 1A), demonstrating that Cdk8 kinase activity is required for repression. Furthermore, coexpression of CDK8 and MED3 resulted in the complete suppression of FLO11 activation, suggesting that Med3 functions downstream of Cdk8 (Fig. 1B). This effect of CDK8 overexpression occurs in cells expressing only endogenous levels of CycC, suggesting that this effect is limited by CycC levels yet still results in repression of FLO11 to background levels. This suppression effect was attributed largely to the kinase function of Cdk8, as coexpression of kinase-inactive CDK8 (D290A) with MED3 did not abolish FLO11 expression; rather, expression was maintained at ∼40% compared with MED3 alone (Fig. 1B). These data imply that the activation potential of Med3 is suppressed by Cdk8 kinase activity. This effect appears due to a direct effect on Med3, as ChIP analysis demonstrated that in the presence of ectopically expressed CDK8 promoter occupancy, by Med3 was reduced by more than 90% compared with Med3 alone (Fig. 1C). This observation suggests that Cdk8 functions to exclude Med3 from the promoter and therefore that it could destabilize Mediator on the promoter of repressed genes, thus preventing activator-dependent transcription.
Cdk8 Interacts with and Phosphorylates Med3.
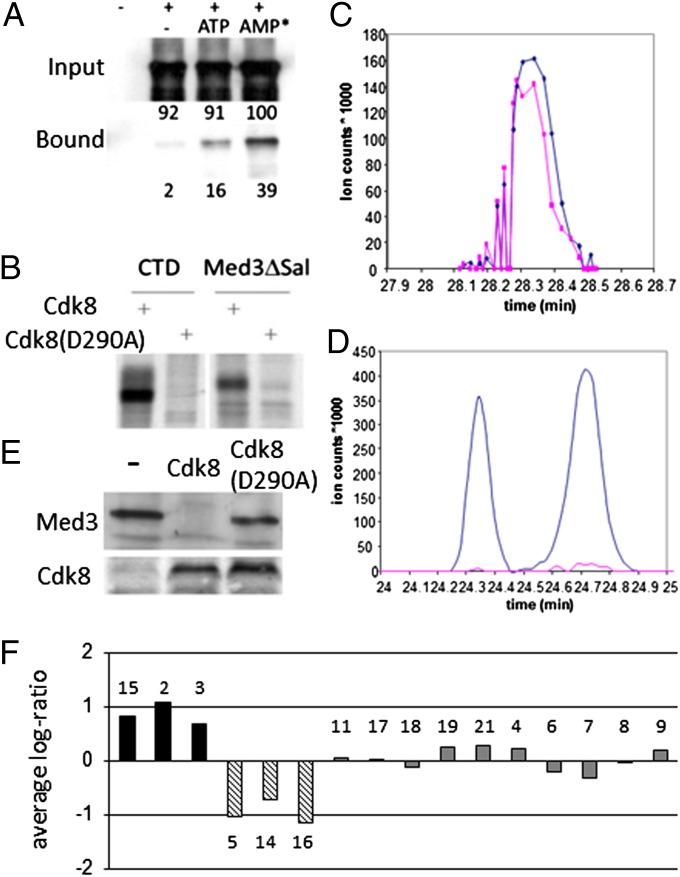
The Cdk8-dependent exclusion of Med3 from the FLO11 promoter led us to determine whether the mechanism modulating Med3 recruitment was due to phosphorylation of Med3 by Cdk8. First, we examined if there was a direct interaction between Med3 and Cdk8 that would facilitate any subsequent phosphorylation event. Using a two-hybrid assay, we found that both full-length Med3 and Med3∆Sal, the N-terminal region (amino acids 1–234; Fig. S1A) of Med3 truncated after a predicted phosphorylation region, interacted in vivo with Cdk8 (Table S1), localizing the interaction between Med3 and Cdk8 to this N-terminal region. To understand whether this represented a direct interaction, we conducted in vitro assays using myc-tagged Cdk8 (8) immune-purified from yeast cells and recombinant Med3∆Sal in the presence of ATP donor molecules. In the absence of ATP, only very low levels of Med3 (2% input) were seen to interact with Cdk8 (Fig. 2A). However, when phosphate donors were used, a significant pull-down of input Med3 (ATP, 16%; AMPNP, 39%) was observed (Fig. 2A). This result suggests that Med3 is likely to be phosphorylated by Cdk8 following the formation of an ATP-dependent complex. Based on our genetic observations, Med3 phosphorylation would be expected to have a negative effect on activation function and would offer an explanation as to how Cdk8 modulates Med3 activity.
Fig. 2.
Med3 interacts with and is phosphorylated by Cdk8. (A) Recombinant Med3ΔSal protein interacts with myc-Cdk8, and this interaction is enhanced in the presence of ATP and AMPNP (AMP*). Purified myc-Cdk8 was incubated with Med3ΔSal protein and either 500 mM ATP or AMPNP. Values indicate percentage protein relative to AMPNP input reaction. (B) Cdk8 phosphorylates Med3. Recombinant histidine-tagged RNA pol II CTD or Med3ΔSal protein was incubated with myc-Cdk8 in the presence of 33P γ-ATP. Extracted ion chromatograms of (C) nonphosphorylated or (D) phosphorylated Med3-peptide SGSTMGTPTVHNSTAAAPIAAPK obtained from WT (blue) and cdk8Δ (pink) strains. (E) Med3 is unstable in the presence of Cdk8 in vivo. Total protein isolated from cells transformed with MED3 and either CDK8 or cdk8 (D290A) were analyzed for Med3 or Cdk8 by immunoblotting with anti-HA or anti-myc, respectively. (F) Mediator stability in absence of Cdk8. Quantitative MS analysis of Tap-Med15 purified Med proteins in the presence or absence of Cdk8. Observed ratios of purified Med proteins (numbered) were normalized so that the average log-ratio equaled 0 between samples.
In vitro kinase assays were carried out using myc-tagged Cdk8 immunoprecipitated from yeast cells (together with the other components of the kinase module) and recombinant Med3∆Sal that contains two putative Cdk8 phosphorylation sites: T149 and T195 (Fig. S1A) (16). High levels of phosphorylation were observed when Med3 was incubated with functional immune-purified kinase but not with the inactive D290A mutant (Fig. 2B and Fig. S2), confirming the notion that Med3 was a Cdk8 target. The RNA pol II CTD control was phosphorylated to a similar level as Med3, suggesting Med3 is a major target of the kinase (Fig. 2B).
To identify Cdk8 phosphorylation sites on Med3, we combined quantitative MS analysis using differentially labeled cell populations with immunoprecipitation of Mediator using a Tap-Med15 strain. Chromatography of a nonphosphorylated Med3 peptide SGSTMGTPTVHNSTAAAPIAAPK demonstrated it was present in both WT (Fig. 2C, blue) and cdk8Δ (Fig. 2C, pink) strains at approximately equal levels. The corresponding phosphorylated form of the Med3 peptide was detected in WT (Fig. 2D, blue) as a peak doublet, suggesting phosphorylation occurred at distinct positions within the peptide. This Med3-specific phosphorylation was essentially absent from cdk8Δ cells (Fig. 2D, pink), confirming the requirement for Cdk8 to phosphorylate Med3 in vivo. MS-MS analysis of phospho-Med3 peptide fractions identified two phosphorylation sites within the peptide (Fig. S1 B and C). One phosphorylation site SGS(phos)TMGTPTVHNSTAAAPIAAPK corresponded to a predicted serine phosphorylation site S191, with a site localization probability of 69%. The second phosphorylation site SGSTMGT(phos)PTVHNSTAAAPIAAPK corresponded to one of the predicted Cdk phosphorylation sites at T195 in Med3, with a localization probability of 88% (Fig. S1A). No phosphorylation was detected at the second putative Cdk site T149; the S191/T195-containing peptide was the only phospho-peptide ever detected for Med3.
To further understand the stability of Med3, in vivo protein extracts were prepared from cells ectopically expressing MED3 and functional or inactive CDK8. Expression of MED3 alone resulted in the accumulation of Med3 protein, whereas when MED3 and CDK8 were coexpressed, no Med3 protein was detected (Fig. 2E). This effect was specific to functional Cdk8, as Med3 was detected at the same levels in the presence of the inactive kinase as with MED3 alone (Fig. 2E). Therefore, it appears that, although phosphorylated Med3 can be detected in vitro, it does not accumulate in vivo, suggesting active turnover of the phosphorylated Med3 protein occurs.
To determine the effect of the loss Cdk8 on the stability of endogenous Med3 and other mediator components, quantitative MS analysis was undertaken. Tap-Med15 was used to purify proteins in the presence or absence of Cdk8. In the absence of Cdk8, levels of Med3 along with the other triad components Med2/15 increased, showing that endogenous Med3 is more stable in the absence of Cdk8 (Fig. 2F) and suggesting that Med3 stabilizes the Mediator tail module. Additionally it appears that Med5/14/16 specifically drop out of the complex, as their relative log ratios become negative values, whereas the tail module stays connected to the head module. This observation implies that Med5/14/16 do not form the bridge between the head and tail, as is often assumed in the standard Mediator model.
Grr1 Modulates Cdk8-Mediated Stability of Med3.
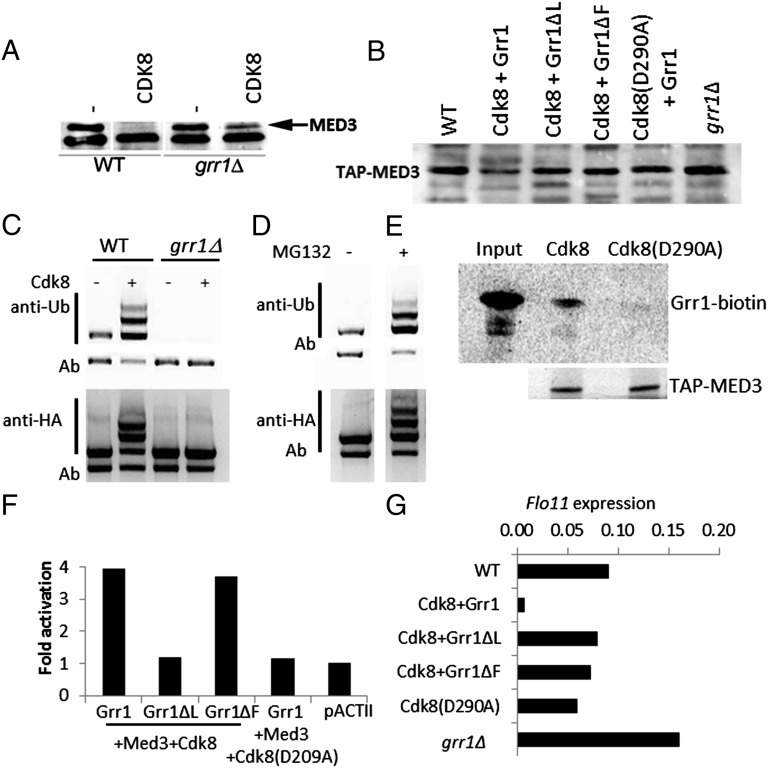
Inactivation of protein function can occur through ubiquitination-mediated proteolysis involving a Skp, Cullin, F-box containing complex (SCF) containing an F-box E3 ubiquitin ligase, which confers specificity. In Saccharomyces cerevisiae, the E3 ligase Grr1 has several roles including morphogenesis and environmental response (17, 18). Several of these functions overlap with those of Cdk8, including response to glucose, osmotic stress, nitrogen starvation, and enhanced filamentous growth (19), which are also linked to FLO11 expression (20). We therefore tested whether loss of Med3 was mediated through Grr1. Med3 protein levels were measured in WT and grr1Δ cells expressing ectopic MED3 and CDK8. In WT cells, we were unable to detect Med3, whereas in grr1Δ cells, Med3 was detected, indicating that Grr1 was involved in the destabilization of Med3 (Fig. 3A). That Med3 was expressed at slightly reduced levels compared with when CDK8 was not coexpressed suggests that this effect cannot be entirely attributed to Grr1. Conversely, ectopic expression of MED3, GRR1, and CDK8 resulted in reduced levels of Med3 but only in the presence of functional Grr1 (Fig. S3A). When either Grr1ΔL or Grr1ΔF, proteins lacking the functional leucine-rich repeat or F-box domains of Grr1 (21), were tested, Med3 levels remained unchanged (Fig. S3A). Similarly, when we tested endogenous Tap-Med3, levels of the Mediator component were reduced substantially in cells expressing GRR1 and CDK8 (Fig. 3B) but remained stable when grr1ΔL or grr1ΔF was used, demonstrating that functional E3 ligase is required to inhibit Med3 activity. Accordingly, Med3 levels increased by 32% in Tap-MED3::grr1Δ cells (compared with Tap-MED3 cells; Fig. 3B). Furthermore, ubiquitination of Med3 was only detectable in WT and not grr1Δ cells (Fig. 3C), demonstrating the requirement for Grr1 in this process. iseΔ cells, which permit the uptake of the 26S proteasome inhibitor MG132 (carbobenzoxy-Leu-Leu-leucinal), overexpressing Med3 were tested to establish the effect of inhibiting the proteasome degradation pathway. In iseΔ cells, ubiquitinated Med3 accumulated (Fig. 3D), indicating that even in cells expressing only endogenous levels of Cdk8, Med3 was being rapidly degraded. This ubiquitination and degradation process appears to be due to the formation of a Med3-Grr1 complex as Med3 was found to directly interact with in vitro synthesized Grr1, but only when Med3 was isolated from cells containing functional Cdk8 (Fig. 3E). In addition, phosphorylated Med3 seems to be a direct Grr1 substrate because it interacts with Grr1 and Grr1ΔF but not with Grr1ΔL (the Grr1 derivative lacking the leucine-reach repeat region known to bind phosphorylated substrates; Fig. 3F).
Fig. 3.
Med3 is targeted for degradation in a Grr1-dependent manner. (A) Loss of Med3 due to Cdk8 activity requires functional Grr1. grr1Δ transformed with MED3 and CDK8 and assayed for Med3 with anti-HA. Arrow indicates position of Med3 protein. (B) Med3 stability is decreased by Grr1 and Cdk8. Total cell extracts were made from cells harboring functional Grr1 or nonfunctional Grr1ΔL or Grr1ΔF derivatives and functional Cdk8 or a kinase inactive variant (D290A), and Med3 levels were assessed using anti-Tap antibody. (C) Grr1 activity mediates Cdk8 regulated Med3 degradation. WT and grr1Δ cells were transformed with CDK8 and MED3 to assess the impact of Grr1 function on Cdk8 regulation of Med3 stability. HA-Med3 was immunoprecipitated using anti-HA, and proteins were analyzed using anti-ubiquitin or anti-HA (for Med3). Ab, nonspecific antibody cross reaction. (D) MG132 blocks Med3 turnover. ise1Δ cells transformed with MED3 were incubated with 100 μM MG132 or DMSO for 1 h. HA-Med3 was immunoprecipitated using anti-HA and analyzed using anti-ubiquitin to recognize ubiquitin residues or with an anti-HA to confirm HA-Med3. (E) Med3 interacts with Grr1 in vitro. Biotin-labeled Grr1 was incubated with Tap-purified Med3 isolated from WT or cdk8 (D290A) cells. Grr1 was detected using streptavidin-HRP and Med3 using an anti-Tap-antibody and chemiluminescent detection. (F) Med3 is a direct Grr1 substrate. Yeast two-hybrid assay was performed with pAS2Grr1, and derivatives Grr1ΔL and Grr1ΔF in Tap-Med3 cells transformed with CDK8 and CDK8 (D290A). Cells were grown to midlog phase and assayed for LacZ expression. pACTII, negative control. (G) GRR1 is required for FLO11 repression. FLO11 transcript levels were measured in WT (Tap-MED3) and grr1Δ cells, WT cells transformed with Grr1 or nonfunctional Grr1ΔL or Grr1ΔF derivatives, and functional Cdk8 or Cdk8 (D290A). Values are given as relative starting quantities and normalized to ACT1.
The consequence on Med3 stability of manipulating Grr1 and Cdk8 is the loss of target gene repression; loss of GRR1 (Fig. 3G) or CDK8 (Fig. 1A) results in FLO11 de-repression. Conversely, repression of FLO11 is established in the presence of functional Grr1, but not when the function of Grr1 (Grr1ΔL or Grr1ΔF) or Cdk8(D290A) becomes compromised (Fig. 3G).
Serine 191 Regulates Med3 Function and Its Interaction with Grr1.
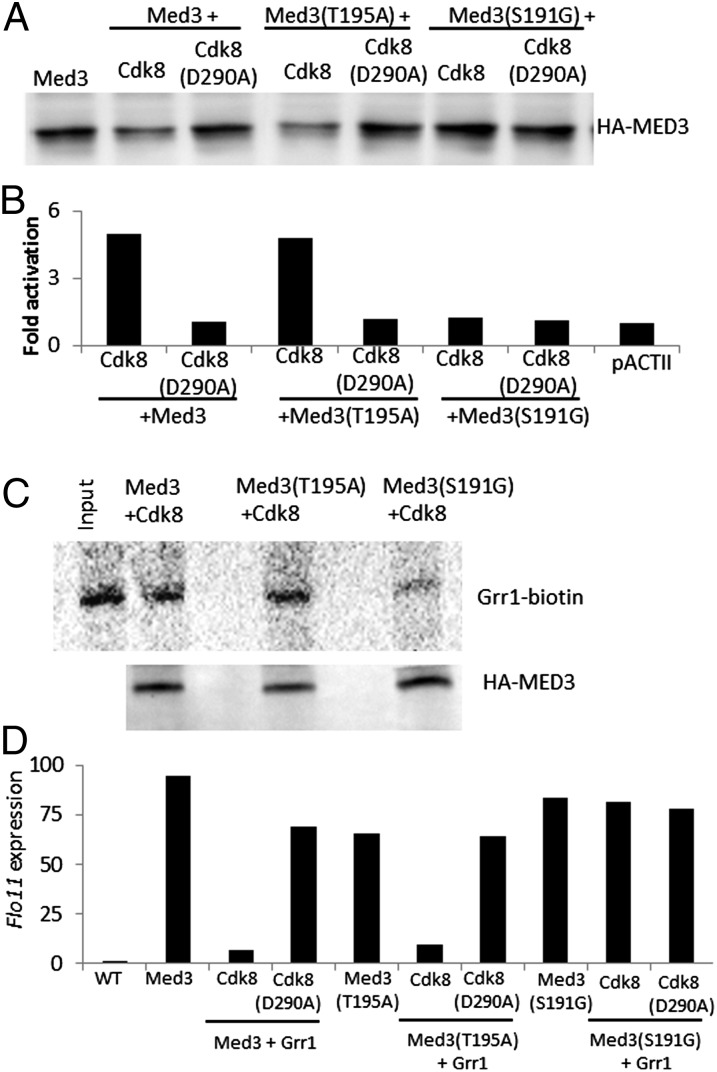
The identification of S191 and T195 as Cdk8-dependent phosphorylation targets in Med3 suggested that their phosphorylation had an important role in regulating Med3 function. S191G and T195A mutations were introduced into Med3, and any functional consequences were investigated. In the presence of GRR1 and CDK8, Med3 levels were decreased, and a similar effect was seen with Med3 containing the T195A mutation (Fig. 4A). However, levels of Med3 harboring the S191G mutation were unchanged, remaining high, suggesting that S191 phosphorylation is the key regulatory step in the modulation of Med3 function. The S191G mutation reduced the level of interaction between Med3 and Grr1 when tested both in vivo (Fig. 4B) and in vitro using biotin-labeled Grr1 and Tap-purified Med3 (Fig. 4C). Blocking the formation of this Med3-Grr1 complex would prevent the subsequent recruitment of an SCF complex to degrade Med3. The downstream effect of inhibiting the interaction between and turnover of Med3 by Grr1, through preventing S191 phosphorylation, is the maintenance of high levels of FLO11 expression by Med3 even in the presence of CDK8 and GRR1 (Fig. 4D).
Fig. 4.
S191 in Med3 is required for interaction with Grr1 and loss of Med3 stability. Med3 stability is increased following mutation on S191. (A) WT cells transformed with MED3 or MED3 (S191G) or MED3 (T195A) together with CDK8 or CDK8 (D290A). Total protein was extracted from cells, and following separation, immunoblots were probed with anti-HA for Med3. Med3 interacts with Grr1. (B) Yeast two-hybrid assay using pASGrr1 and HA-Med3 (WT or S191G/T195A variants) in the presence of Cdk8 or Cdk8 (D290A). Cells were grown to midlog phase and assayed for LacZ expression. pACTII, negative control. (C) Phosphorylation of S191 in Med3 is needed for Med3-Grr1 interaction. Biotin-labeled Grr1 was incubated with Med3 immuno-precipitated from cells expressing HA-Med3 (WT or S191G/T195A variants) and cdk8. Grr1 was visualized using streptavidin-HRP and Med3 using αHA-antibody and chemiluminescent detection. (D) Med3(S191G) cannot repress FLO11 expression. Cells were transformed with MED3 (WT or S191G/T195A variants), CDK8 or Cdk8 (D290A), and GRR1 (where indicated). Total RNA was isolated and FLO11 expression levels determined using qRT-PCR. Values are MED3 set at 100% and normalized to ACT1.
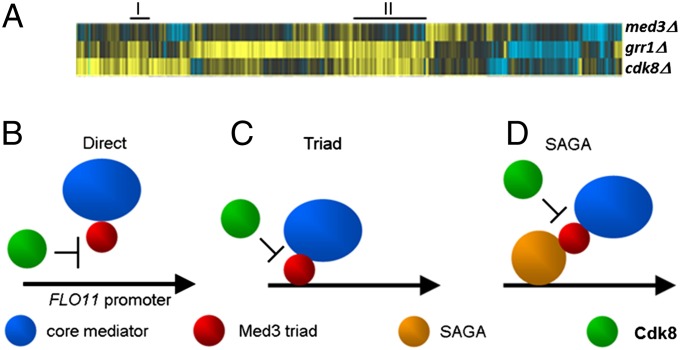
This interplay between MED3, GRR1, and CDK8 appears to be a widespread mechanism as transcriptome microarray analysis revealed a substantial set of genes that require MED3 for their expression and that are de-repressed in the absence of either GRR1 or CDK8 (Fig. 5A and Table S2).
Fig. 5.
Targets and mechanisms of Med3 Mediator regulation by Cdk8-Grr1. MED3 is required for the expression of a distinct set of gene that are repressed by CDK8 and GRR1. (A) Microarray analysis of mRNA extracted from med3Δ, cdk8Δ, and grr1Δ cells. Treeview showing all genes that changed at least 1.7-fold in any single deletion mutant vs. WT with P ≤ 0.05. Black, no significant difference in expression values between yeast strains vs. WT; yellow, up vs. WT; blue, down vs. WT. Examples of groups of genes going up in both cdk8Δ and grr1Δ and down/unchanged in med3Δ are indicated (solid black line). Phosphorylation of Med3 by Cdk8 and the subsequent degradation of Med3 is consistent with three existing models for Mediator function. (B) Phosphorylation and turnover of Med3 by Cdk8 away from a promoter prevents Mediator recruitment to target promoters. (C) Recruitment of core Mediator to a promoter by the Triad of Gal11, Med2, and Med3 is prevented when Med3 is degraded. (D) Recruitment of Mediator by SAGA is prevented by Med3 degradation, which is required for the interaction with SAGA.
Discussion
The regulation of Mediator function provides a means by which transcription can be controlled independently of chromatin modifications. Here we demonstrate a molecular mechanism for the active repression of Med3-mediated transcription where phosphorylation of Med3 by Cdk8 results in the Grr1-dependent turnover of the coactivator. Blocking this process appears to result in the formation of a Mediator complex that contains the tail module triad Med2/3/15, but lacks Med5/14/16, suggesting a specific form of Mediator with a specialized tail module, stabilized through Med3, could be recruited to specific promoters in response to Cdk8 loss or inactivation.
We propose a model in which the regulated turnover of Med3 at specific promoters is triggered through phosphorylation of Med3 by Cdk8, thereby inhibiting its activation function. This Med3-specific regulation of Mediator appears likely to be coordinated with the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex function (22, 23) as Spt8, a component of SAGA, is also required for transcription derepression by Cdk8 (Fig. 1A). This mode of SAGA-dependent Mediator-regulated activation of gene expression is one of two modalities that have emerged for transcription involving these two regulatory complexes (15).
Such a mechanism seems likely to be used by transcription corepressors as exemplified by Ssn6-Tup1 which requires Cdk8 for its repression function. Under normal growth conditions, transcription of genes would be repressed through an active mechanism involving the direct interaction between a corepressor, Cdk8, and Med3 (3) and modulation of Cdk8 activity. Transcription cannot initiate as Med3, which is essential for Mediator recruitment and/or activation function (11, 24), is marked for degradation by Cdk8 phosphorylation. Under appropriate conditions such as heat shock (25) or diauxic shift (26), the Cdk8 module becomes inactivated, through loss of the associated cyclin, leading to Mediator recruitment and transcription de-repression. The stability of Med3 is regulated by the ubiquitin ligase Grr1, because in the absence of Grr1, Med3 remains at high levels in cells expressing CDK8. Although an indirect effect involving Grr1 targeting the CDK associated cyclin cannot be ruled out (27), this is unlikely as our experiments were conducted under normal temperature conditions and in high levels of glucose, conditions in which the Cdk8-associated cyclin is known to be stable (25, 26).
The dynamic interplay between Cdk8 and Med3 is consistent with each of the different models that have been proposed for activation of gene transcription by Mediator. Where Cdk8 interaction with Mediator occurs away from the promoter (10), turnover of Med3 would prevent Mediator recruitment (Fig. 5B). Where the tail-module or its triad subcomplex associates with promoters independently from core Mediator (7), which is subsequently recruited, regulation of Med3 stability would modulate this process (Fig. 5C). Alternatively, transient interactions between Cdk8 and promoter-bound Mediator containing Med3 (11) could modulate an interaction with SAGA (via Spt8) through the degradation of Med3, explaining how the functional status of positioned yet inactive Mediator can be switched (Fig. 5D). In each model, the repression mechanism described here allows for modulation of Mediator activity (Fig. 5) and serves to explain how Cdk8 functions as a negative regulator in the repression of distinct sets of genes via different molecular mechanisms.
Materials and Methods
Yeast Strains and Plasmids.
All strains and plasmids used in this study are detailed in Tables S3 and S4.
ChIP Assays.
Assays were performed essentially as described in ref. 28, using cells transformed with e2p6MED3 (3) and pGIG2 (29) or pGIG2-3 containing a D290A point mutation in the CDK8 (Quick Change; Stratagene). Chromatin lysate was precleared with rabbit IgG (Santa Cruz Biotechnology) and incubated with either no antibody or 10 µL anti-hemagglutinin (HA) (Santa Cruz Biotechnology).
RNA Analysis.
RNA was extracted from 3 × 108 yeast cells, and 100 ng RNA was used to prime reverse transcription reactions (Ambion; RetroScript). The resulting cDNA was diluted 1:100 for analysis by real-time PCR (Biorad SYBR Green). Primer information is given in Table S5.
Recombinant Protein Synthesis.
Histidine-tagged Med3ΔSal was purified as previously described (3). Med3 was purified using either anti-tandem affinity purification (TAP) antibody (Pierce) or anti-HA (Santa Cruz Biotechnology). For interaction assays, biotinylated-Grr1 was synthesized using an in vitro Transcription/Translation system and Transcend biotinylated tRNA (TNT T7 Quick System; Promega) as previously described (5). Following SDS/PAGE and electroblotting, biotinylated Grr1 was visualized by streptavidin-HRP and chemiluminescent detection.
In Vitro Interaction and Kinase Assays.
Myc-tagged Cdk8 complexes were immunopurified from yeast strains YC7 and YC17 as described previously (8). For interaction assays, myc-Cdk8 complexes were incubated with 500 ng Med3ΔSal protein resuspended in a final volume of 10 μL kinase assay buffer containing either 500 mM ATP or AMPNP. For kinase assays, 25 μCi 33P γ-ATP was included in the reactions. Samples were incubated for 15 min at room temperature before being analyzed by SDS/PAGE and immunoblotting or autoradiography.
Ubiquitination Assays.
WT and grr1Δ yeast cells were transformed with pGIG2 and pe2p6MED3. ise1 yeast cells, used to assess 26S proteasome function (30), were treated with 100 μM MG132 (in DMSO) or DMSO for 1 h as described in SI Methods.
LC and MS.
WT and cdk8Δ Tap-Med15 strains were differentially labeled by culturing in 14N- and 15N-containing media (31). Tap-purified proteins were analyzed as detailed in SI Methods and data are given in Dataset S1.
Yeast Two-Hybrid Assays.
The yeast two-hybrid screen was conducted as previously described (5) using reporter plasmids harboring either LexA or Gal4 binding sites upstream of a minimal promoter and the LacZ reporter gene. β-galactosidase activity was determined using o-nitrophenyl galctopyranoside (OPNG; Sigma). Color development was quantified by measuring absorbance at 420 nm.
Microarray Analysis.
Array experiments and analysis were performed as previously described (32) using deletion strains from the Saccharomyces genome deletion consortium (33). Outputs were visualized using Treeview (jtreeview.sourceforge.net).
Supplementary Material
Acknowledgments
We thank Andres Aguilera, Darius Balciunas, Reymond Deshesia, Mark Johnston, Mark Solomon, Dimitris Tzamarias, and Richard Young for plasmids and strains. This work was supported by Cancer Research UK Grant C10323 (to R.S.C.). T.N. was sponsored by the European Community Action Scheme for the Mobility of University Students (ERASMUS) scheme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in www.ebi.ac.uk/arrayexpress under accession numbers E-TABM-1210 and E-TABM-1074.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307525111/-/DCSupplemental.
References
- 1.Karijolich JJ, Hampsey M. The Mediator complex. Curr Biol. 2012;22(24):R1030–R1031. doi: 10.1016/j.cub.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Herschbach BM, Arnaud MB, Johnson AD. Transcriptional repression directed by the yeast alpha 2 protein in vitro. Nature. 1994;370(6487):309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- 3.Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J Biol Chem. 2000;275(12):8397–8403. doi: 10.1074/jbc.275.12.8397. [DOI] [PubMed] [Google Scholar]
- 4.Wu R-C, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129(6):1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol. 2007;27(15):5306–5315. doi: 10.1128/MCB.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BA, Reinberg D. The mediator coactivator complex: Functional and physical roles in transcriptional regulation. J Cell Sci. 2003;116(Pt 18):3667–3675. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol. 2004;24(15):6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi Y, et al. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15(9):1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421(6919):187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 10.Hengartner CJ, et al. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2(1):43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 11.Andrau JC, et al. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22(2):179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Nemet J, Jelicic B, Rubelj I, Sopta M. The two faces of Cdk8, a positive/negative regulator of transcription [published online ahead of print October 15, 2013] Biochimie. 2013 doi: 10.1016/j.biochi.2013.10.00. [DOI] [Google Scholar]
- 13.Conlan RS, Tzamarias D. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J Mol Biol. 2001;309(5):1007–1015. doi: 10.1006/jmbi.2001.4742. [DOI] [PubMed] [Google Scholar]
- 14.Reeves WM, Hahn S. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol Cell Biol. 2003;23(1):349–358. doi: 10.1128/MCB.23.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari SA, et al. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 2012;31(1):44–57. doi: 10.1038/emboj.2011.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 17.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9(4):399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 18.Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153(4):1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.La Rue J, Tokarz S, Lanker S. SCFGrr1-mediated ubiquitination of Gis4 modulates glucose response in yeast. J Mol Biol. 2005;349(4):685–698. doi: 10.1016/j.jmb.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 20.Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18(5):1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16(18):5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18(3):333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu H, et al. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol. 2005;25(9):3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Reese JC. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J Biol Chem. 2004;279(38):39240–39250. doi: 10.1074/jbc.M407159200. [DOI] [PubMed] [Google Scholar]
- 25.Cooper KF, Strich R. Saccharomyces cerevisiae C-type cyclin Ume3p/Srb11p is required for efficient induction and execution of meiotic development. Eukaryot Cell. 2002;1(1):66–74. doi: 10.1128/EC.01.1.66-74.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95(5):717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 27.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91(2):209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 28.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399(6736):609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 29.Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23(21):4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271(44):27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 31.Gouw JW, Tops BBJ, Krijgsveld J. Metabolic labeling of model organisms using heavy nitrogen (15N) Methods Mol Biol. 2011;753:29–42. doi: 10.1007/978-1-61779-148-2_2. [DOI] [PubMed] [Google Scholar]
- 32.van de Peppel J, et al. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19(4):511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.