Significance
Liver kinase B1 (LKB1) is a serine/threonine kinase often inactivated in human cancer. We demonstrate here that loss of LKB1 expression in cancer cells promotes a progrowth metabolic profile that enables increased cell growth and proliferation. Loss of LKB1 promotes increased tumor cell metabolism through mammalian target of rapamycin complex 1- and reactive oxygen species-dependent increases in hypoxia-inducible factor-1α (HIF-1α). LKB1-null cells are dependent on HIF-1α to maintain cellular ATP and viability under poor nutrient conditions, raising the possibility of targeting HIF-1α for synthetic lethality in LKB1-deficient tumors. Together, our data reveal that regulation of cellular metabolism is a key function of LKB1 that may contribute to its tumor-suppressor function in human cancer.
Keywords: HIF-1alpha, cancer metabolism, Warburg effect, Peutz-Jeghers Syndrome, PJS, glutamine metabolism
Abstract
One of the major metabolic changes associated with cellular transformation is enhanced nutrient utilization, which supports tumor progression by fueling both energy production and providing biosynthetic intermediates for growth. The liver kinase B1 (LKB1) is a serine/threonine kinase and tumor suppressor that couples bioenergetics to cell-growth control through regulation of mammalian target of rapamycin (mTOR) activity; however, the influence of LKB1 on tumor metabolism is not well defined. Here, we show that loss of LKB1 induces a progrowth metabolic program in proliferating cells. Cells lacking LKB1 display increased glucose and glutamine uptake and utilization, which support both cellular ATP levels and increased macromolecular biosynthesis. This LKB1-dependent reprogramming of cell metabolism is dependent on the hypoxia-inducible factor-1α (HIF-1α), which accumulates under normoxia in LKB1-deficient cells and is antagonized by inhibition of mTOR complex I signaling. Silencing HIF-1α reverses the metabolic advantages conferred by reduced LKB1 signaling and impairs the growth and survival of LKB1-deficient tumor cells under low-nutrient conditions. Together, our data implicate the tumor suppressor LKB1 as a central regulator of tumor metabolism and growth control through the regulation of HIF-1α–dependent metabolic reprogramming.
Although unchecked cell proliferation and aberrant survival are hallmark features of cancer, tumor cells must also engage pathways of cellular metabolism to generate the energy and biosynthetic intermediates required to support increased cell division (1). To meet increased energetic and biosynthetic demand, cancer cells often display fundamental changes in their cellular metabolism, including a switch to aerobic glycolysis, a phenomenon known as the “Warburg effect” (2). Increased use of glutamine (“glutaminolysis”) for mitochondrial-dependent ATP production and cellular biosynthesis is also a key feature of many tumor cells (3).
Many of the predominant driver mutations observed in cancer alter tumor-cell metabolism as part of their mode of action (4). For example, loss of the tumor suppressor PTEN can promote increased glucose uptake through elevated PI3K/Akt/mTOR signaling (5) while loss of the Von-Hippel-Lindau (VHL) tumor suppressor promotes a similar metabolic phenotype through stabilization of the hypoxia inducible factor (HIF)-1α (6). HIF-1α and HIF-2α are transcription factors whose activity is regulated by oxygen availability. HIF-1α and HIF-2α protein expression is normally stabilized only under hypoxic conditions; however, the HIFs are commonly expressed in human cancers even in the absence of hypoxia (7). Importantly, elevated expression of both HIF-1α and HIF-2α has been demonstrated in many cases of non-small cell lung cancer (NSCLC) (8), and HIF-2α has been linked to poor prognosis in lung-cancer patients (9).
The liver kinase B1 (LKB1) is a serine/threonine kinase encoded by STK11, the tumor suppressor gene responsible for Peutz-Jeghers Syndrome (PJS) (10, 11). LKB1 is a unique serine-threonine kinase, in that inactivation, rather than activation, of its kinase activity is associated with tumorigenesis. Somatic STK11 mutations are associated with a number of human cancers including lung, breast, and cervical cancer (12–15), and genetic ablation of LKB1 in mice promotes tumorigenesis in a variety of tissues (16). LKB1 is involved in a diverse array of cellular processes, including cell polarity, apoptosis, and cell growth (17, 18). All these processes play a role in cancer initiation and progression, and as such their relative contribution to LKB1-mediated tumor suppression remains unclear.
Although LKB1 is widely accepted as a regulator of cell growth control, the impact of LKB1 on tumor metabolism has remained unclear. Benign tumors haploinsufficient for LKB1 can be visualized using 18F-deoxyglucose-positron emission tomography (FDG-PET) imaging (19), suggesting that loss of LKB1 can promote increased glucose uptake by tumor cells. LKB1 may also influence ATP consumption by limiting mTORC1-dependent mRNA translation (20, 21). In this study, we have characterized the impact of LKB1 loss on cellular metabolism in both transformed and nontransformed cells. We find that silencing LKB1 in tumor cells increases glucose and glutamine consumption and promotes a metabolic switch to aerobic glycolysis. We demonstrate that HIF-1α drives the metabolic shift induced by LKB1 loss and that ablation of HIF-1α reverses the metabolic advantage of LKB1-deficient cells. Together, our data implicate LKB1 loss as a key regulator of tumor-cell metabolism and growth through regulation of HIF-1α–dependent metabolic reprogramming.
Results
Loss of LKB1 Promotes Enhanced Glucose and Glutamine Metabolism.
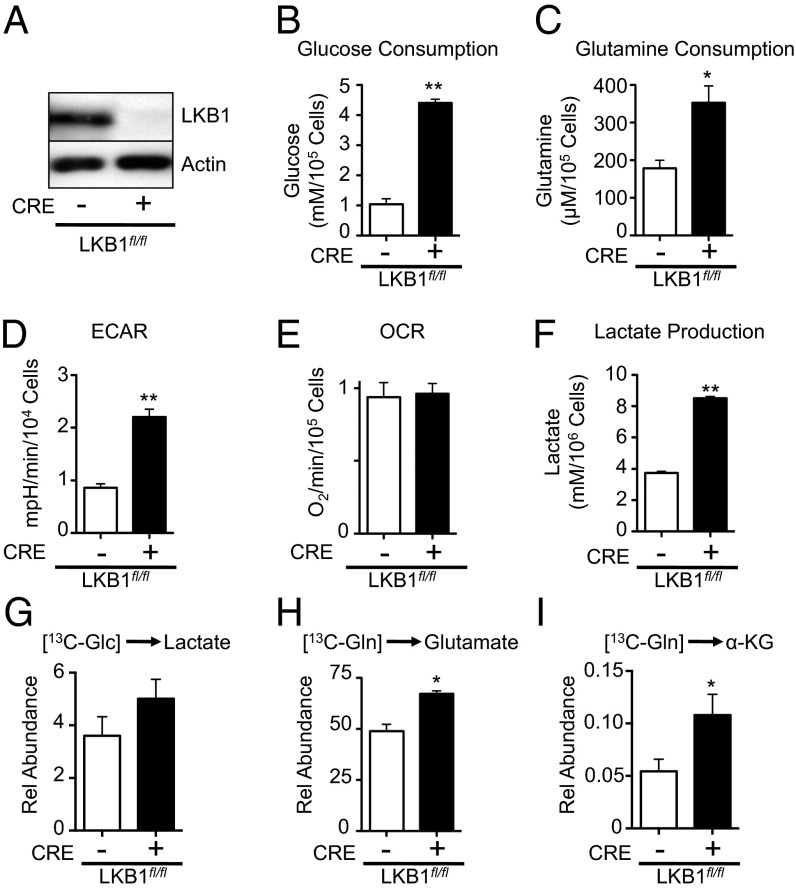
To examine the metabolic consequences of LKB1 loss, we manipulated LKB1 expression in mouse embryonic fibroblasts (MEFs) harboring a conditional mutation in the stk11 gene (LKB1fl/fl). We used Cre recombinase to generate isogenic MEFs expressing (Cre−) or lacking (Cre+) LKB1 expression (Fig. 1A) and then examined the effect of LKB1 loss on nutrient uptake. LKB1-deficient MEFs displayed increased glucose (Fig. 1B) and glutamine consumption (Fig. 1C) relative to control cells expressing LKB1. We next measured the basal extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) for control or LKB1-deficient MEFs using a flux analyzer (22). Cells lacking LKB1 displayed a twofold increase in ECAR (Fig. 1D), with no significant change in oxygen consumption (Fig. 1E). This change in ECAR displayed by LKB1-deficient MEFs correlated with increased lactate production by these cells (Fig. 1F).
Fig. 1.
Loss of LKB1 promotes enhanced glucose and glutamine metabolism. (A) LKB1 immunoblot on lysates from LKB1fl/fl MEFs transduced with control retrovirus (Cre−) or a retrovirus expressing Cre recombinase (Cre+). (B and C) Glucose and glutamine consumption by LKB1-deficient MEFs. LKB1fl/fl MEFs expressing empty vector (open bar) or Cre recombinase (filled bar) were grown for 72 h, and glucose consumption (B) and glutamine consumption (C) were determined by enzymatic assay. (D and E) ECAR (D) and OCR (E) for LKB1fl/fl MEFs with (+) or without (−) Cre expression. (F) Lactate production by LKB1-deficient MEFs. Cells were treated as in B, and extracellular lactate in the culture medium was measured via enzymatic assay. (G–I) Metabolic processing of glucose and glutamine by LKB1-null MEFs. LKB1-null (filled bar) or control (open bar) MEFs were pulsed with 13C-glucose or 13C-glutamine for 1 h, and 13C incorporation into lactate (G), glutamate (H), and α-ketoglutarate (I) was determined by GC-MS. *P < 0.05; **P < 0.01.
We next cultured control or LKB1-null MEFs with uniformly labeled (U-13C) glucose or glutamine, and examined the total 13C contribution of these carbon sources to intracellular metabolite pools. Cells lacking LKB1 (Fig. 1G, filled bar) displayed a slight increase in intracellular lactate levels derived from glucose (Fig. 1G). The strong increase in ECAR (Fig. 1D) and extracellular lactate (Fig. 1F) associated with LKB1 loss suggests that intracellular lactate is rapidly exported once generated. Using 13C-glutamine, we observed increased glutamine conversion to glutamate and α-ketoglutarate (Fig.1 H and I), suggesting an increase in glutaminolysis in LKB1-deficient MEFs.
LKB1-Deficient Tumor Cells Display Enhanced Glycolytic and TCA Cycle Flux.
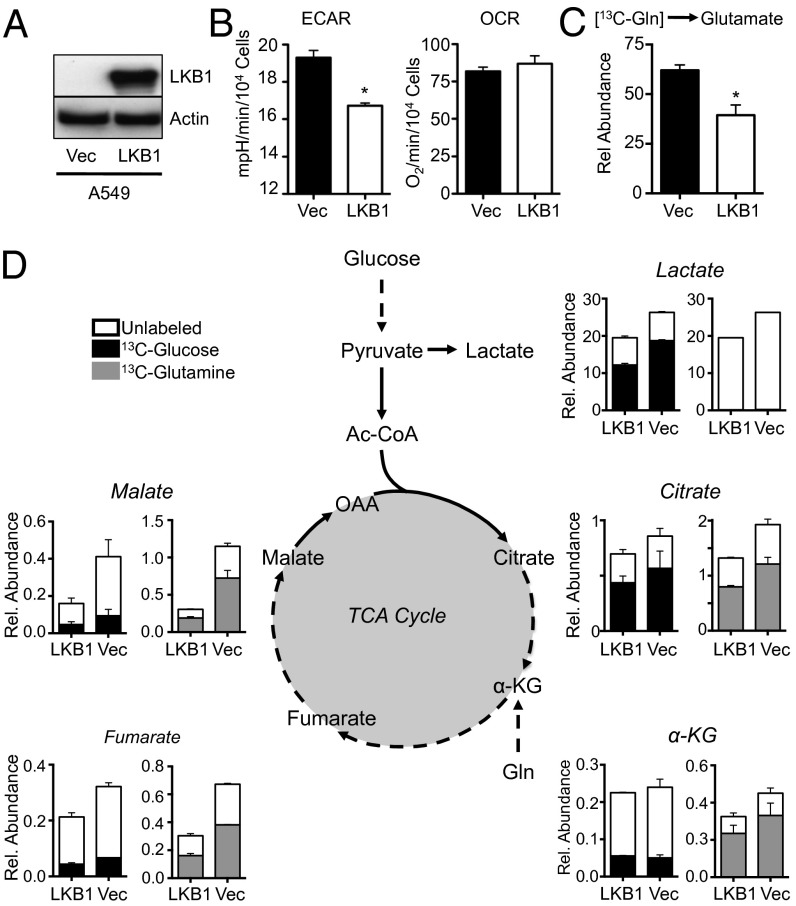
To investigate the role of LKB1 loss on tumor-cell metabolism, we examined the metabolic activity of A549 NSCLC cells, which naturally lack LKB1 expression (Fig. 2A and Fig. S1A). Reexpression of LKB1 in A549 cells (A549/LKB1) promoted an ∼20% decrease in ECAR relative to control cells lacking LKB1 expression (A549/Vec) whereas OCR was unaffected (Fig. 2B). A549 cells reexpressing LKB1 also displayed reduced production of glutamine-derived glutamate compared with control cells, consistent with a reduction in glutaminolysis in these cells (Fig. 2C). We observed similar effects of LKB1 expression on glycolysis using an independent LKB1-deficient NSCLC cell line (A427) (23). A427/Vec cells lacking LKB1 displayed an ∼twofold higher ECAR relative to A427 cells reexpressing LKB1 (Fig. S1B).
Fig. 2.
LKB1-deficient tumor cells display enhanced glycolytic and TCA cycle flux. (A) LKB1 immunoblot on lysates from A549 cells transduced with empty vector (Vec) or LKB1 cDNA. (B) ECAR and OCR of A549 cells expressing empty vector (filled bar) or LKB1 cDNA (open bar). (C) Intracellular glutamate levels derived from 13C-glutamine in A549 cells expressing empty vector (Vec, filled bar) or LKB1 (LKB1, open bar) as measured by GC-MS. (D) Metabolic flux analysis of LKB1-deficient A549 cells. A549 cells expressing empty vector (Vec) or LKB1 cDNA (LKB1) were pulsed with 13C-glucose or 13C-glutamine for 1 h, and 13C incorporation into lactate and TCA cycle metabolites were determined by GC-MS. Relative incorporation of 13C into total metabolite pools is indicated by shaded bars for glucose (black) and glutamine (gray). Metabolite abundance is expressed relative to basal levels in A549/LKB1 cells. *P < 0.05.
We next examined the metabolic fate of glucose and glutamine in A549 cells using 13C-labeled glucose and glutamine. A549/Vec cells lacking LKB1 displayed an increase in the total abundance of metabolites derived from both glycolysis (lactate) and the tricarboxylic acid (TCA) cycle (citrate, alpha-ketoglutarate, fumarate, and malate) relative to A549 cells reexpressing LKB1 (A549/LKB1) (Fig. 2D). Moreover, the proportion of these metabolites containing 13C label was also elevated in LKB1-null A549 cells. A549 cells displayed enrichment of 13C-glucose carbon in lactate (Fig. 2D, black bars), consistent with the increased glycolytic activity of these cells. However, the relative proportion of 13C-glucose–derived carbon in TCA metabolites downstream of citrate was low for both cell lines, despite the total metabolite levels remaining higher in A549 cells lacking LKB1. The decreased labeling from glucose in TCA cycle metabolites was compensated for by an increase in glutamine-derived carbons entering the TCA cycle at alpha-ketoglutarate, providing the majority of carbon pools for the TCA cycle (Fig. 2D, gray bars). Again, LKB1-deficient A549 cells displayed increased labeling from 13C-glutamine in alpha-ketoglutarate, fumarate, malate, and citrate (Fig. 2D, gray bars).
Although overall lactate and TCA cycle metabolite abundance, as well as flux into these pathways from glucose or glutamine, was elevated in LKB1-null cells, the relative distribution of mass isotopomers in the metabolite pools did not vary dramatically in A549 cells regardless of LKB1 status. One consistent difference was a small elevation in glutamine-dependent reductive carboxylation in A549 cells lacking LKB1, which was apparent by the slight increase in m+5 citrate production from 13C-glutamine (Fig. S2A). Similar trends in metabolite abundance and labeling patterns from glucose and glutamine were observed in LKB1-deficient MEFs relative to controls (Fig. S1B). Thus, under normal growth conditions, loss of LKB1 in tumor cells appears to enhance glucose and glutamine flux but does not dramatically alter the metabolic fate of these carbon sources.
LKB1-Null Cells Display Enhanced Growth and Biosynthetic Capacity.
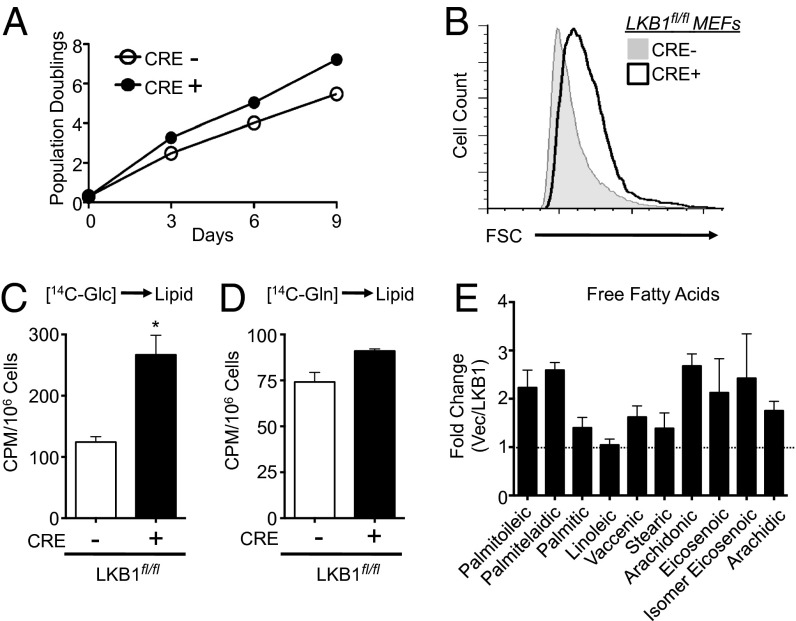
Given the observation of increased carbon flow of glucose and glutamine in LKB1-null cells, we examined the impact of LKB1 loss on proliferation and de novo lipid biosynthesis. LKB1-deficient MEFs displayed increased rates of proliferation (Fig. 3A) and displayed a 15% increase in cell size (Fig. 3B) relative to control cells. Given the enhanced carbon flow from glucose and glutamine observed in LKB1-null cells, we measured levels of glucose- and glutamine-derived lipid synthesis in these cells. MEFs were pulsed with radioactively labeled (14C) glucose or glutamine, and 14C-labeling in lipids was measured. LKB1-null cells displayed a twofold increase in glucose-dependent lipid biosynthesis relative to control cells (Fig. 3C) whereas no appreciable difference in 14C-glutamine incorporation to fatty acids was observed between cell lines (Fig. 3D). We next used GC-MS to measure the total abundance of fatty acids in LKB1-deficient cells. The overall abundance of several fatty acid species was increased when LKB1 was absent in A549 cells (Fig. 3E). Cumulatively, these data indicate that loss of LKB1 enhances biosynthetic pathways that support cell growth and proliferation.
Fig. 3.
LKB1-null cells display enhanced growth and biosynthetic capacity. (A) Growth curve of LKB1fl/fl MEFs expressing empty vector (CRE−, open circle) or Cre recombinase (CRE+, filled circle) following a 3T3 passage protocol. (B) Size of control (Cre−, gray histogram) or LKB1-deficient (Cre+, open histogram) MEFs as determined by forward scatter (FSC) of cells via flow cytometry. (C and D) Glucose- and glutamine-dependent lipid biosynthesis by LKB-null MEFs. Control (Cre−,open bar) or LKB1-null (Cre+, closed bar) MEFs were incubated with uniformly labeled 14C-glucose (C) or 14C-glutamine (D) for 72 h, and radioactive counts in extracted lipids were measured. Data are expressed as cpm per 106 cells (mean ± SEM) for samples in triplicate. (E) Free fatty acid (FFA) levels in LKB1-null cancer cells. FFAs in cell extracts from A549/Vec or A549/LKB1 cells were measured by GC-MS following 3 d of growth. Data are expressed as the ratio of FFA species in A549/Vec to A549/LKB1 cells. *P < 0.05.
LKB1 Deletion Promotes HIF-1α Protein Expression in Cancer Cells Under Normoxia.
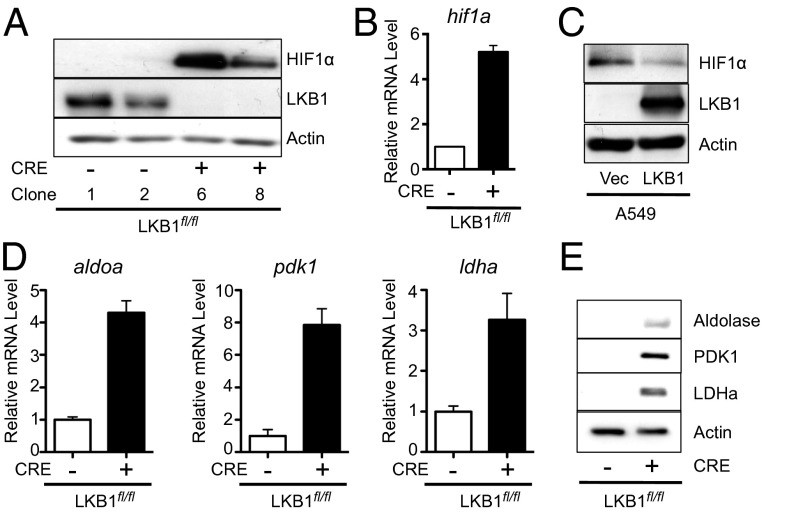
It has previously been shown that LKB1-deficient MEFs display enhanced HIF-1α protein levels under normoxia (19). Acute deletion of LKB1 in MEFs also resulted in increased HIF-1α protein levels under normoxic conditions (Fig. 4A). HIF-1α mRNA levels were also elevated fivefold in LKB1-deficient MEFs relative to isogenic controls (Fig. 4B). Similar to LKB1-null MEFs, A549 cells lacking LKB1 displayed elevated levels of HIF-1α protein expression under normoxia, which was reduced by reexpression of LKB1 (Fig. 4C). Similarly, HIF-1α protein was detectable in A427 cells under normoxic conditions and was reduced upon ectopic expression of LKB1 in these cells (Fig. S3A). Furthermore, reducing LKB1 expression by siRNA treatment promoted an increase in HIF-1α protein levels in U20S cells (Fig. S2B) and HCT116 cells (Fig. S3C). We next examined the expression of several HIF-1α target genes involved in metabolic control, assessing both mRNA (Fig. 4D) and protein (Fig. 4E) levels. The expression levels of Aldolase A, pyruvate dehydrogenase kinase 1 (PDK1), and lactate dehydrogenase A (LDHA) were all specifically elevated in LKB1-null MEFs (Fig. 4E).
Fig. 4.
LKB1 loss promotes HIF-1α protein expression under normoxic conditions. (A) Immunoblot for HIF-1α protein expression in whole-cell lysates from control (Cre−) or LKB1-null (Cre+) MEFs grown under 20% O2. (B) Relative expression of hif1a mRNA by control (Cre−, open bar) or LKB1-null (Cre+, filled bar) MEFs as determined by qPCR. Data were expressed relative to actin mRNA levels for triplicate samples and normalized relative to control (Cre−) cells. (C) Immunoblot of HIF-1α protein in lysates from A549/Vec or A549/LKB1 cells grown under 20% O2. (D) Relative expression of aldoa, ldha, and pdk1 mRNA levels in control (Cre-, open bar) or LKB1-null (Cre+, filled bar) MEFs as determined by qPCR. (E) Immunoblot for Aldolase, PDK1, and LDHA expression in lysates from cells as in D.
LKB1-Dependent HIF1α Expression Is Regulated by mTORC1 and ROS.
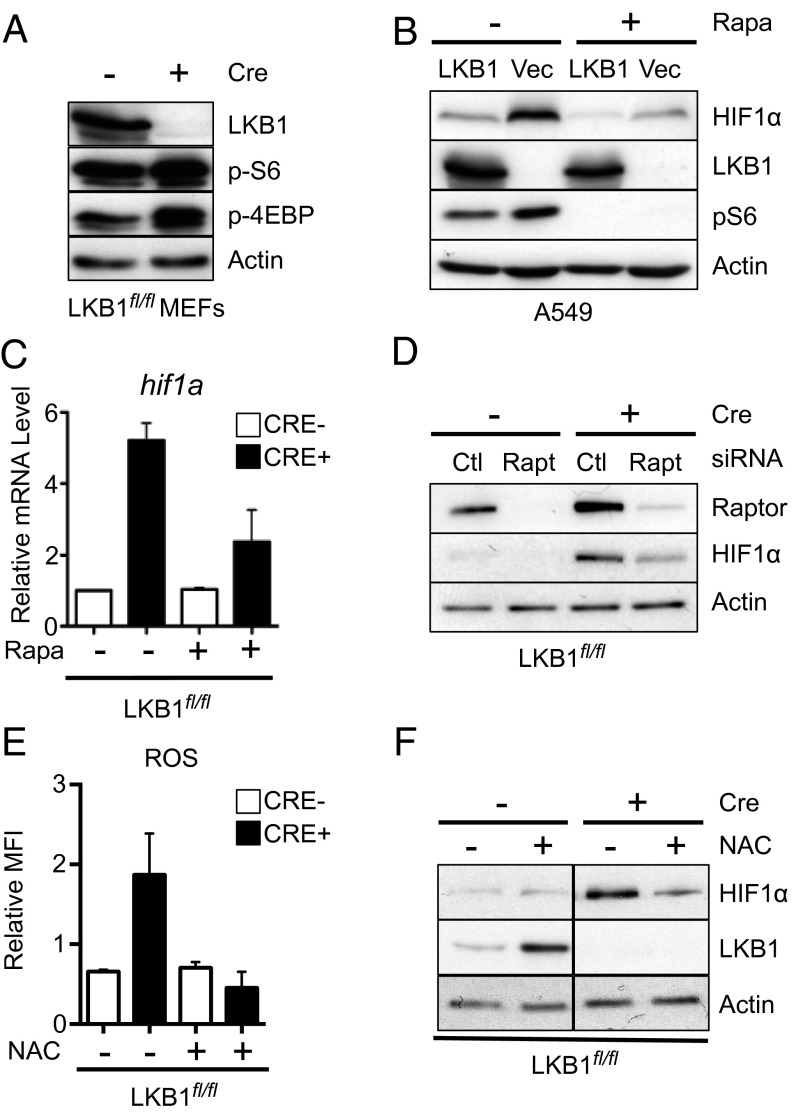
Aberrant mTOR signaling has been linked to deregulated HIF-1α protein expression under normoxic conditions (19, 24). Consistent with previous reports (21), acute deletion of LKB1 in MEFs promoted heightened activation of mTORC1 signaling marked by increased rS6 phosphorylation and hyperphosphorylation of 4E-BP1 (Fig. 5A). A549 cells lacking LKB1 also displayed increased rS6 phosphorylation (Fig. 5B). To test whether elevated HIF-1α protein levels in LKB1-null cells were supported by mTOR activity, we treated cells with the mTORC1 inhibitor rapamycin and measured HIF-1α protein levels in cell lysates. Rapamycin treatment reduced HIF-1α protein expression in LKB1-deficient A549 cells under normoxic conditions (Fig. 5B). Moreover, HIF-1α mRNA levels in LKB1-null cells were reduced in response to rapamycin treatment (Fig. 5C). Finally, we examined HIF-1α protein expression in LKB1-deficient MEFs with specific ablation of mTORC1 signaling by reducing expression of the mTORC1 complex component Raptor using RNAi. Similar to rapamycin, knockdown of Raptor ablated normoxic HIF-1α protein expression in LKB1-null MEFs (Fig. 5D).
Fig. 5.
LKB1-dependent HIF1α expression is regulated by mTORC1 and ROS. (A) Immunoblot for LKB1, pS6, p4EBP, and actin protein levels in whole-cell lysates from control (Cre−) or LKB1-null (Cre+) MEFs. (B) Immunoblot for HIF-1α protein levels in A549/Vec or A549/LKB1 cells cultured with (+) or without (−) 25 nM rapamycin for 24 h before cell lysis. Levels of LKB1, pS6, and actin are shown. (C) Relative HIF-1α mRNA expression in MEFs cells from control (Cre−) or LKB1-deficient (Cre+) MEFs treated with 25 nM rapamycin or vehicle control for 24 h. (D) Immunoblot for Raptor and HIF-1α protein levels in whole-cell lysates from control (Cre−) and LKB1-null (Cre+) MEFs treated with control (Ctl) or Raptor-specific (Rapt) siRNA. (E) Relative mean fluorescence intensity (MFI) of DFC-DA staining in LKB1fl/fl cells with (+) or without (−) Cre expression. Cells were treated with or without 10mM N-acetyl cysteine (NAC) for 1 h before ROS measurements. (F) Representative immunoblot of HIF-1α protein expression for cells treated as in E.
Elevated levels of mitochondrial reactive oxygen species (ROS) have been shown to promote increased HIF-1α activity (25–28). Consistent with recent results in LKB1-null tumor cells (23), LKB1-deficient MEFs displayed an ∼twofold increase in ROS levels that could be reduced via addition of the ROS scavenger N-acetyl-cysteine (NAC) (Fig. 5E). The addition of NAC also reduced HIF-1α protein levels in LKB1-null MEFs back to levels observed in control MEFs (Fig. 5F). Together, these data suggest that both mTOR signaling and cellular ROS levels contribute to increased HIF-1α protein expression in cells lacking LKB1.
HIF-1α Drives the Metabolic Phenotype Induced by LKB1 Loss.
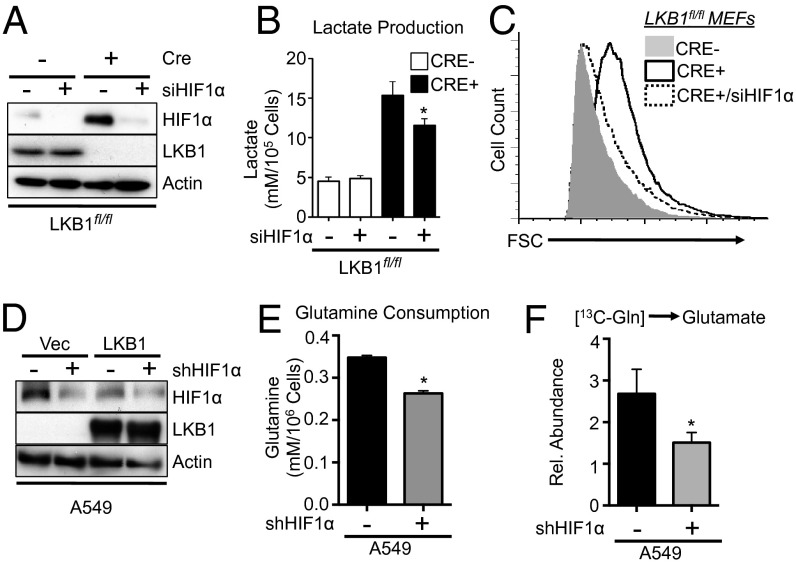
HIF-1α has well-established roles in redirecting metabolism in response to stress (29). To assess the contribution of HIF-1α to the metabolic phenotypes induced by LKB1 loss, we used siRNA to knock down HIF-1α protein expression in LKB1-null MEFs (Fig. 6A). Knockdown of HIF-1α had no effect on the level of lactate production by control cells but specifically reduced the level of lactate produced by MEFs lacking LKB1 (Fig. 6B). Reductions in lactate production were also observed in LKB1-null MEFs (Fig. S4A) and A549 cells (Fig. S4B) treated with rapamycin. Reducing HIF-1α expression decreased the size of LKB1-deficient MEFs, restoring cell size to control levels (Fig. 6C). Next we reduced HIF-1α expression in A549 cells via stable expression of shRNA specific for HIF-1α (Fig. 6D). Knockdown of HIF-1α in A549 cells lacking LKB1 promoted an ∼30% decrease in glutamine consumption by these cells (Fig. 6E). Glutaminolysis, as measured by 13C-glutamine conversion to 13C-glutamate, was similarly reduced in A549 cells when HIF-1α signaling was ablated (Fig. 6F).
Fig. 6.
HIF-1α promotes the metabolic program induced by LKB1 loss. (A) Immunoblot of HIF-1α protein expression in lysates from control (Cre−) or LKB1-deficient (Cre+) MEFs treated with control or HIF-1α siRNA. LKB1 and actin levels are shown. (B) Lactate production by cells treated as in A after 72 h of growth. (C) Forward scatter (FSC) of control (gray histogram), LKB1-deficient (open histogram), or LKB1-deficient MEFs expressing HIF-1α siRNA (hatched histogram). (D) Immunoblot of HIF-1α protein levels in lysates from A549/Vec or A549/LKB1 cells expressing control (−) or HIF-1α–specific (+) shRNAs. LKB1 and actin levels are shown. (E) Glutamine consumption by A549 cells expressing control (black bar) or HIF-1α–specific (gray bar) shRNAs as determined by enzymatic assay. (F) Glutamine-derived glutamate levels in A549 cells expressing control (−) or HIF-1α–specific (+) shRNA. 13C incorporation into intracellular glutamate following 1 h of culture with 13C-glutamine was determined by GC-MS. *P < 0.05.
HIF-1α Promotes the Growth and Survival of LKB1-Deficient Cells Under Conditions of Nutrient Limitation.
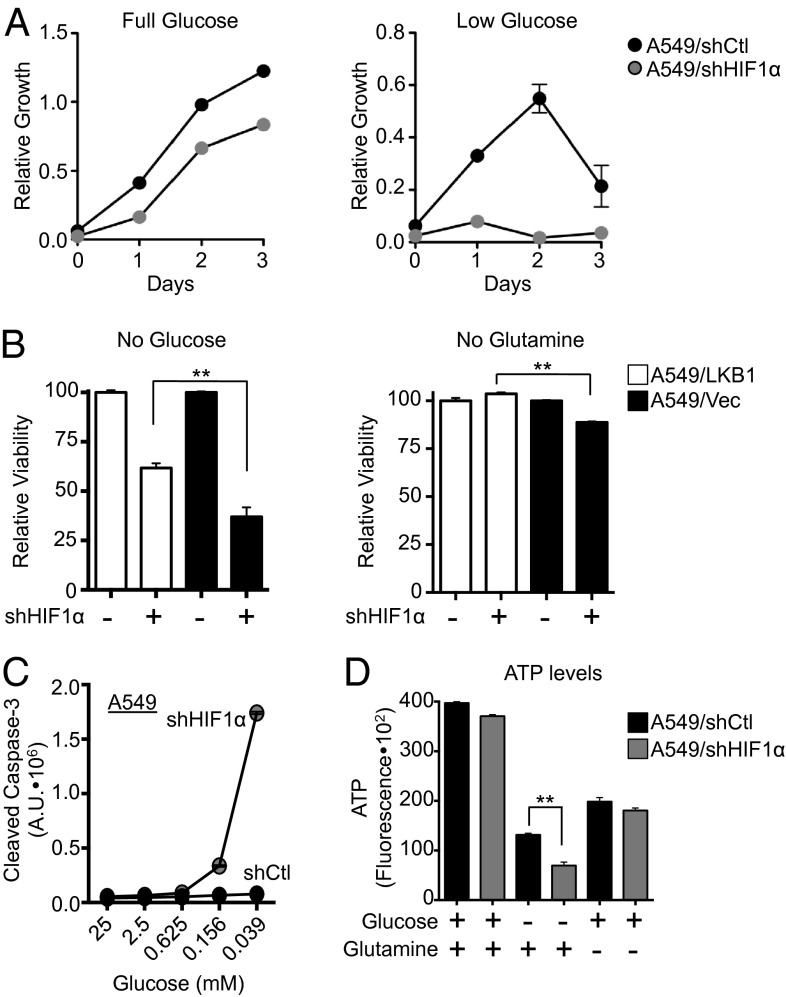
Data presented in Figs. 1–3 indicate that loss of LKB1 promotes increased nutrient acquisition and processing, and ultimately increased cell growth. Given the importance of HIF-1α in directing metabolism and bioenergetics in the absence of LKB1, we next assessed the requirement of HIF-1α in regulating the growth and survival of LKB1-deficient tumor cells. A549 cells expressing control or HIF-1α shRNAs were grown under full (25 mM) or low (0.04 mM) glucose conditions, and cell counts were measured over 72 h. A549 cells lacking HIF-1α displayed a slight reduction in proliferative rate compared with control cells under full-glucose conditions (Fig. 7A, Left). However, under low-glucose conditions, the proliferative capacity of A549 cells expressing HIF-1α shRNA was significantly impaired (Fig. 7A, Right).
Fig. 7.
HIF-1α is required for the growth and survival of LKB1-deficient cells in response to nutrient limitation. (A) Growth curves of A549 cells expressing control (black circle) or HIF-1α (gray circle) shRNA grown under full (25 mM) or low (0.4 mM) glucose conditions. (B) Viability of A549/LKB1 (white bars) or A549/Vec (black bars) cells expressing control (−) or HIF-1α–specific (+) shRNA following culture in glucose- or glutamine-free media. Cell viability was measured after 48 h by propidium iodide uptake. (C) Caspase-3 activation in A549 cells expressing control (Vec, black circles) or HIF-1α–specific (gray circles) shRNA following culture in decreasing concentrations of glucose. (D) Relative ATP levels of A549 cells expressing control (black bars) or HIF-1α–specific (gray bars) shRNA following culture in glucose- or glutamine-free media. **P < 0.01.
To further assess the dependence of A549 cells on HIF-1α for cell growth, we measured the proliferation of A549/Vec or A549/shHIF-1α cells under low-glucose conditions along with serum starvation and/or hypoxia (1% O2). A549 cells cultured under low glucose displayed considerable blocks in cell growth when serum or oxygen was limiting (Fig. S5A). Interestingly, A549 cells lacking HIF-1α displayed increased sensitivity to combined glucose and serum starvation, but not glucose starvation combined with hypoxia (Fig. S5A). In addition, A549 cells expressing HIF-1α shRNA displayed reduced viability under glucose and glutamine withdrawal relative to A549 cells expressing control shRNA (Fig. 7B). A549 cells lacking HIF-1α displayed increased caspase-3 activation at low-glucose concentrations (Fig. 7C), indicating the induction of apoptosis in these cells.
To investigate whether the increased apoptosis of A549 cells lacking HIF-1α was due to defects in cellular bioenergetics, we characterized the bioenergetic profile of LKB1-deficient tumor cells lacking HIF-1α. A549 cells with reduced HIF-1α displayed a modest increase in oxygen consumption under low-glucose conditions (Fig. S5B). However, these cells also displayed a 50% reduction in spare respiratory capacity (SRC) (Fig. S5C), suggesting reduced mitochondrial fitness in these cells (30). We also measured the ATP content of A549 cells with or without HIF-1α expression (Fig. 7D). Under basal growth conditions (25 mM glucose, 4 mM glutamine), silencing HIF-1α had little effect on cellular ATP levels. However, following overnight glucose withdrawal, A549 cells expressing HIF-1α shRNA displayed a significant drop in cellular ATP levels relative to control tumor cells. Although glutamine starvation also stimulated a decrease in cellular ATP levels, this drop appeared to be HIF-1α-independent (Fig. 7D). Together, these data suggest that LKB1-null tumor cells require HIF-1α to maintain mitochondrial respiratory capacity, ATP levels, and cell viability in response to nutrient limitation.
Discussion
In this study, we provide evidence that the tumor suppressor LKB1 promotes a metabolic checkpoint that regulates carbon utilization in proliferating cells. To date, the main link between LKB1 and tumor metabolism has been the observation of increased FDG-PET signal in vivo in benign LKB1+/− tumors (19). Here, we show that silencing LKB1 is sufficient to promote both aerobic glycolysis and glutaminolysis (Figs. 1 and 2) and that this increase in glucose and glutamine metabolism fuels cell growth and lipid biosynthesis in cells lacking LKB1 (Fig. 3). The progrowth metabolic program induced by LKB1 loss is mediated by the transcription factor HIF-1α, which displays increased protein stabilization under normoxia when LKB1 is deleted (Fig. 4). We find that the metabolic and biosynthetic phenotypes of LKB1-null cells are dependent upon HIF-1α (Fig. 6) and that targeting HIF-1α impairs the growth and survival of LKB1-deficient tumor cells (Fig. 7). This work highlights the existence of a metabolic circuit regulated by HIF-1α that coordinates cellular bioenergetics when LKB1 activity is suppressed.
Our data suggest that LKB1 loss disrupts normal metabolic homeostasis in cells, which paradoxically has a net positive effect on cell growth and proliferation. Glucose-derived citrate is a key intermediate in lipid biosynthesis (31). Flux of glucose-derived pyruvate into citrate is enhanced by LKB1 loss, as is glucose-dependent lipid biosynthesis and overall lipid content. This change occurs despite HIF-1α-dependent elevation of PDK1, which has been shown to negatively regulate pyruvate entry into the mitochondrion under hypoxic conditions (32, 33). Interestingly, although glutaminolysis is increased in LKB1-deficient cells, LKB1 loss did not appear to promote increased glutamine-dependent lipid biosynthesis or the differential use of glutamine in pathways such as reductive carboxylation of alpha-ketoglutarate (34–36). LKB1-null cells appear to use glutamine as an anaplerotic substrate to support mitochondrial metabolism.
Our work here identifies HIF-1α as a key mediator of the metabolic transformation triggered by LKB1 loss. Using multiple cell systems, we demonstrate that acute down-regulation of LKB1 is sufficient to increase HIF-1α protein levels under normoxic conditions. Reducing HIF-1α levels reverses the metabolic effects triggered by LKB1 loss in cells (Fig. 6). We show here that targeting the mTORC1 complex, either by using rapamycin or through Raptor knockdown, reduces HIF-1α protein expression in LKB1-null cells, suggesting that deregulated mTORC1 activity links LKB1 loss to elevated HIF-1α activity. However, we also demonstrate that elevated ROS levels may contribute to HIF-1α protein expression in LKB1-null cells. LKB1 loss has previously been reported to promote enhanced levels of intracellular ROS in A549 cells (23). Here, we observe a similar trend in MEFs lacking LKB1. Reducing ROS levels with NAC abrogated the increase in HIF-1α levels in LKB1-null cells. It is unclear whether these two systems (mTORC1 and ROS) work separately or in concert to affect HIF-1α protein expression. One possibility is that increased metabolic activity of LKB1-deficient cells is driven by mTORC1 and that mitochondrial ROS generated as a consequence of this increased metabolic activity promotes HIF-1α expression, thus reinforcing the progrowth metabolic program induced by LKB1 deletion.
Disruption of the downstream LKB1 effectors AMPK (37) or TSC2 (24) promotes elevated mTORC1 activity and increased HIF-1α protein levels under normoxia. We have recently demonstrated that loss of AMPK activity is sufficient to promote the Warburg effect in tumor cells (37), suggesting that LKB1 may be linked to metabolic control through its upstream regulation of AMPK (16). However, LKB1 and AMPK appear to influence HIF-1α protein expression through different mechanisms. Silencing LKB1 promotes both increased transcription and translation of HIF-1α, events which are sensitive to mTORC1 inhibition. In contrast, loss of AMPK results in increased HIF-1α protein levels with no discernible changes in mRNA levels (37). Moreover, mTORC1 inhibition has little effect on HIF-1α protein levels when AMPK is silenced (37). These data suggest the existence of both AMPK-dependent and -independent mechanisms linking LKB1 to HIF-1α and metabolic reprogramming.
Our observation that LKB1 loss promotes a progrowth metabolic profile in tumor cells raises the prospect that there may be selective pressure for tumors to lose or silence LKB1-AMPK signaling (38), as suggested by the frequent inactivation of LKB1 in NSCLC (14). We speculate that the metabolic effects of LKB1 inactivating mutations may also synergize with other genetic lesions, ultimately favoring the selection of tumor cells with distinct metabolic advantage. For example, oncogenic K-ras mutations (G12D) in pancreatic ductal carcinoma have been shown to redirect glucose metabolism to fuel increased pentose phosphate shunt activity and ribose biosynthesis (39). Interestingly, comutation of LKB1 and K-ras is frequently observed in NSCLC (40), and LKB1 inactivating mutations synergize with oncogenic K-ras to accelerate tumorigenesis in mouse models of lung cancer (41). Thus, LKB1 loss may augment the metabolic activities of other driver mutations in cancer by enhancing their ability to promote nutrient acquisition and utilization by tumor cells. However, although loss of LKB1 reprograms cancer-cell metabolism, it also confers a dependence on HIF-1α, rendering LKB1-null tumor cells more susceptible to apoptosis under poor nutrient conditions. This observation raises the possibility of targeting HIF-1α for synthetic lethality in LKB1-deficient tumors. Given that mTORC1 inhibition affects both aberrant mTORC1 signaling and HIF-1α expression in LKB1-deficient cancer cells, mTORC1-targeting compounds may be particularly effective for treating tumors with somatic LKB1 mutations or cancers associated with PJS.
Materials and Methods
Full methods are available as SI Materials and Methods. Primary mouse embryonic fibroblasts (MEFs) conditional for stk11 (LKB1-fl/fl) were generated by timed mating and immortalized with SV40 Large T Antigen as previously described (42). A549 and A427 NSCLC cell lines expressing LKB1 have been previously described (23). OCR and ECAR were measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience), and GC-MS analysis of metabolites was conducted using established protocols (37). Statistics were determined using paired Student t test, ANOVA, or Log-rank (Mantel–Cox) test using Prism software (GraphPad). Statistical significance is represented in figures as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
Acknowledgments
We acknowledge Ralph DeBerardinis, Arnim Pause, and members of the R.G.J. laboratory for critical reading of this manuscript. We acknowledge Marie-Claude Gingras, as well as Gäelle Bridon and Luc Choinière of the Goodman Cancer Research Centre Metabolomics Core Facility (McGill University) for technical assistance. B.F. was funded by a fellowship from the Canadian Institutes of Health Research (CIHR). T.G. was funded by the McGill Integrated Cancer Research Training Program. This work was supported by grants to R.G.J. from the CIHR (MOP-93799), the Canadian Cancer Society (2010-700586), and the Terry Fox Research Foundation (TEF-116128).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312570111/-/DCSupplemental.
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: Metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan JG, Mak TW. Metabolic targeting as an anticancer strategy: Dawn of a new era? Sci STKE. 2007;2007(381):pe14. doi: 10.1126/stke.3812007pe14. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365(6):537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- 7.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giatromanolaki A, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85(6):881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy BC, et al. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010;70(2):136–145. doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Hemminki A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391(6663):184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 11.Avizienyte E, et al. LKB1 somatic mutations in sporadic tumors. Am J Pathol. 1999;154(3):677–681. doi: 10.1016/S0002-9440(10)65314-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contreras CM, et al. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer Res. 2008;68(3):759–766. doi: 10.1158/0008-5472.CAN-07-5014. [DOI] [PubMed] [Google Scholar]
- 13.Wingo SN, et al. Somatic LKB1 mutations promote cervical cancer progression. PLoS ONE. 2009;4(4):e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Cespedes M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62(13):3659–3662. [PubMed] [Google Scholar]
- 15.Hearle N, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res. 2006;12(10):3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 16.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw RJ. LKB1: Cancer, polarity, metabolism, and now fertility. Biochem J. 2008;416(1):e1–e3. doi: 10.1042/BJ20082023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boudeau J, Sapkota G, Alessi DR. LKB1, a protein kinase regulating cell proliferation and polarity. FEBS Lett. 2003;546(1):159–165. doi: 10.1016/s0014-5793(03)00642-2. [DOI] [PubMed] [Google Scholar]
- 19.Shackelford DB, et al. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci USA. 2009;106(27):11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18(13):1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6(1):91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Xu Q, Vu H, Liu L, Wang TC, Schaefer WH. Metabolic profiles show specific mitochondrial toxicities in vitro in myotube cells. J Biomol NMR. 2011;49(3-4):207–219. doi: 10.1007/s10858-011-9482-8. [DOI] [PubMed] [Google Scholar]
- 23.Shackelford DB, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4(2):147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 25.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1(6):393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Horak P, et al. Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc Natl Acad Sci USA. 2010;107(10):4675–4680. doi: 10.1073/pnas.0907705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37(Pt 6):1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- 31.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8(4):311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17(1):113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardie DG, Alessi DR. LKB1 and AMPK and the cancer-metabolism link: Ten years after. BMC Biol. 2013;11:36. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makowski L, Hayes DN. Role of LKB1 in lung cancer development. Br J Cancer. 2008;99(5):683–688. doi: 10.1038/sj.bjc.6604515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 42.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.