Abstract
Intracellular pathogens have developed elaborate mechanisms to exploit the different cellular systems of their unwilling hosts to facilitate their entry, replication and survival. In particular, a diverse range of bacteria and viruses have evolved unique strategies to harness the power of Arp2/3-mediated actin polymerization to enhance their cell-to-cell spread. In this review, we discuss how studying these pathogens has revolutionized our molecular understanding of Arp2/3-dependent actin assembly, and revealed key signalling pathways regulating actin assembly in cells. Further studies with known and newly emerging pathogens will undoubtedly continue to enhance our understanding of the role of the actin cytoskeleton during pathogenesis. Moreover, looking back over the last 20 years, it would be surprising if future analyses of microbe-host interactions did not continue to uncover new mechanisms regulating actin assembly and dynamics, as well as unexpected cellular functions for actin.
Introduction
The host cell actin cytoskeleton is a key target of microbial pathogens. Bacterial pathogens frequently inhibit cellular processes by disabling the cytoskeleton using secreted toxins that target actin or its regulators (Aktories, 2011; Aktories et al., 2011). Alternatively, many bacterial pathogens and most viruses use actin assembly to promote their invasion or uptake enabling cellular colonization or replication (Carabeo, 2011; Taylor et al., 2011). Many pathogens have also evolved a capacity to hijack the force generating capacity of actin polymerization to power intracellular or surface-associated motility. The frequent occurrence of pathogen exploitation of host cell actin has led to the proposal that perturbing actin may be a hallmark of infection or “pattern of pathogenesis” (Vance et al., 2009).
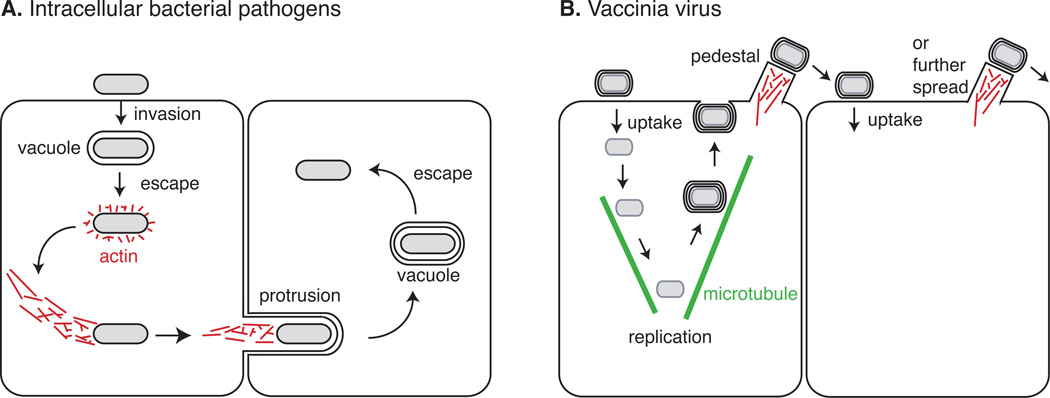
For pathogens such as Listeria monocytogenes and Shigella flexneri, which use actin from within the cytosol, motility promotes collision with the plasma membrane, formation of a membranous protrusion containing the pathogen, and engulfment of the protrusion by adjoining cells, enabling cell-cell spread (Figure 1A). For pathogens such as vaccinia that induce actin polymerization when bound to the outside of the cell, motility promotes spread to adjacent cells (Figure 1B). Hijacking actin from inside or outside the cell represent topologically and biochemically distinct challenges that involve interfacing with components of signaling pathways that regulate actin assembly. Because pathogens exploit distinct layers of the actin regulatory machinery of their hosts, studies examining the mechanisms by which pathogens impact actin have illuminated key pathways of actin regulation.
Figure 1. Schematic of Arp2/3-induced actin polymerization in pathogen spread from within or on the surface of a cell.
(A) Following invasion and escape from the vacuole, actin-based bacterial motility promotes collision with the plasma membrane, protrusion formation, and protrusion engulfment by an adjacent cell. (B) After being transported to the cell periphery on microtubules, vaccinia fuses with the plasma membrane and induces an outside-in signaling cascade to stimulate actin polymerization, which propels the virus onto neighboring cells. If the adjacent cell is already infected, the virus does not enter but again induces actin polymerization, which propels the virion across the cell surface enhancing its chances of reaching a non-infected cell.
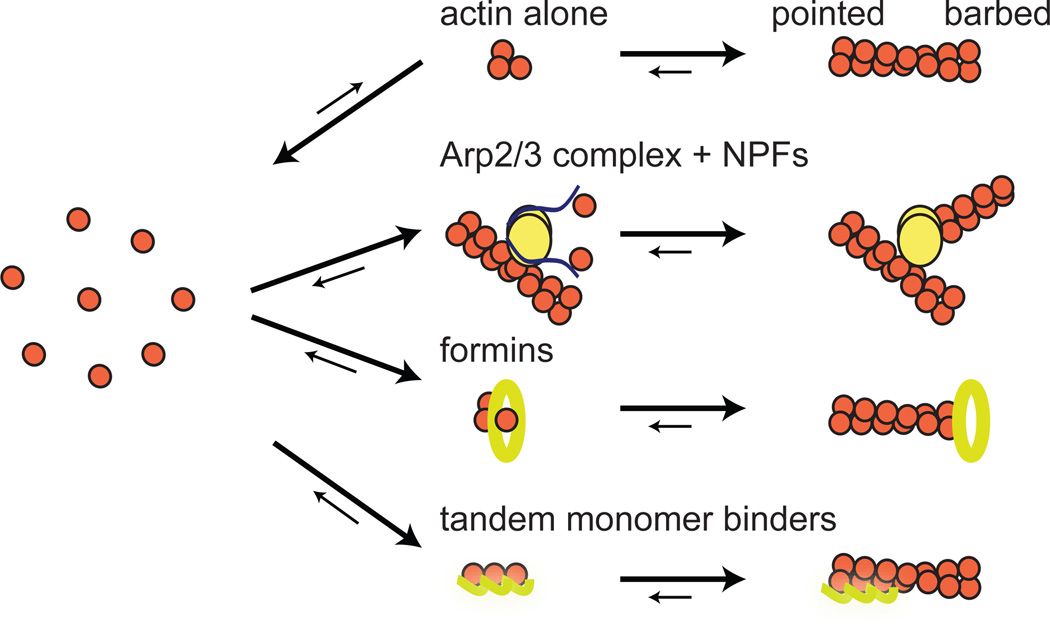
Mobilizing the host actin cytoskeleton requires that pathogens deploy proteins that interact with actin or mimic regulators that mediate or control its polymerization. Actin assembly is regulated by a variety of signaling molecules including tyrosine kinases, adapter proteins such as Nck and Grb2, and Rho-family GTPases including Rho, Rac and Cdc42. These factors act upstream of three major classes of proteins that nucleate actin filaments (F-actin) from actin monomers (G-actin) (Figure 2). One class, the formins, nucleate actin and processively associate with the fast growing barbed end of the filament (Chesarone et al., 2010). A second class, the tandem-monomer-binding family, nucleate actin but do not associate with growing filament ends (Qualmann and Kessels, 2009). Although both of these classes are exploited by pathogens, here we will focus on the third class, the Arp2/3 complex and its activators the nucleation promoting factors (NPFs) (Rotty et al., 2013), as this is the primary class used by pathogens to promote actin-based motility during infection.
Figure 2. The three different types of actin nucleation mechanisms.
Spontaneous actin nucleation is the rate-limiting step in actin filament assembly. There are three major classes of host actin-nucleating factors that accelerate nucleation. The Arp2/3 complex is activated by NPFs, and nucleates a new filament from the side of an existing filament, linking the two filaments into a Y-branch. Formins nucleate a filament and then remain processively associated with the fast-growing barbed end as it elongates. Tandem-monomer-binding proteins nucleate a filament and can remain attached to the slow-growing pointed end.
The Arp2/3 complex is a weak actin nucleator, but when activated by NPFs it binds to an actin filament and robustly nucleates a new filament that emerges to form a Y-branch (Campellone and Welch, 2010; Rotty et al., 2013). Mammalian class I NPFs include the WASP/N-WASP, WAVE/Scar, WHAMM, WASH and JMY proteins, each of which exhibits a specific localization and activates actin assembly during distinct cellular processes including lamellipodia protrusion (WAVE/Scar), endocytosis and endosome remodeling (WASP/N-WASP, WASH), and anterograde transport (WHAMM) (Campellone and Welch, 2010; Rotty et al., 2013). Once nucleated, actin filament elongation provides the driving force for cellular movements.
Many unrelated bacterial and viral pathogens mobilize the Arp2/3 complex to nucleate actin by mimicking or exploiting molecules ranging from tyrosine kinase substrates to NPFs, proving the flexibility of evolutionary strategies to hijack the cytoskeleton (Haglund and Welch, 2011) (Figure 3). As it is unfortunately impossible to cover every pathogen that takes advantage of the Arp2/3 complex in the space available, we have focused our attention on Listeria monocytogenes, Shigella flexneri, vaccinia virus and baculoviruses, each of which has developed the capacity to use Arp2/3 mediated actin polymerization to enhance their spread (Figure 3). We include early studies for historical perspective, and contemporary studies for current information and future directions. Developments in the field over the past 25 years have highlighted that studying how pathogens exploit actin has enhanced our understanding of pathogenesis, and revolutionized our knowledge of the host pathways that regulate actin assembly in uninfected cells.
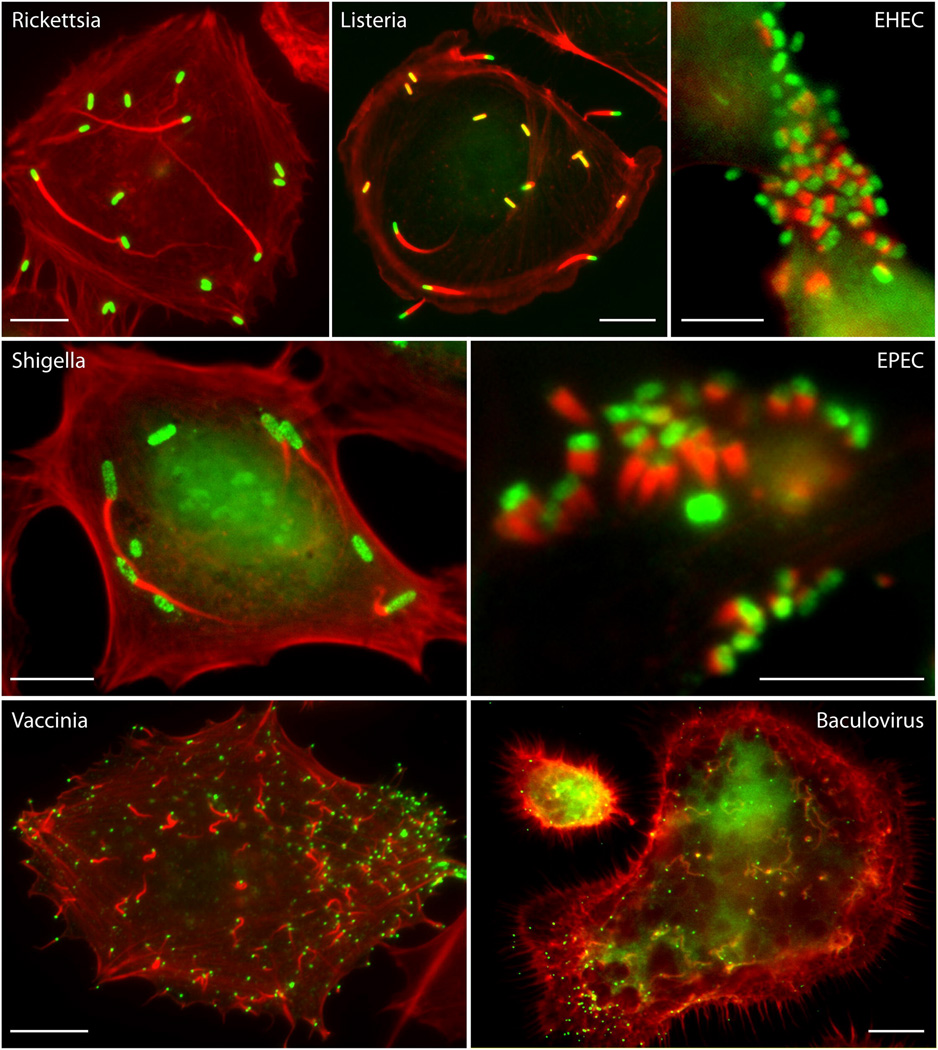
Figure 3. Immunofluorescence images of actin tails or EPEC and EHEC pedestals polymerized by the indicated pathogen.
F-actin, red; pathogens, green. All scale bars = 10 µm.
The discovery of bacterial actin-based motility
Intracellular bacterial movement was first observed in the 1950’s by timelapse microscopy of cells infected with Rickettsia rickettsii (Schaechter et al., 1957). Motility resulted in the interaction of bacteria with the host cell plasma membrane, the formation of bacteriacontaining protrusions, and the release of bacteria from the cell. A similar phenomenon was subsequently described for Shigella in the 1960’s, suggesting a role for bacterial movement in cell-to-cell spread (Ogawa et al., 1968). The direct association of protrusions with cell-to-cell spread was confirmed using ultra-structural analysis of epithelial cells infected with Listeria, where bacteria-containing membrane protrusions were seen to extend into invaginations and become internalized by adjoining cells (Racz et al., 1972; Racz et al., 1970). Nevertheless, the molecular basis of intracellular motility and its functional connection with spread remained mysterious for nearly two decades.
Insights into the mechanism of motility came in the late 1980’s from seminal studies on Listeria, Shigella and Rickettsia rickettsii, which documented the association of cytoplasmic bacteria with the host actin cytoskeleton (Bernardini et al., 1989; Heinzen et al., 1993; Mounier et al., 1990; Tilney and Portnoy, 1989) (Figure 3). The first comprehensive study focused on Listeria, which was shown to associate with actin in stages, with the bacteria initially surrounded by an actin cloud, and then trailed by an actin comet tail (Tilney and Portnoy, 1989). Bacteria with actin tails often extended into protrusions of the host cell plasma membrane, with the bacterium at the tip. Some protrusions were internalized by neighboring cells, resulting in a double-membrane vacuole from which the bacterium eventually escaped. Subsequent studies confirmed this same pathway for Shigella and Rickettsia conorii (Gouin et al., 1999). In support of a function for actin, treatment of infected cells with cytochalasin D, an inhibitor of actin assembly, prevented protrusion formation and spread of Listeria and Shigella (Bernardini et al., 1989; Tilney and Portnoy, 1989), as well as release of Rickettsia rickettsii from host cells (Heinzen et al., 1993). Together these data supported a model in which the actin cytoskeleton promotes intracellular bacterial movement, protrusion formation, and penetration into neighboring cells.
Timelapse imaging confirmed this model and revealed for the first time the kinetics of bacterial movement and spread. All three pathogens moved at rates ranging from 2–60 µm/min, with variations between individual bacteria in a single cell and between bacteria in different cell types (Dabiri et al., 1990; Goldberg and Theriot, 1995; Sanger et al., 1992). The relationship between movement and spread was later directly observed for Listeria and Shigella. Moving bacteria collide with the plasma membrane, and either ricochet back into the cytosol or enter into protrusions, with relative frequencies that depend on the strain and age of the cell monolayer (Monack and Theriot, 2001; Robbins et al., 1999). Protrusions that extend into the neighboring cell can be internalized, resolved into a vacuole, and then disrupted as bacteria escape into the cytosol. It is likely that this pathway of cell-to-cell spread is common to most intracellular bacterial pathogens that undergo actin-based motility. An exception are Burkholderia species, which have the intriguing ability to induce host cell-cell fusion to enable direct access between cells, bypassing reliance on protrusion formation and uptake (Stevens and Galyov, 2004).
The organization and dynamics of actin also enable the role of actin polymerization in driving motility. Actin filaments are orientated with their fast growing barbed ends facing the bacterium surface (Gouin et al., 1999; Tilney et al., 1992a; Tilney et al., 1992b), and actin assembly at the surface is coupled to bacterial movement, resulting in the formation of the characteristic comet tails (Sanger et al., 1992; Theriot et al., 1992). Actin filaments in Listeria actin tails are organized into a dendritic network of Y-branches (Cameron et al., 2001), similar to the organization of actin in cellular lamellipodia (Svitkina and Borisy, 1999). Filaments in the comet tail remain fixed in place and are depolymerized with a half-life of 30 s for Listeria actin tails (Theriot et al., 1992) or 100 s for Rickettsia rickettsii tails (Heinzen et al., 1993), remarkably similar to actin dynamics in motile eukaryotic cells (Theriot and Mitchison, 1991; Theriot et al., 1992). Actin depolymerization is also crucial for bacterial movement as it replenishes the G-actin pool to fuel further actin assembly (Carlier et al., 1997; Rosenblatt et al., 1997). Based on the similarities in actin organization and dynamics in bacterial comet tails and cellular structures including lamellipodia, Listeria and Shigella motility have been used as a model to study the molecular mechanisms that control actin dynamics in cells.
Bacterial proteins important for actin assembly
The identification of bacterial proteins required for actin assembly was first accomplished for Listeria and Shigella, where genes encoding these proteins were identified in screens for transposon mutants deficient in cell-to-cell spread and plaque formation. The discovery of these proteins initiated a revolution in our understanding of the molecular mechanisms of actin assembly by pathogens, and by the host cell in the absence of infection.
IcsA of Shigella (also called VirG), which is encoded by the icsA locus on the virulence plasmid pWR100, was the first protein identified (Bernardini et al., 1989; Lett et al., 1989; Makino et al., 1986). IcsA is a member of the autotransporter (AT) family (or Type Va secretion system), and features an N-terminal signal sequence, central passenger domain, and C-terminal translocation domain that mediates insertion into the outer membrane of the Gram-negative bacterium (Figure 4). Although passenger domain sequences are essential for actin assembly (Suzuki et al., 1998), other than a series of glycine-rich repeats, the passenger domain shows minimal sequence similarity with other proteins. For this reason, the molecular mechanism of IcsA function remained unclear for years after its implication in actin assembly.
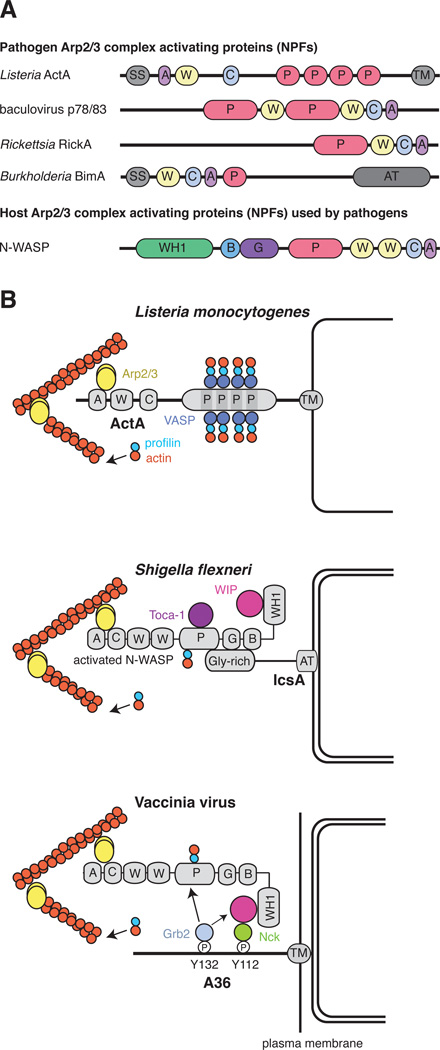
Figure 4. NPFs and their role in pathogen actin assembly.
(A) Schematic representation of the domains and motifs in pathogen NPFs and N-WASP, which is shown in its open conformation. SS = signal sequence; A = acidic; W = WASP homology 2 (WH2); C = central; P = proline rich; TM = transmembrane; AT = autotransporter; WH1 = WASP homology 1; B = basic; G = GTPase-binding. (B) Schematic representation of the components that Listeria, Shigella and Vaccinia use to stimulate Arp2/3-mediated actin polymerization. ActA binds and activates the Arp2/3 complex, while recruitment of VASP:profilin:actin complexes by its proline-rich repeats enhances actin filament elongation and Listeria actin-based motility. Shigella and Vaccinia use different strategies to recruit host N-WASP to stimulate Arp2/3-induced actin polymerization. In the case of Shigella, the glycine-rich repeats of IcsA interact directly with the N-WASP:WIP complex, which also requires Toca-1 for N-WASP activation and actin polymerization. In contrast, vaccinia recruits N-WASP via a signalling network involving Nck and WIP, downstream of Src and Abl-mediated phosphorylation of tyrosine 112 of the integral viral membrane protein A36. Grb2 recruitment is not essential, but its interaction with the proline-rich regions of WIP and N-WASP enhances actin tail formation.
Listeria ActA was identified later than IcsA, and is encoded by actA on the bacterial chromosome (Kocks et al., 1992). ActA has an N-terminal signal sequence, a surface exposed region, and a single C-terminal transmembrane domain that inserts into the cytoplasmic membrane of the Gram-positive bacterium. The surface exposed region consists of sequence motifs including acidic (A), central (C), and actin-binding (AB) motifs that are crucial for actin assembly and exhibit some similarity to sequences in Wiskott-Aldrich Syndrome family proteins (Gouin et al., 2005) (Figure 4). Moreover, ActA contains 3–4 proline-rich repeat motifs that are not essential for actin assembly but are important for enhancing its efficiency (Lasa et al., 1995; Niebuhr et al., 1997; Skoble et al., 2000; Smith et al., 1996). As with IcsA, a lack of extensive sequence similarity with other proteins slowed progress in identifying its molecular mechanism of action.
Both IcsA and ActA are sufficient for actin assembly and motility, as expression of these proteins on the surface of bacteria that are unable to polymerize actin, for example E. coli for IcsA (Goldberg and Theriot, 1995), or Listeria innocua or Streptococcus pneumonia for ActA (Kocks et al., 1995; Smith et al., 1995), enables motility. ActA is also sufficient to direct motility in the absence of other bacterial proteins (Cameron et al., 1999). Demonstrating this relied on the ability to reconstitute motility in cell cytoplasmic extracts (Theriot et al., 1994), which were subsequently shown to support the motility of plastic beads coated with ActA (Cameron et al., 1999). Thus motility only requires display of ActA or IcsA on the bacterial surface, and the bacteria are otherwise passive participants, expending little of their own energy for movement and spread.
Although IcsA and ActA were the first pathogen actin assembly proteins to be identified, later studies revealed many such proteins that are expressed by numerous bacterial pathogens, including NPFs from Rickettsia and Burkholderia (Figure 4), and other actin polymerizing proteins from Rickettsia, Chlamydia, Mycobacterium, and Vibrio species. These proteins have been shown to promote actin assembly by mimicking or hijacking representatives of all three major classes of actin assembly proteins in host cells (Haglund and Welch, 2011) (Figure 2).
Listeria monocytogenes and the rise of the host Arp2/3 complex
Bacteria expressing ActA or IcsA polymerize actin in cell extracts (Goldberg and Theriot, 1995; Marchand et al., 1995). However, ActA and IcsA themselves were unable to directly induce actin filament assembly, suggesting additional host proteins are required for actin nucleation (Loisel et al., 1999; Welch et al., 1997). Identification of the host actin assembly factor was made possible by the reconstitution of motility in cytoplasmic extracts, which could be fractionated and assayed for an activity that promotes actin assembly on the Listeria surface (Theriot et al., 1994; Welch et al., 1997). A host factor sufficient for actin assembly was purified and identified as the Arp2/3 complex. Arp2/3 had previously been identified in amoebae, and was proposed to function in actin nucleation (Machesky et al., 1994), although no activity was detected in in vitro actin assembly assays (Kelleher et al., 1995). The Arp2/3 complex has since been shown to be necessary for Listeria actin assembly (Loisel et al., 1999; May et al., 1999; Yarar et al., 1999). Notably, although Arp2/3 could promote the assembly of actin by bacteria, it was not sufficient to enable motility, indicating that additional host components were required for full reconstitution of motility (Welch et al., 1997). Moreover, actin polymerization by Arp2/3 complex at the Listeria surface required ActA (Welch et al., 1997), consistent with the essential nature of ActA in actin assembly during infection.
The fact that actin assembly by Listeria requires both ActA and Arp2/3 complex suggested that these factors act together to nucleate actin assembly. Subsequent experiments using purified ActA and Arp2/3 complex demonstrated that, although neither factor alone was sufficient, together the proteins formed an efficient nucleator (Welch et al., 1998). Based on the subunit composition of Arp2/3 complex and the presence of actin related proteins Arp2 and Arp3, it was proposed that ActA is an activator or NPF for Arp2/3. Subsequent work showed that ActA was indeed the first identified member of a broad class of NPF proteins, which are characterized by the presence of actin-binding WH2 domains (W), along with Arp2/3-binding C and A motifs (collectively called WCA) (Campellone and Welch, 2010). The WCA domain is the minimal region of NPF proteins that stimulates Arp2/3-dependent actin nucleation. The NPF family also includes other pathogen proteins such as baculovirus p78/83 (see later), Rickettsia spp. RickA (Gouin et al., 2004; Jeng et al., 2004) and Burkholderia thailandensis BimA (Sitthidet et al., 2010) (Figure 4). Thus, expressing proteins that mimic NPFs is a conserved mechanism of pathogenesis, and studying how pathogens deploy their NPFs will shed light on both pathogenic strategies as well as the function and regulation of actin assembly in uninfected cells.
It is noteworthy that Listeria actin-based motility appears to occur largely independently of regulation by host signaling pathways (tyrosine kinases and GTPases) that control actin assembly (Ebel et al., 1999; Marchand et al., 1995). However, it has been shown that the serine-threonine kinase CK2 phosphorylates ActA, enhancing Arp2/3 binding and Listeria motility, similar to the function for CK2 in phosphorylating host NPFs WASP and WAVE (Chong et al., 2009). Notably, despite the activity of CK2, bacterially expressed and purified ActA is active (Skoble et al., 2000; Welch et al., 1998). Thus, Listeria has evolved the ability to bypass the requirement for many host cell actin regulatory pathways, which differs from the behavior of other pathogens including Shigella and vaccinia virus.
Shigella and the discovery of host NPFs
The discovery that ActA is an NPF for Arp2/3 complex suggested that Shigella IcsA might possess a similar activity. However, biochemical studies indicated that this is not the case (Egile et al., 1999). The mechanism that IcsA employs to promote actin nucleation emerged from a seminal study that implicated N-WASP in Shigella motility (Suzuki et al., 1998). This study reported that N-WASP localizes at the site on Shigella from which the actin tail emerges (Figure 4). In contrast, N-WASP is not recruited by Listeria. Interestingly, Shigella specifically engages N-WASP but cannot recruit other NPFs, including the closely related WASP that is expressed in hematopoietic cell lineages (Suzuki et al., 2002). Consistent with this, Shigella cannot undergo actin-based motility in macrophages, which express WASP but not N-WASP (Suzuki et al., 2002). Moreover, they do not move in cells expressing dominant negative variants of N-WASP or in N-WASP −/− cells (Lommel et al., 2001; Snapper et al., 2001; Suzuki et al., 1998). Notably, N-WASP was implicated in Shigella motility before it was demonstrated to be an NPF for Arp2/3, and thus studies with Shigella were among the first to implicate N-WASP in actin nucleation.
The mechanism of N-WASP recruitment to Shigella involves direct binding to the IcsA protein. In particular, the glycine-rich repeats of IcsA, which are implicated in actin assembly, bind N-WASP in vitro (Suzuki et al., 1998) (Figure 4). In an influential study, it was shown that IcsA binding causes N-WASP to shift from an inactive auto-inhibited conformation to an open state where the WCA domain of N-WASP activates Arp2/3 complex (Egile et al., 1999) (Figure 4). Consistent with this, Arp2/3 complex was required for Shigella motility. This finding was remarkable because it built on a contemporary report that the host signaling molecules Cdc42 and PIP2 also bind to N-WASP and cause a change from an inactive to an active NPF (Rohatgi et al., 1999). Thus, it appears that Shigella IcsA evolved as a mimic of host signaling pathways that recruit and activate N-WASP, in contrast with Listeria ActA, which mimics activated N-WASP.
IcsA is not sufficient to activate N-WASP in host cells entirely independently of host signaling proteins or pathways. It has been reported that the activities of Abl kinase (Burton et al., 2005) and Bruton’s tyrosine kinase (Btk) (Dragoi et al., 2013) are important for N-WASP phosphorylation and Shigella motility. N-WASP activation and actin assembly by Shigella also requires Toca-1 (Leung et al., 2008), a cellular cofactor needed for N-WASP activation by Cdc42 and PIP2 (Ho et al., 2004) (Figure 4). However, N-WASP activation is independent of other cellular factors needed for N-WASP activity including Cdc42 or the WASP-interacting protein (WIP) (Moreau et al., 2000). Thus, IcsA bypasses the need for Cdc42 and PIP2 in N-WASP activation, but not the requirement for other N-WASP activating inputs.
Reconstitution of actin-based motility
The study of ActA and IcsA was instrumental in defining the involvement of the NPF-Arp2/3 pathway in actin nucleation by Listeria and Shigella. However, it was clear that additional factors beyond Arp2/3 and NPFs were required for motility, as purified Arp2/3 only promoted actin assembly but not movement (Welch et al., 1997). Just ten years after the initial discovery of bacterial actin-based motility, a crowning achievement in the field was the reconstitution of this process using purified proteins (Loisel et al., 1999). Surprisingly, the minimal reconstitution mix required for bacterial motility consisted of the NPF tethered to the bacterial surface and four other components: actin, Arp2/3 complex, capping protein, and ADF/cofilin. In the presence of these core factors, slow motility occurred (0.5 µm/min). The role of actin and Arp2/3 in motility was already discussed above. Two mechanisms have been proposed for the function of capping protein in the reconstituted motility mix. The funneling hypothesis proposes that capping older filaments and inhibiting their growth increases the concentration of available actin monomers, leading to more rapid elongation of new uncapped filaments at the bacterial surface (Carlier et al., 1997). The monomer-gating hypothesis proposes that capping acts as a switch that gates actin monomers to the Arp2/3 complex, enhancing the rate of actin nucleation (Akin and Mullins, 2008). Resolving these hypotheses will await further experimentation. The remaining essential factor, ADF/cofilin, severs and depolymerizes actin filaments, enabling monomer recycling for further polymerization (Carlier et al., 1997; Rosenblatt et al., 1997). Thus, motility requires factors that both enhance actin assembly and disassembly.
Although motility can be reconstituted with only four components, further addition of the actin monomer-binding protein profilin and the adapter protein Ena/VASP (for Listeria only) increases the speed of movement (to ~ 3 µm/min). Profilin is thought to promote filament elongation at barbed ends through several mechanisms including enhancing ADP/ATP exchange, displacing monomers from sequestering proteins, and enhancing the local concentration of actin monomers available for assembly. Moreover, it has been shown to be important for enhancing actin elongation during Listeria motility (Grenklo et al., 2003). Ena/VASP proteins, which specifically bind to the proline-rich repeat regions of ActA (but not IcsA or N-WASP), enhance motility by recruiting profilin (Auerbuch et al., 2003; Geese et al., 2002) (Figure 4) and by enabling processive barbed end elongation while antagonizing the activity of capping proteins (Breitsprecher et al., 2011; Hansen and Mullins, 2010). These factors synergize with Arp2/3 to enable rapid filament elongation and actin-based motility.
The reconstitution of actin-based bacterial motility demonstrates that the process is driven by a core set of proteins that regulate actin assembly dynamics. It also provides insights into the basic biochemical mechanisms that underlie host processes like lamellipodia protrusion during cell migration, which are driven by the same set of factors (Campellone and Welch, 2010). The reconstitution approach also highlights the fact that Listeria and Shigella motility rely on a short list of components, whereas the corresponding processes in host cells are more complex. Because they represent a stripped-down system, these pathogens have served as very useful models for cell biologists and biophysicists to study the basic mechanisms that control actin-based movement. Future studies will continue to make use of pathogens to study the biochemical basis of actin-based movement and how the biochemical properties of the system enable force generation to drive motility.
Pathogen actin assembly and autophagy
It was presumed for many years that the central function of actin assembly by Listeria and Shigella was to enable cell-to-cell spread. More recently, a new role for actin polymerization has emerged. Listeria actA mutants that fail to recruit the host Arp2/3 complex or Ena/VASP proteins are more readily modified by ubiquitination, resulting in recruitment of the autophagy machinery (Yoshikawa et al., 2009). These results suggest that recruitment of the actin assembly machinery inhibits bacterial destruction by autophagy. Interestingly, actin recruitment appears to play the opposite role during Shigella infection, as inhibition of actin assembly by cytochalasin D or N-WASP depletion reduces targeting of Shigella by the autophagy machinery (Mostowy et al., 2010; Mostowy et al., 2011). Thus, the recruitment of actin nucleation factors and actin itself modulates autophagy, but the precise effect of actin differs between pathogens. Future studies aimed at understanding how pathogens exploit actin should focus on the potential role for actin in autophagy, and may reveal unexpected connections between autophagy and the cytoskeleton in uninfected cells.
Vaccinia - a virus stimulates actin polymerization
Like intracellular bacteria, most viruses manipulate or use the actin cytoskeleton at some stage during their entry, replication and spread (Taylor et al., 2011). One of the most striking examples is vaccinia virus, a large double-stranded DNA virus that is the most studied member of the Orthopoxviridae. Vaccinia was used as the vaccine in the WHO global vaccination programme against small pox, a disease induced by its close relative variola virus. Vaccinia promotes its entry into cells by stimulating actin-dependent macropinocytosis (Mercer and Helenius, 2008). Once inside, the virus quickly establishes a complex replication and assembly program in viral factories located near the microtubule-organizing centre of the cell (Roberts and Smith, 2008; Smith et al., 2002) (Figure 1B).
The first suggestion that vaccinia might use the actin cytoskeleton came from electron microscopy studies showing virions on the tips of large microvilli projecting from infected cells (Stokes, 1976). These viral-tipped projections, which appear late in infection, contained actin, a-actinin, fimbrin and filamin but not tropomyosin or myosin (Hiller et al., 1981; Hiller et al., 1979; Krempien et al., 1981). These initial studies, were, however, largely forgotten until Cudmore et al., (1995) reported that like Listeria and Shigella, Vaccinia is moved by the power of actin polymerization on the tips of actin tails before extending out into adjacent non-infected cells (Cudmore et al., 1995; Cudmore et al., 1996) (Figure 3 and Figure 5, Movie 1). As with Listeria and Shigella, actin tail assembly involved the polarized nucleation of actin on the virus and the filaments were orientated with their fast-growing ends towards the virion (Cudmore et al., 1996).
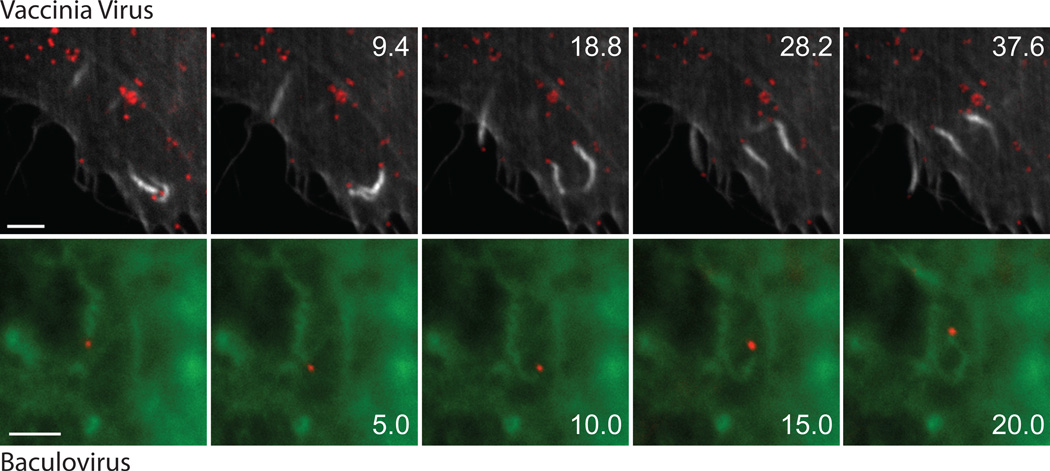
Figure 5. Viral-induced actin-based motility.
The panels taken from supplemental movies 1 and 2 show the actin-based motility of vaccinia virus (top) and baculovirus (bottom) at 8 and 1 hour post infection respectively. The scale bars represent 3 µm and the time is indicated in seconds.
From the start it was clear that not all viruses in an infected cell induce actin tails (Cudmore et al., 1995). In this initial study using widefield fluorescent imaging, it was thought that cytoplasmic intracellular enveloped virions (IEV) induced actin tails as the latter were absent when IEV assembly was inhibited (Cudmore et al., 1995). The first suggestion that this might not be the case came when it was found that cell-associated enveloped virions (CEV) attached to the outside of the cell also induce actin tails (Cudmore et al., 1996) (Figure 1B). Subsequent studies demonstrated that actin tails are only induced by CEV when the IEV fuse with the plasma membrane after being transported from their peri-nuclear site of assembly on microtubules by kinesin-1 (Dodding and Way, 2011).
A viral protein that induces actin assembly
The correlation between actin tail assembly and the presence of IEV, which are required for CEV formation, suggested that an IEV protein initiated actin tail formation. In 1996 only six viral proteins (A33, A34, A36, A56, B5 and F13) were known to be associated with the IEV membrane (Smith et al., 2002). Deletion of the genes encoding A33, A34, A36, B5 or F13 resulted in a loss of actin tails and a small plaque phenotype, which is indicative of defects in cell-to-cell spread (Roper et al., 1998; Röttger et al., 1999; Sanderson et al., 1998; Wolffe et al., 1997; Wolffe et al., 1998). However, only A36, an integral membrane protein, was required for actin tail formation and not IEV assembly (Röttger et al., 1999; Sanderson et al., 1998; Wolffe et al., 1998). Nevertheless, the proposed type II membrane topology of A36 (Parkinson and Smith, 1994) meant that it was not exposed on the surface of IEV, raising the question of how it could recruit the host proteins to stimulate actin polymerization. This conundrum was resolved when it was demonstrated that A36 has a type Ib membrane topology that exposes a cytoplasmic domain of ~ 195 residues on the IEV surface (Röttger et al., 1999; van Eijl et al., 2000) (Figure 4). Moreover, when IEV fuse with the plasma membrane A36 becomes localized in the membrane beneath CEV (Smith et al., 2002; van Eijl et al., 2000).
A36 was in the right place for the job, but in the absence of any obvious domains or sequence homologies to any other protein it was not clear how it stimulated actin polymerization. Immunofluorescence analysis of infected cells, however, suggested, that like Listeria and Shigella, Vaccinia-induced actin polymerization was likely to involve the Arp2/3 complex (Frischknecht et al., 1999a). This study also revealed the presence of a phosphotyrosine signal at the site of vaccinia, but not bacterial actin tail assembly. Furthermore, microinjection of anti-phosphotyrosine antibodies into infected cells inhibited vaccinia but not Listeria actin tail formation (Frischknecht et al., 1999a). During vaccinia infection, three proteins at 200 (EGF-receptor), 80/85 (cortactin) and 50 kDa consistently become tyrosine phosphorylated (Frischknecht et al., 1999b). Furthermore, pTyr50 was absent in cells infected with the ΔA36R virus suggesting that A36 was the unknown phosphorylated protein required for actin tail formation. Expression of A36 tyrosine (Y) to phenylalanine (F) mutants in cells infected with the ΔA36R virus revealed that only changing Y112 dramatically reduced actin tail formation (Frischknecht et al., 1999b). Inhibition of Src family kinases also abrogated actin tail formation, consistent with the sequence surrounding Y112 conforming to a consensus for Src phosphorylation. The sequence also suggested that when phosphorylated, Y112 was likely to bind the SH2 domain of Nck. Immunofluorescence analysis confirmed Nck was recruited to the virus, while in vitro peptide pull down assays demonstrated that it bound phosphorylated Y112 (Frischknecht et al., 1999b) (Figure 4). Furthermore, expression of the SH2 domain of Nck largely inhibited actin tail formation. As seen with Shigella (Suzuki et al., 1998), N-WASP was also recruited to vaccinia and expression of N-WASP lacking its C-terminal Arp2/3 binding site inhibited actin tail formation (Frischknecht et al., 1999b).
Curiously, the complete loss of actin tails and A36 phosphorylation was only observed when Y112 and Y132 were both mutated, even though mutation of Y132 alone had no impact on actin tail formation (Frischknecht et al., 1999b). However, the sequences surrounding Y132 matched the consensus-binding motif for the SH2 domain of Grb2 (Scaplehorn et al., 2002). Consistent with their predicted binding, Grb2 and Nck interacted with phosphorylated Y132 and Y112 of A36, respectively (Scaplehorn et al., 2002). Using recombinant viruses encoding A36 mutants, it was shown that phosphorylation of Y112 is essential for actin tail formation. In contrast, Y132 only enhances the number of actin tails (Scaplehorn et al., 2002). Vaccinia-induced actin polymerization had strong parallels with receptor tyrosine kinase signaling cascades, which also frequently involve multiple phosphorylation sites and adaptor proteins.
The ability to quantify actin tail formation meant that vaccinia was an excellent model to investigate how N-WASP couples Nck to actin polymerization. Unexpectedly, the WH1 domain and not the SH3 adaptor binding proline-rich region of N-WASP was recruited to the virus (Moreau et al., 2000). This result was striking, as the majority of mutations leading to Wiskott-Aldrich syndrome are found in the WH1 domain of WASP (Jin et al., 2004). This observation led to the realization that vaccinia also recruits WIP, which interacts with the WH1 domain of WASP and Nck (Anton et al., 1998; Moreau et al., 2000; Ramesh et al., 1997; Zettl and Way, 2002) (Figure 4). Overexpression of the WASP binding domain (WBD) of WIP inhibited actin tails by blocking viral recruitment of N-WASP (Moreau et al., 2000; Zettl and Way, 2002). In contrast, even though WIP is recruited to the bacterium, expression of WBD did not inhibit Shigella actin tail formation (Moreau et al., 2000). Moreover, the WBD also inhibited the recruitment of endogenous WIP to vaccinia, suggesting that WIP and N-WASP are recruited as a complex (Moreau et al., 2000). Subsequent studies using N-WASP −/− fibroblasts confirmed that WIP requires N-WASP for its recruitment to vaccinia (Snapper et al., 2001; Weisswange et al., 2009). There is also no recruitment of Grb2 in the absence of N-WASP (Weisswange et al., 2009). In contrast, Nck is still recruited to the virus, suggesting its recruitment is independent of WIP and N-WASP. Furthermore, the absence of WIP and its homologue WIRE results in the failure to recruit N-WASP but not Nck (Donnelly et al., 2013). Thus, Nck recruits a complex of WIP:N-WASP to the virus, which then associates with Grb2 interacting with phosphorylated Y132 of A36. This explains why Nck, but not Grb2, is essential for vaccinia to induce actin tails.
Molecular dissection of the vaccinia signalling network
Unravelling how a signalling cascade stimulates actin polymerization requires detailed knowledge of the interactions, dynamics and stoichiometry of the proteins in the network. Unfortunately, many signalling networks controlling actin polymerization are not amenable to such quantitative analyses as their components and/or activation is often transient and dispersed. In contrast, the signalling pathway used by vaccinia to induce actin polymerization is localized and sustained. Taking advantage of this, fluorescence recovery after photobleaching (FRAP) was used to analyse the dynamics of GFP-tagged Nck, Grb2, WIP and N-WASP during vaccinia actin-based motility (Weisswange et al., 2009). All four proteins undergo rapid exchange, with a half time of recovery of 0.14, 0.8, 0.8 and 2.7 s for Grb2, Nck, WIP and N-WASP respectively. The turnover of Nck, WIP and N-WASP increases significantly in the absence of Grb2, demonstrating that it enhances vaccinia actin tail formation by stabilising the signalling complex. It was surprising that N-WASP exchanges ~ 3.5 times slower than Nck and WIP, given they are responsible for its recruitment. N-WASP turnover also did not occur in the absence of Arp2/3 recruitment, suggesting that active actin polymerization promotes exchange of the vaccinia-signalling complex. Consistent with this, the turnover rate of N-WASP depends on its interaction with both Grb2 and the growing plus ends of actin filaments. Loss of either of these interactions led to a faster rate of N-WASP exchange and virus movement. This suggests that N-WASP not only activates the Arp2/3 complex, but also modulates the rate of actin-based motility by regulating the extent of actin polymerization, possibly by antagonizing filament capping.
Nck and N-WASP play a key role in connecting phosphotyrosine-based signalling to Arp2/3-mediated actin polymerization during a wide variety of cellular processes, in addition to driving actin-based motility of intracellular pathogens (Campellone and Welch, 2010; Dodding and Way, 2009; Lommel et al., 2001; Rotty et al., 2013; Snapper et al., 2001; Weisswange et al., 2009). What is less clear is the precise role played by WIP within Nck and N-WASP signalling networks. WIP inhibits the ability of N-WASP to activate the Arp2/3 complex until it receives the right signalling input (Ho et al., 2004; Martinez-Quiles et al., 2001; Takano et al., 2008). Recent analysis using MEFs lacking WIP, which have also been treated with RNAi against the WIP homologue WIRE, has now demonstrated that an interaction of WIP with the second SH3 domain of Nck is essential for vaccinia actin tail formation, as it is required to recruit the WIP:N-WASP complex (Donnelly et al., 2013). Furthermore, the recruitment of N-WASP depends on its interaction with WIP rather than Nck. Finally, the first and third SH3 domains of Nck are not involved in recruiting the WIP:N-WASP complex but are essential to stimulate actin assembly. Vaccinia has thus provided essential insights into the connectivity within this important signalling network (Donnelly et al., 2013). WBD over expression studies and infection of WIP −/− cells suggest that WIP is not required for Shigella actin tail formation (Garber et al., 2012; Moreau et al., 2000). However, it still remains to be determined whether Shigella can still recruit N-WASP and induce actin polymerization in the absence of both WIP and WIRE.
Role of Src and Abl kinases in vaccinia actin polymerization and spread
A36 only promotes actin tail formation beneath CEV, even though it is exposed on the surface of IEV. The reason is that Src activation and phosphorylation of A36 only occurs after the virus fuses with the plasma membrane (Newsome et al., 2004). This suggested that one or more of the four integral viral membrane proteins (A33, A34, A56 and B5) on the surface of CEV induces an outside-in signal to activate Src. The molecular mechanism still remains to be established, but the SCR4 domain of B5 is required to activate Src, phosphorylate A36 and induce actin tail formation (Newsome et al., 2004). Phosphorylation of A36 also promotes the release of kinesin-1 after the virus fuses with the plasma membrane.
Src is the prototypic member of a family of non-receptor tyrosine kinases that play redundant roles in regulating a wide variety of cellular processes. It was not surprising then, that vaccinia recruits multiple Src family kinases (Src, Fyn, and Yes) as well as the related Abl family kinases (Abl and Arg) to promote actin tail formation (Newsome et al., 2006; Reeves et al., 2005). While additional tyrosine kinases may also phosphorylate A36, inhibition of both Src and Abl family kinases is sufficient to inhibit vaccinia actin tail formation (Reeves et al., 2005). Consistent with this, in vitro kinase assays demonstrate that Abl, Arg, Fyn, Src and Yes can phosphorylate Y112 of A36 (Newsome et al., 2006). Interestingly, Abl and Arg but not Src-family kinases are also required to promote the release of CEV from infected cells (Reeves et al., 2005). Moreover, treatment of infected mice with Gleevec/STI-571/Imatinib, an Abl-family kinase inhibitor used to treat chronic myelogenous leukemia, reduces viral spread and promotes survival from an otherwise lethal infection (Reeves et al., 2005; Reeves et al., 2011).
More than 30 years ago Payne and Kristensson (1982) demonstrated that inhibition of actin polymerization blocked release of vaccinia from infected cells (Payne and Kristensson, 1982). Recently, studies have provided some molecular insights into this old observation (Horsington et al., 2013). The ability of Abl to promote virus release is independent of its ability to phosphorylate A36. Nevertheless, A36 phosphorylation and actin polymerization are required to drive CEV out of plasma membrane invaginations. In the absence of actin polymerization, CEV remain trapped within these invaginations and are not released unless the functionality of A34 or B5 is compromised. Consistent with this, structured illumination microscopy reveals that induction of actin polymerization polarizes A36 on the virus (Horsington et al., 2013). Actin is not the only cellular factor contributing to A36 polarization. Following their fusion with the plasma membrane, but before actin tail formation, vaccinia recruits clathrin in an AP-2 dependent fashion (Humphries et al., 2012). The clathrin is, however, left behind when CEV stimulate actin polymerization. Nevertheless, in the absence of clathrin recruitment, it takes longer for the virus to induce actin polymerization and fewer actin tails are formed. Clathrin appears to have an organizational role, promoting clustering of A36, which in turn helps polarize and stabilize N-WASP, making initiation of actin-based motility easier (Humphries et al., 2012).
The activity of CK2 also enhances vaccinia actin tail formation and cell-to-cell spread (Alvarez and Agaisse, 2012). Loss of CK2 does not affect CEV formation, but does reduce the ability to recruit and activate Src. While the localization of CK2 during infection and the molecular basis for these observations remain to be established, it is curious that A36, which is heavily serine phosphorylated, contains several predicted CK2 phosphorylation sites (Alvarez and Agaisse, 2012; Wolffe et al., 2001).
It is not just kinases that impact viral spread. The phosphoinositide 5-phosphatase SHIP2 acts as a negative regulator of viral release, although it is not required for actin tail formation (McNulty et al., 2011). The basis of this regulation remains to be established. However, interestingly, SHIP2 recruitment to actin tails is dependent on its SH2 domain and N-WASP. Future analysis will confirm whether the activity of SHIP2 is related to the recent observations of Horsington et al., (2013).
Actin tail formation enhances the cell-to-cell spread of vaccinia. However, it was unclear how the virus can spread faster in a cell monolayer than its replication cycle would allow. Live cell imaging of viral spread during plaque formation has now answered this mystery (Doceul et al., 2010). The authors found that if a virus (CEV or EEV) lands on a recently infected neighbouring cell that lacks a virus factory, it forms a new actin tail without being internalized (Figure 1B). Motility then drives the virus across the surface of the infected cell and onto adjacent non-infected cells, where uptake occurs. In this way, vaccinia ignores already-infected cells, enhancing the rate of spread through the cell monolayer. This “super repulsion” is mediated by A33 and A36 on the surface of recently-infected cells. The SCR4 domain of B5 is also required (Doceul et al., 2012). It remains to be established whether “super repulsion” depends on Src and Abl mediated phosphorylation of A36 and its downstream signalling network. However, the striking similarities between these two actin dependent events would suggest this is almost certainly the case.
A common mechanism to promote the spread of poxvirus infection
Actin driven cell-to-cell spread of orthopoxviruses is likely to be common, as A36 is highly conserved (www.poxvirus.org). Consistent with this notion, variola and monkeypox viruses induce Abl and Src family kinase dependent actin tails, while the A36 homologue of ectromelia virus, the causative agent of mousepox, is required for viral spread and actin tail formation (Lynn et al., 2012; Reeves et al., 2011). Actin tail formation is, however, not restricted to orthopoxviruses, as Yaba-Like Disease virus (YLDV; yatapoxvirus) and myxoma (leporipoxvirus) also induce actin tails (Duteyrat et al., 2006; Law et al., 2004), even though they lack an obvious A36 orthologue. Using a complementation approach, YL126 of YLDV was found to promote Nck and N-WASP dependent actin polymerization despite having less than 15% sequence identity with Vaccinia A36 (Dodding and Way, 2009). Five phosphorylated tyrosines in YL126 can recruit Nck to promote actin polymerization. However, YL126-mediated actin tail formation, like that of A36, is also enhanced by the recruitment of Grb2 by a single phosphorylated tyrosine. Highly divergent YL126 orthologues in other vertebrate poxviruses, with as little as 6% homology to each other, can also induce Nck and N-WASP dependent actin polymerization. Actin-based motility thus appears to be a common mechanism used by vertebrate poxviruses (Chordopoxviridae) to enhance cell-to-cell spread (Dodding and Way, 2009).
Baculoviruses use the Arp2/3 pathway for motility and nuclear actin assembly
Poxviruses are not the only viruses to hijack the Arp2/3 complex to promote actin polymerization. More than 20 years ago, it was observed that the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) induces thick actin cables in the cytoplasm of its host (Charlton and Volkman, 1991). These actin structures appear from 30 min post infection and were tipped by a single nucleocapsid, suggesting the virus induced actin polymerization (Charlton and Volkman, 1993). Intriguingly, AcMNPV also stimulates the assembly of actin filaments in the nucleus (Charlton and Volkman, 1991, 1993; Volkman et al., 1992), a phenomenon that is essential for nucleocapsid assembly and progeny virus production (Hess et al., 1989; Ohkawa and Volkman, 1999; Volkman, 1988; Volkman et al., 1987; Volkman et al., 1992). Subsequent in vitro assays demonstrated that purified AcMNPV nucleocapsids are capable of weakly stimulating actin polymerization (Lanier and Volkman, 1998). Furthermore, two viral capsid proteins, p39 and p78/83, were found to bind directly to actin. Examining the sequence of p78/83 suggested that it might represent a viral WASP-like protein, as it contained proline rich region, as well as a WCA motif that is indicative of G-actin and Arp2/3 binding (Machesky et al., 2001) (Figure 4).
Fifteen years after the discovery of baculovirus interactions with actin, p78/83 was shown to stimulate Arp2/3-dependent actin polymerization in vitro. Recent live cell imaging shows that p78/83-mediated Arp2/3-induced actin polymerization propels AcMNPV nucleocapsids throughout the cytoplasm at rates of 7–22 µm/min as early as 5 min post infection (Ohkawa et al., 2010). Actin-based motility promotes nuclear collisions, a process that enables rapid nucleocapsid transit into the nucleus to speed the onset of early gene expression. Interestingly, once early gene expression is established, but before new viral progeny are produced, remaining cytoplasmic nucleocapsids accumulate in actin rich protrusions at the cell surface, presumably ready to bud and spread to neighbouring cells. This behavior may allow the virus to spread to neighbouring cells before the initially infected cell is removed by apoptosis and sloughing, allowing the virus to establish an infection and enhancing its rapid dissemination through the epithelium of the caterpillar midgut (Ohkawa et al., 2010).
Interestingly, p78/83 and Arp2/3-dependent actin polymerization is also required later in infection for nuclear actin assembly (Goley et al., 2006). However, the role of nuclear actin remains unclear, and may include virus assembly, nuclear egress, and nuclear envelope remodeling during viral envelopment. Actin also plays a role in the nucleus of uninfected cells, yet the precise form and function of nuclear actin remains unclear (Weston et al., 2012). Baculoviruses represent an emerging system for studying how pathogens hijack actin in the nucleus, and for studying the normal nuclear roles of actin.
What can the study of pathogen actin assembly still tell us?
Three decades after the discovery that Listeria and Shigella use actin-based motility to promote their cell-to-cell spread, we have achieved an understanding of how this process works at the molecular level. Key proteins involved in Arp2/3-dependent motility have been identified, and reconstitution of the process has been achieved in vitro using purified proteins. These studies have revolutionized our understanding of how pathogens exploit actin, and have revealed essential molecular pathways involved in actin regulation in host cells.
Nevertheless, much still remains to be discovered. With regard to Listeria and Shigella, we do not yet know at a biophysical level how Arp2/3-dependent actin polymerization is coupled to force generation to drive motility. Reconstitution of bacterial motility represents an experimentally accessible system to address this question, and what is learned will also apply to force-generating mechanisms involved in host cellular and intracellular motility. We also do not understand how host motility is coupled with cell-to-cell spread, what membrane trafficking factors might be important for this process, or how membrane-cytoskeleton linkers like Ezrin and CD44 promote cell-to cell spread (Pust et al., 2005). Moreover, we do not know how the ability to recruit Arp2/3 and actin plays other roles during infection, for example in avoidance of autophagy. The information gained from studying Listeria and Shigella motility and cell-to-cell spread will continue to be of major importance in uncovering basic cell biological principles related to cytoskeletal function and regulation. It is also important to note that other bacteria, for example Rickettsia and Burkholderia spp., are likely to exploit distinct host actin polymerization pathways involving formins or tandem-monomer-binding nucleators (Haglund and Welch, 2011). Thus, the study of how evolutionarily diverse bacterial pathogens usurp actin will undoubtedly shed light on the function and regulation of all three major host actin assembly pathways.
Vaccinia virus has also been a powerful model, in particular for understanding how a signalling network activated by Src and Abl family kinases functions to stimulate actin polymerization. However, as with bacterial pathogens, many outstanding questions remain. Vaccinia-induced actin polymerization is dependent on the activation of Src and Abl family kinases (Frischknecht et al., 1999b; Newsome et al., 2004; Newsome et al., 2006; Reeves et al., 2005). Nevertheless, the molecular basis of how CEV activate Src and Abl-family kinases remains to be determined. The temporal aspects of vaccinia-induced kinase activation as the virus fuses with the plasma membrane (or lands on another cell during “super repulsion”) and its relationship to and/or role in the release of kinesin-1 and recruitment of clathrin also need to be established. The virus is amenable to live imaging, including FRAP and FRET-based approaches, to address these important questions and also provides a great system for the development of new sensors to monitor the activity and interactions of these proteins together with components in the vaccinia-signalling cascade.
We also still lack a detailed understanding of how the initial level of A36 tyrosine phosphorylation determines the final output of the vaccinia-signalling network. Addressing this question is not easy for most phospho-based signalling networks. However, the ability to manipulate the number of phosphorylation competent A36 molecules beneath CEV (Humphries et al., 2012), combined with quantitative live imaging (Weisswange et al., 2009), provides a unique opportunity to determine how actin-based motility and dynamics of a signalling network changes in response to the level of tyrosine phosphorylation. Within the signalling network itself, we still lack such basic information as the stoichiometry of components and how many A36 molecules are actually recruiting Nck. Does the slow exchange rate of N-WASP compared to Nck and WIP mean there is an additional binding partner in the system? We also need to understand the exact sequence of events in the Nck-mediated recruitment and activation of the WIP:N-WASP complex, ideally at the single molecule level. This will allow us to address such fundamental questions as whether each N-WASP molecule activates single or multiple Arp2/3 complexes before it dissociates from the virus. We also have no information on the number and organization of actin filaments in the tail and why loss of clathrin recruitment results in the slower disassembly of the tail.
Many of the imaging approaches and quantitative aspects that are possible with Vaccinia and other intracellular pathogens would be difficult, if not impossible, in other systems. Furthermore, the quantitative data derived from pathogens can be used to develop mathematical models that will provide additional molecular insights into the emergent properties and co-operative nature of signalling networks and how they regulate Arp2/3-dependent actin polymerization. The study of pathogens also has the potential to reveal insights into new functions for actin. One uncharted frontier in actin relates to its function in the nucleus. Nuclear actin is thought to participate in RNA biology, chromatin remodelling, nuclear shape, and cell differentiation (Weston et al., 2012). However, the regulation, state, dynamics and roles of nuclear actin are poorly understood. Because of their ability to polymerize and harness actin in the nucleus, baculoviruses represent an outstanding model for understanding the regulation and function of nuclear actin. Finally, it should not be forgotten that pathogens cause a wide range of diseases that can have serious social and economic consequences. Understanding exactly how pathogens subvert cell signalling and the host actin cytoskeleton thus offers the potential to identify new therapeutic drug targets to combat infection that in the case of bacteria are independent of any acquired antibiotic resistance.
Supplementary Material
Acknowledgements
We would like to thank David Barry (Way lab) for Vaccinia movie and Ashley Humphries (Way lab) for Vaccinia infected cell image; Theresia Stradal (University of Munster, Germany) and Klemens Rottner (Bonn University, Germany) for images of Shigella, EPEC and EHEC infected cells; Taro Ohkawa (University of California, Berkeley) for the image of a baculovirus infected cell, and Rebecca Lamason (University of California, Berkeley) for the image of a Rickettsia infected cell. We also thank Klemens Rottner, Jasmine Abella (Way lab) and Taro Ohkawa (Welch Lab) for comments on the text. Matt Welch is supported by NIH grants R01 GM059609 and R01 AI074760, and Michael Way is supported by Cancer Research UK.
References
- Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nature reviews Microbiology. 2011;9:487–498. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- Aktories K, Lang AE, Schwan C, Mannherz HG. Actin as target for modification by bacterial protein toxins. The FEBS journal. 2011;278:4526–4543. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- Alvarez DE, Agaisse H. Casein kinase 2 regulates vaccinia virus actin tail formation. Virology. 2012;423:143–151. doi: 10.1016/j.virol.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Anton IM, Lu W, Mayer BJ, Ramesh N, Geha RS. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adapter protein Nck. J Biol Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- Auerbuch V, Loureiro JJ, Gertler FB, Theriot JA, Portnoy DA. Ena/VASP proteins contribute to Listeriamonocytogenes pathogenesis by controlling temporal and spatial persistence of bacterial actin-based motility. Molecular microbiology. 2003;49:1361–1375. doi: 10.1046/j.1365-2958.2003.03639.x. [DOI] [PubMed] [Google Scholar]
- Bernardini ML, Mounier J, d’Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TE, Small JV, Curth U, Dickinson RB, Faix J. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton EA, Oliver TN, Pendergast AM. Abl kinases regulate actin comet tail elongation via an N-WASP-dependent pathway. Molecular and cellular biology. 2005;25:8834–8843. doi: 10.1128/MCB.25.20.8834-8843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Nat Acad Sci U S A. 1999;96:4906–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Svitkina TM, Vignjevic D, Theriot JA, Borisy GG. Dendritic organization of actin comet tails. Curr Biol. 2001;11:130–135. doi: 10.1016/s0960-9822(01)00022-7. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nature reviews Molecular cell biology. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo R. Bacterial subversion of host actin dynamics at the plasma membrane. Cellular microbiology. 2011;13:1460–1469. doi: 10.1111/j.1462-5822.2011.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton CA, Volkman LE. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J Virol. 1991;65:1219–1227. doi: 10.1128/jvi.65.3.1219-1227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton CA, Volkman LE. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf 21 cells induces actin cable formation. Virology. 1993;197:245–254. doi: 10.1006/viro.1993.1585. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chong R, Swiss R, Briones G, Stone KL, Gulcicek EE, Agaisse H. Regulatory mimicry in Listeria monocytogenes actin-based motility. Cell host & microbe. 2009;6:268–278. doi: 10.1016/j.chom.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Sanger JM, Portnoy DA, Southwick FS. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V, Hollinshead M, Breiman A, Laval K, Smith GL. Protein B5 is required on extracellular enveloped vaccinia virus for repulsion of superinfecting virions. J Gen Virol. 2012;93:1876–1886. doi: 10.1099/vir.0.043943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP, Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell host & microbe. 2009;6:536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly SK, Weisswange I, Zettl M, Way M. WIP Provides an Essential Link between Nck and N-WASP during Arp2/3-Dependent Actin Polymerization. Curr Biol. 2013;23:999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragoi AM, Talman AM, Agaisse H. Bruton’s tyrosine kinase regulates Shigella flexneri dissemination in HT-29 intestinal cells. Infection and immunity. 2013;81:598–607. doi: 10.1128/IAI.00853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteyrat JL, Gelfi J, Bertagnoli S. Ultrastructural study of myxoma virus morphogenesis. Arch Virol. 2006;151:2161–2180. doi: 10.1007/s00705-006-0791-2. [DOI] [PubMed] [Google Scholar]
- Ebel F, Rohde M, von Eichel-Streiber C, Wehland J, Chakraborty T. The actin-based motility of intracellular Listeria monocytogenes is not controlled by small GTP-binding proteins of the Rho- and Ras-subfamilies. FEMS Microbiol Lett. 1999;176:117–124. doi: 10.1111/j.1574-6968.1999.tb13651.x. [DOI] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin- based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht F, Cudmore S, Moreau V, Reckmann I, Rottger S, Way M. Tyrosine phosphorylation is required for actin-based motility of vaccinia but not Listeria or Shigella. Curr Biol. 1999a;9:89–92. doi: 10.1016/s0960-9822(99)80020-7. [DOI] [PubMed] [Google Scholar]
- Frischknecht F, Moreau V, Röttger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M. Actin based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999b;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- Garber JJ, Takeshima F, Anton IM, Oyoshi MK, Lyubimova A, Kapoor A, Shibata T, Chen F, Alt FW, Geha RS, et al. Enteropathogenic Escherichia coli and vaccinia virus do not require the family of WASP-interacting proteins for pathogen-induced actin assembly. Infection and immunity. 2012;80:4071–4077. doi: 10.1128/IAI.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS. Contribution of Ena/VASP proteins to intracellular motility of listeria requires phosphorylation and proline-rich core but not F-actin binding or multimerization. Molecular biology of the cell. 2002;13:2383–2396. doi: 10.1091/mbc.E02-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Theriot JA. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc Natl Acad Sci USA. 1995;92:6572–6576. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D’Alessio JA, Cande WZ, Volkman LE, Welch MD. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science. 2006;314:464–467. doi: 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, Li R, Cossart P. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- Gouin E, Gantelet H, Egile C, Lasa I, Ohayon H, Villiers V, Gounon P, Sansonetti PJ, Cossart P. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci. 1999;112:1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Grenklo S, Geese M, Lindberg U, Wehland J, Karlsson R, Sechi AS. A crucial role for profilin-actin in the intracellular motility of Listeria monocytogenes. EMBO Rep. 2003;4:523–529. doi: 10.1038/sj.embor.embor823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. The Journal of cell biology. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. Directional actin polymerization associated with spotted fever group rickettsia infection of vero cells. Infect Immun. 1993;61:1926–1935. doi: 10.1128/iai.61.5.1926-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess RT, Goldsmith PA, Volkman LE. Effect of cytochalasin D on cell morphology and AcMNPV replication in a Spodoptera frugiperda cell line. J Invertebr Pathol. 1989;53:169–182. doi: 10.1016/0022-2011(89)90005-0. [DOI] [PubMed] [Google Scholar]
- Hiller G, Jungwirth C, Weber K. Fluorescence microscopical analysis of the life cycle of vaccinia virus in the chick embryo fibroblasts. Exp Cell Res. 1981;132:81–87. doi: 10.1016/0014-4827(81)90085-9. [DOI] [PubMed] [Google Scholar]
- Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Horsington J, Lynn H, Turnbull L, Cheng D, Braet F, Diefenbach RJ, Whitchurch CB, Karupiah G, Newsome TP. A36-dependent actin filament nucleation promotes release of vaccinia virus. PLoS pathogens. 2013;9:e1003239. doi: 10.1371/journal.ppat.1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AC, Dodding MP, Barry DJ, Collinson LM, Durkin CH, Way M. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell host & microbe. 2012;12:346–359. doi: 10.1016/j.chom.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Jeng RL, Goley ED, D’Alessio JA, Chaga OY, Svitkina TM, Borisy GG, Heinzen RA, Welch MD. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cellular microbiology. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, Gandellini F, Stewart DM, Zhu Q, Nelson DL, et al. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–4019. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba . J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kocks C, Marchand JB, Gouin E, d’Hauteville H, Sansonetti PJ, Carlier MF, Cossart P. The unrelated surface proteins ActA of Listeria monocytogenes and IcsA of Shigella flexneri are sufficient to confer actin-based motility on Listeria innocua and Escherichia coli respectively. Molecular microbiology. 1995;18:413–423. doi: 10.1111/j.1365-2958.1995.mmi_18030413.x. [DOI] [PubMed] [Google Scholar]
- Krempien U, Schneider L, Hiller G, Weber K, Katz E, Jungwirth C. Conditions for pox virus-specific microvilli formation studied during synchronized virus assembly. Virology. 1981;113:556–564. doi: 10.1016/0042-6822(81)90183-5. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Volkman LE. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology. 1998;243:167–177. doi: 10.1006/viro.1998.9065. [DOI] [PubMed] [Google Scholar]
- Lasa I, Violaine D, Gouin E, Marchand J, Cossart P. The amino-terminal part of ActA is critical for the actin-based motility of Listeria monocytogenes; the central proline-rich region acts as a stimulator. Molec Microbiol. 1995;18:425–436. doi: 10.1111/j.1365-2958.1995.mmi_18030425.x. [DOI] [PubMed] [Google Scholar]
- Law M, Hollinshead M, Lee HJ, Smith GL. Yaba-like disease virus protein Y144R, a member of the complement control protein family, is present on enveloped virions that are associated with virus-induced actin tails. J Gen Virol. 2004;85:1279–1290. doi: 10.1099/vir.0.79863-0. [DOI] [PubMed] [Google Scholar]
- Lett MC, Sasakawa C, Okada N, Sakai T, Makino S, Yamada M, Komatsu K, Yoshikawa M. virG, a plasmid-coded virulence gene of Shigella flexneri: identification of the virG protein and determination of the complete coding sequence. Journal of bacteriology. 1989;171:353–359. doi: 10.1128/jb.171.1.353-359.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y, Ally S, Goldberg MB. Bacterial actin assembly requires toca-1 to relieve N-wasp autoinhibition. Cell host & microbe. 2008;3:39–47. doi: 10.1016/j.chom.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kuhn R. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn H, Horsington J, Ter LK, Han S, Chew YL, Diefenbach RJ, Way M, Chaudhri G, Karupiah G, Newsome TP. Loss of cytoskeletal transport during egress critically attenuates ectromelia virus infection in vivo. J Virol. 2012;86:7427–7443. doi: 10.1128/JVI.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH, Volkman LE. WASP homology sequences in baculoviruses. Trends Cell Biol. 2001;11:286–287. doi: 10.1016/s0962-8924(01)02009-8. [DOI] [PubMed] [Google Scholar]
- Makino S, Sasakawa C, Kamata K, Kurata T, Yoshikawa M. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell. 1986;46:551–555. doi: 10.1016/0092-8674(86)90880-9. [DOI] [PubMed] [Google Scholar]
- Marchand JB, Moreau P, Paoletti A, Cossart P, Carlier MF, Pantaloni D. Actin-based movement of Listeria monocytogenes: actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J Cell Biol. 1995;130:331–343. doi: 10.1083/jcb.130.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Quiles N, Rohatgi R, Anton IM, Medina M, Saville SP, Miki H, Yamaguchi H, Takenawa T, Hartwig JH, Geha RS, et al. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat Cell Biol. 2001;3:484–491. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- May RC, Hall ME, Higgs HN, Pollard TD, Chakraborty T, Wehland J, Machesky LM, Sechi AS. The Arp2/3 complex is essential for the actin-based motility of Listeria monocytogenes. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- McNulty S, Powell K, Erneux C, Kalman D. The host phosphoinositide 5-phosphatase SHIP2 regulates dissemination of vaccinia virus. J Virol. 2011;85:7402–7410. doi: 10.1128/JVI.02391-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- Monack DM, Theriot JA. Actin-based motility is sufficient for bacterial membrane protrusion formation and host cell uptake. Cellular microbiology. 2001;3:633–647. doi: 10.1046/j.1462-5822.2001.00143.x. [DOI] [PubMed] [Google Scholar]
- Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, Tham TN, Mallet A, Lelek M, Gouin E, Demangel C, Brosch R, Zimmer C, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell host & microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. The Journal of biological chemistry. 2011;286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J, Ryter A, Coquis-Rondon M, Sansonetti PJ. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infection and immunity. 1990;58:1048–1058. doi: 10.1128/iai.58.4.1048-1058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Scaplehorn N, Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Weisswange I, Frischknecht F, Way M. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cellular microbiology. 2006;8:233–241. doi: 10.1111/j.1462-5822.2005.00613.x. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Nakamura A, Nakaya R. Cinemicrographic study of tissue cell cultures infected with Shigella flexneri. Japanese journal of medical science & biology. 1968;21:259–273. doi: 10.7883/yoken1952.21.259. [DOI] [PubMed] [Google Scholar]
- Ohkawa T, Volkman LE. Nuclear F-actin is required for AcMNPV nucleocapsid morphogenesis. Virology. 1999;264:1–4. doi: 10.1006/viro.1999.0008. [DOI] [PubMed] [Google Scholar]
- Ohkawa T, Volkman LE, Welch MD. Actin-based motility drives baculovirus transit to the nucleus and cell surface. The Journal of cell biology. 2010;190:187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Smith GL. Vaccinia virus gene A36R encodes a Mr. 43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- Payne LG, Kristensson K. The effect of cytochalasin D and monensin on enveloped vaccinia virus release. Arch Virol. 1982;74:11–20. doi: 10.1007/BF01320778. [DOI] [PubMed] [Google Scholar]
- Pust S, Morrison H, Wehland J, Sechi AS, Herrlich P. Listeria monocytogenes exploits ERM protein functions to efficiently spread from cell to cell. EMBO J. 2005;24:1287–1300. doi: 10.1038/sj.emboj.7600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Racz P, Tenner K, Mero E. Experimental Listeria enteritis. I. An electron microscopic study of the epithelial phase in experimental listeria infection. Lab Invest. 1972;26:694–700. [PubMed] [Google Scholar]
- Racz P, Tenner K, Szivessy K. Electron microscopic studies in experimental keratoconjunctivitis listeriosa. I. Penetration of Listeria monocytogenes into corneal epithelial cells. Acta Microbiol Acad Sci Hung. 1970;17:221–236. [PubMed] [Google Scholar]
- Ramesh N, Anton IE, Hartwig JH, Geha RS. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, et al. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med. 2005;11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- Reeves PM, Smith SK, Olson VA, Thorne SH, Bornmann W, Damon IK, Kalman D. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on abl and SRC family tyrosine kinases. J Virol. 2011;85:21–31. doi: 10.1128/JVI.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JR, Barth AI, Marquis H, de Hostos EL, Nelson WJ, Theriot JA. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J Cell Biol. 1999;146:1333–1350. doi: 10.1083/jcb.146.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008;10:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Roper RL, Wolffe EJ, Weisberg A, Moss B. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J Virol. 1998;72:4192–4204. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Agnew BJ, Abe H, Bamburg JR, Mitchison TJ. Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttger S, Frischknecht F, Reckmann I, Smith GL, Way M. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J Virol. 1999;73:2863–2875. doi: 10.1128/jvi.73.4.2863-2875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Sanderson CM, Frischknecht F, Way M, Hollinshead M, Smith GL. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- Sanger JM, Sanger JW, Southwick FS. Host cell actin assembly is necessary and likely to provide the propulsive force for intracellular movement of Listeria monocytogenes . Infect Immun. 1992;60:3609–3619. doi: 10.1128/iai.60.9.3609-3619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaplehorn N, Holmstrom A, Moreau V, Frischknecht F, Reckmann I, Way M. Grb2 and nck act cooperatively to promote actin-based motility of vaccinia virus. Curr Biol. 2002;12:740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Bozeman FM, Smadel JE. Study on the growth of Rickettsiae. II. Morphologic observations of living Rickettsiae in tissue culture cells. Virology. 1957;3:160–172. doi: 10.1016/0042-6822(57)90030-2. [DOI] [PubMed] [Google Scholar]
- Sitthidet C, Stevens JM, Field TR, Layton AN, Korbsrisate S, Stevens MP. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. Journal of bacteriology. 2010;192:5249–5252. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. Journal of Cell Biology. 2000;150:527–538. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Portnoy DA, Theriot JA. Asymetric distribution of the Listeria monocytogenes ActA protein is required and sufficient to direct actin-based motility. Molecular microbiology. 1995;17:945–951. doi: 10.1111/j.1365-2958.1995.mmi_17050945.x. [DOI] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Vanderplasschen A, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002;83:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Takeshima F, Anton I, Liu CH, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Galyov EE. Exploitation of host cells by Burkholderia pseudomallei. Int J Med Microbiol. 2004;293:549–555. doi: 10.1078/1438-4221-00292. [DOI] [PubMed] [Google Scholar]
- Stokes GV. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miki H, Takenawa T, Sasakawa C. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 1998;17:2767–2776. doi: 10.1093/emboj/17.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Mimuro H, Suetsugu S, Miki H, Takenawa T, Sasakawa C. Neural Wiskott-Aldrich syndrome protein (N-WASP) is the specific ligand for Shigella VirG among the WASP family and determines the host cell type allowing actin-based spreading. Cellular microbiology. 2002;4:223–233. doi: 10.1046/j.1462-5822.2002.00185.x. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nature reviews Microbiology. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature (Lond) 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature (Lond) 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Rosenblatt J, Portnoy DA, Goldschmidt CP, Mitchison TJ. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Tilney LG, DeRosier DJ, Tilney MS. How Listeria exploits host cell actin to form its own cytoskeleton. I. Formation of a tail and how that tail might be involved in movement. J Cell Biol. 1992a;118:71–81. doi: 10.1083/jcb.118.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, DeRosier DJ, Weber A, Tilney MS. How Listeria exploits host cell actin to form its own cytoskeleton. II. Nucleation, actin filament polarity, filament assembly, and evidence for a pointed end capper. J Cell Biol. 1992b;118:83–93. doi: 10.1083/jcb.118.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijl H, Hollinshead M, Smith GL. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology. 2000;271:26–36. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell host & microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman LE. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology. 1988;163:547–553. doi: 10.1016/0042-6822(88)90295-4. [DOI] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA, Hess RT. Evidence for microfilament involvement in budded Autographa californica nuclear polyhedrosis virus production. Virology. 1987;156:32–39. doi: 10.1016/0042-6822(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Volkman LE, Talhouk SN, Oppenheimer DI, Charlton CA. Nulcear F-actin: a functional component of baculovirus-infected lepidopteran cells? J Cell Sci . 1992;103:15–22. [Google Scholar]
- Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by the Arp2/3 protein complex at the surface of Listeria monocytogenes . Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy D, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Weston L, Coutts AS, La Thangue NB. Actin nucleators in the nucleus: an emerging theme. J Cell Sci. 2012;125:3519–3527. doi: 10.1242/jcs.099523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Weisberg AS, Moss B. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology. 1998;25:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- Wolffe EJ, Weisberg AS, Moss B. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J Virol. 2001;75:303–310. doi: 10.1128/JVI.75.1.303-310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch DM. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature cell biology. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Zettl M, Way M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr Biol. 2002:1617–1622. doi: 10.1016/s0960-9822(02)01112-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.