Abstract
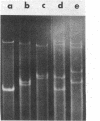
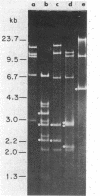
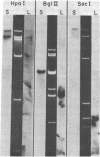
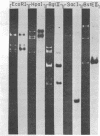
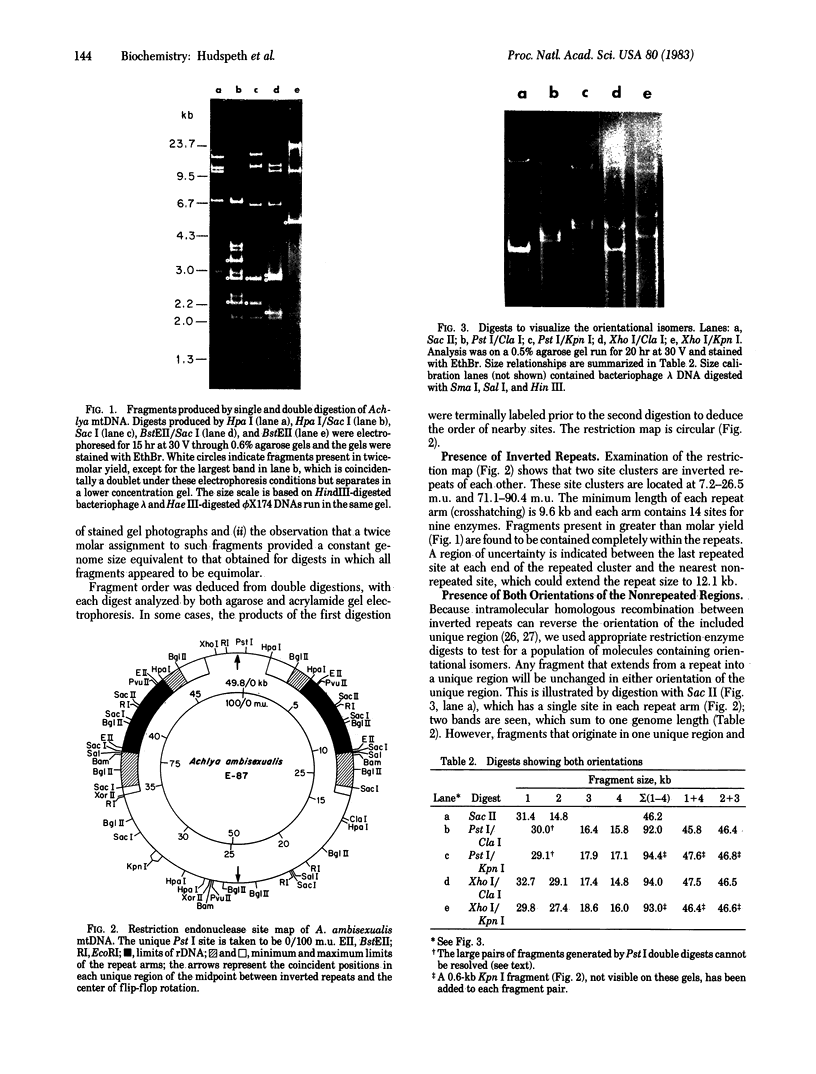
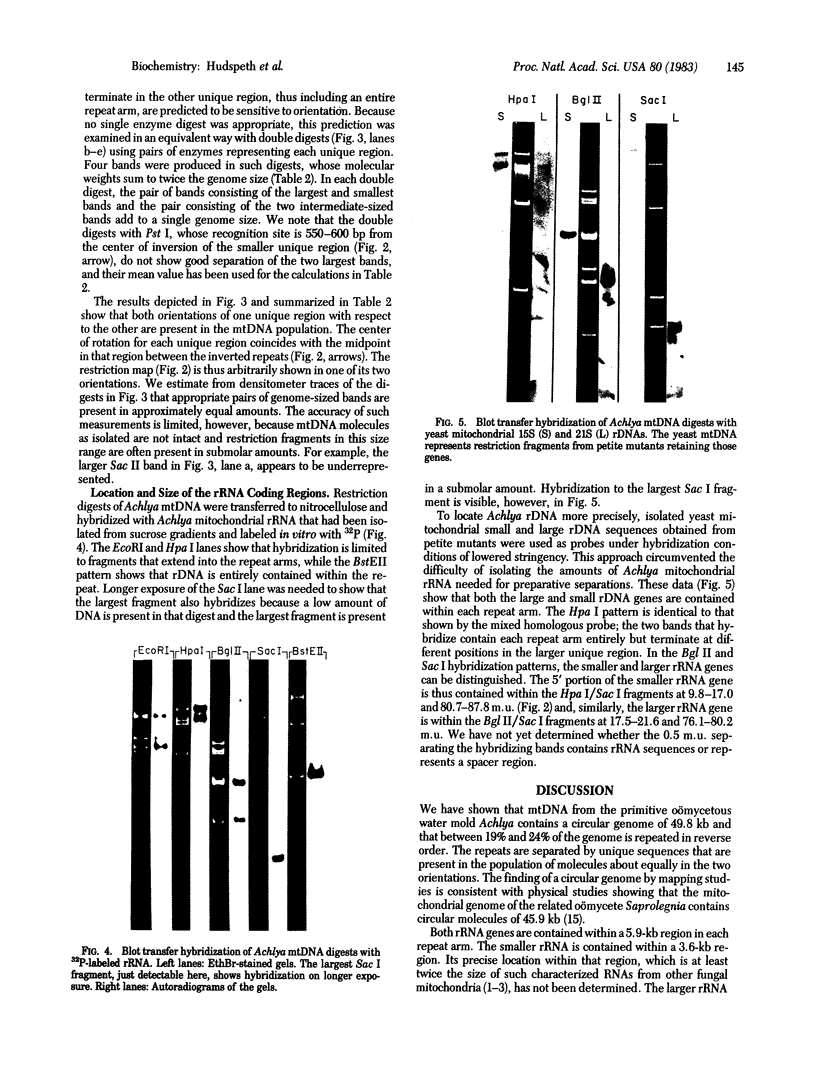
We have investigated mtDNA organization in the oömycetous water mold; Achlya, and report here that this primitive organism contains a circular mitochondrial genome of 49.8 kilobase pairs. Extensive restriction endonuclease analysis indicates that a significant portion of the genome is present as an inverted repeat. Of 52 restriction sites for 14 enzymes thus far mapped, 28 sites cluster in two 9.6-kilobase-pair regions; within these regions, the sequence of sites is inverted but the spacing between analogous sites is identical. The repeat arms have a maximum length of 12.1 kilobase pairs and are separated by 4.6-8.4 and 21.0-22.3 kilobase pairs of unique sequences. Transfer hybridization experiments show that genes for both the large and the small rRNAs are contained within each repeat. Restriction endonuclease analysis shows that the unique regions between the inverted repeats are present in both possible orientations with respect to each other and in approximately equal proportions. These orientational, or "flip-flop," isomers of the unique regions are postulated to occur by intramolecular homologous recombination between the repeated regions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelberg E. A., Bergquist P. The stabilization of episomal integration by genetic inversion: a general hypothesis. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2061–2065. doi: 10.1073/pnas.69.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., Heyting C., Borst P., Arnberg A. C., Van Bruggen E. F. An insert in the single gene for the large ribosomal RNA in yeast mitochondrial DNA. Nature. 1978 Sep 28;275(5678):336–338. doi: 10.1038/275336a0. [DOI] [PubMed] [Google Scholar]
- Browning K. S., RajBhandary U. L. Cytochrome oxidase subunit III gene in Neurospora crassa mitochondria. Location and sequence. J Biol Chem. 1982 May 10;257(9):5253–5256. [PubMed] [Google Scholar]
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Partial duplication of the large ribosomal RNA sequence in an inverted repeat in circular mitochondrial DNA from Kloeckera africana. Implications for mechanisms of the petite mutation. J Mol Biol. 1981 Apr 15;147(3):399–415. doi: 10.1016/0022-2836(81)90492-7. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Goldbach R. W., Bollen-de Boer J. E., van Bruggen E. F., Borst P. Conservation of the sequence and position of the ribosomal RNA genes in Tetrahymena pyriformis mitochondrial DNA. Biochim Biophys Acta. 1978 Nov 21;521(1):187–197. doi: 10.1016/0005-2787(78)90261-7. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. Mitochondrial genome diversity and the evolution of mitochondrial DNA. Can J Biochem. 1982 Mar;60(3):157–171. doi: 10.1139/o82-022. [DOI] [PubMed] [Google Scholar]
- Griffin D. H., Breuker C. Ribonucleic acid synthesis during the differentiation of sporangia in the water mold Achlya. J Bacteriol. 1969 May;98(2):689–696. doi: 10.1128/jb.98.2.689-696.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau M., Grandchamp C., Slonimski P. P. Circular DNA of a yeast episome with two inverted repeats: structural analysis by a restriction enzyme and electron microscopy. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3030–3034. doi: 10.1073/pnas.73.9.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., Underbrink-Lyon K., Martin N. C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982 May 25;257(10):5921–5928. [PubMed] [Google Scholar]
- Lovett J. S., Leaver C. J. High-molecular-weight artifacts in RNA extracted from Blastocladiella at elevated temperatures. Biochim Biophys Acta. 1969 Dec 16;195(2):319–327. doi: 10.1016/0005-2787(69)90639-x. [DOI] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system. The DNA sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. J Biol Chem. 1979 Jun 10;254(11):4617–4623. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moore K. H., Johnson P. H., Chandler S. E., Grossman L. I. A restriction endonuclease cleavage map of mouse mitochondrial DNA. Nucleic Acids Res. 1977;4(5):1273–1289. doi: 10.1093/nar/4.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Thompson W. F. Rearrangements in the chloroplast genomes of mung bean and pea. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5533–5537. doi: 10.1073/pnas.78.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Séquence nucléotidique du gène de l'ARN ribosomique 15S mitochondrial de la levure. C R Seances Acad Sci D. 1980 Dec 8;291(12):933–936. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Structure and evolution of organelle genomes. Microbiol Rev. 1982 Jun;46(2):208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young I. G., Anderson S. The genetic code in bovine mitochondria: sequence of genes for the cytochrome oxidase subunit II and two tRNAs. Gene. 1980 Dec;12(3-4):257–265. doi: 10.1016/0378-1119(80)90108-0. [DOI] [PubMed] [Google Scholar]