Abstract
Axonal protein synthesis has been shown to play a role in developmental and regenerative growth, as well as in cell body responses to axotomy. Recent studies have begun to identify the protein products that contribute to these autonomous responses of axons. In the peripheral nervous system, intra-axonal protein synthesis has been implicated in the localized in vivo responses to neuropathic stimuli, and there is emerging evidence for protein synthesis in CNS axons in vivo. Despite that hundreds of mRNAs have now been shown to localize into the axonal compartment, knowledge of what RNA binding proteins are responsible for this is quite limited. Here, we review the current state of knowledge of RNA transport mechanisms, and highlight recently uncovered mechanisms for dynamically altering the axonal transcriptome. Both changes in the levels or activities of components of the RNA transport apparatus and alterations in transcription of transported mRNAs can effectively shift the axonal mRNA population. Consistent with this, the axonal RNA population shifts with development, with changes in growth state, and in response to extracellular stimulation. Each of these events must impact the transcriptional and transport apparatuses of the neuron, thus directly and indirectly modifying the axonal transcriptome.
Keywords: RNA transport, RNA binding protein, ribonuclear particle, axonal transport
Polarized cells can target mRNAs to subcellular domains, and this provides a means to spatially and temporally control protein levels in these regions (Martin and Ephrussi, 2009). This is particularly needed in neurons that extend their cytoplasmic processes for long distances from the cell body as dendrites and axons. This RNA targeting essentially represents a localized regulation of neuronal gene expression, enabling dendrites and axons to rapidly and autonomously respond to their environment and injury. Intuitively, this would bring an advantage to the axonal processes that can extend a meter or more in large vertebrate organisms. Protein synthesis in dendrites has long been recognized as a means for modulating synaptic function. A variety of stimuli, including neural activity, have since been shown to modify protein synthesis in dendrites, particularly at or near the dendritic spine (Liu-Yesucevitz et al., 2011). Though early ultrastructural studies of rodent hippocampus suggested that axons do not contain any mRNAs or translational machinery (Steward and Levy, 1982), a steady stream of publications up to the late 1990’s suggested that axons could have protein synthetic activity under at least some circumstances (see (Twiss and van Minnen, 2006; Jung et al., 2012) and references within). Advances in nucleic acid detection and reporter systems for visualizing mRNAs and newly synthesized proteins in subcellular locations were since used to definitively show that axons, even those of mammals, contain mRNAs and translational machineries and locally generate new proteins (Jung et al., 2012).
Advances in RNA detection have also allowed investigators to profile axons for their mRNA content. This has uncovered an unexpected complexity in how many different transcripts can localize into axons. DNA microarray approaches from several laboratories using different neuronal preparations have now shown that axons can contain hundreds of different mRNAs (Willis et al., 2007; Taylor et al., 2009; Zivraj et al., 2010; Gumy et al., 2011). There is also increasingly conclusive evidence for axonal mRNA localization in vivo, both in the PNS and CNS (Brittis et al., 2002; Sotelo-Silveira et al., 2008; Donnelly et al., 2011; Willis et al., 2011; Ben-Yaakov et al., 2012; Walker et al., 2012; Merianda et al., 2013a and 2013b), but the in vivo profiles of axonal mRNAs have not been determined thus far. However, recent deep sequencing for RNA profiles of synaptic neuropil segments from hippocampus, where investigators used filtering to remove glial, vascular, interneuron, nuclear and mitochondrial RNA profiles, suggests that similarly complex populations of mRNAs can localize into neuronal processes in vivo (Cajigas et al., 2012).
The recently uncovered complexity of the axonal transcriptome emphasizes how much we do not know about the mechanisms of RNA transport, even for cultured neurons. mRNAs are transported as RNA-protein complexes composed of RNA binding proteins (RBP) and other proteins that afford interaction with motor proteins for microtubule-based transport. Microfilament-based transport likely contributes to short-range movement of mRNAs in axons (e.g., within the growth cone – see Yao et al., 2006), so constituents of the RNA-protein complex presumably must also allow for interaction with actin-myosin motor proteins (Sotelo-Silveira et al., 2004; Sotelo-Silveira et al., 2006; Donnelly et al., 2010). As we outline below, only a few RBPs are known to localize into axons and it is not clear how many different axonal mRNAs a single RBP interacts with. It is clear that axonal mRNA transport can be regulated through multiple different stimuli including the differentiation and growth status of the neuron (Willis et al., 2007; Taylor et al., 2009; Gumy et al., 2011; Merianda et al., 2013a and 2013b), but the molecular mechanisms underlying this regulation are not clear. RBPs are prime candidates for modulating post-transcriptional control at multiple levels and multiple functions have been ascribed to individual RBPs. In the paragraphs below, we will focus on how this RBP exerts multifunctionality and how RBP-mRNA interactions help to determine the axonal transcriptome.
mRNAs are transported as RNA-protein complexes
RBPs play central roles in post-transcriptional processes that ultimately modulate protein expression. In the nucleus, newly transcribed RNAs undergo processing to splice out introns and add 3’ poly-adenylate tails and 5’ methyl-guanosine caps. Various RBPs, such as the multitude of heterogeneous nuclear ribonucleoproteins (hnRNP), facilitate this RNA processing in the nucleus. However, many of these RBPs can also be found in the cytoplasm where they can play a role in mRNA stability, transport, and/or translational regulation (Agnes and Perron, 2004). Oftentimes, the same RBP can exert multiple functions on an mRNA. For example, the same RBP that is needed for axonal transport of β-actin mRNA can also stabilize mRNAs (Neilsen et al., 2004; Weidensdorfer et al., 2009)
RBPs contain one or more RNA-binding domains known as RNA recognition motifs (RRM) or other classic RNA binding motifs (e.g., ‘KH domain’), through which they bind to mRNA(s) to form a ribonucleoprotein (RNP) complex (Glisovic et al., 2008). Sometimes a single domain is sufficient to specify the RNA recognition ability of a given protein. Oftentimes a single RNA binding domain does not function as an independent RNA recognition unit, rather multiple domains contribute to define their mRNA specificity (Maris et al., 2005). The RBPs also interact with other proteins in the RNP complexes. For instance, zip code binding protein 1 (ZBP1; also called Vg1 RNA binding protein [Vg1RBP], insulin like growth factor II mRNA binding protein [IMP1] and coding region determinant binding protein [CRD-BP]) is needed for localization of β-actin mRNA (Ross et al., 1997). ZBP1 interacts with the KH-type splicing regulatory protein (KSRP; also called zipcode binding protein 2 [ZBP2], far-upstream element binding protein [FBP2]), HuC, and hnRNP E1, E2 and L in lysates from rat brain and HEK cells (Snee et al., 2002; Jonson et al., 2007). ZBP1’s interaction with hnRNP Q, hnRNP R, G3BP1, and HuD has also been suggested from analyses of different localizing mRNAs (Atlas et al., 2007; Jonson et al., 2007; Glinka et al., 2010; Yoo et al., 2013). These different RBPs may bring additional properties to the RNP. For example, HuD has classically been linked to stabilization of mRNAs with adenine-uridine rich elements (ARE) in their 3’ untranslated regions (UTR) (Bolognani et al., 2009), but HuD has also been shown to play a role in translational regulation (Fukao et al., 2009; Sosanya et al., 2013). On the other hand, a single RBP may be involved in multiple steps of RNA processing. For instance, the Drosophila hnRNP A/B ortholog, Hrp48, which contributes to RNA splicing (Hammond et al., 1997), has also been shown to play roles in transport and translational repression of Oskar RNA in oocytes (Yano et al., 2004). Sequestering transported mRNAs from the translational machinery seems to be a theme with RBPs, providing further evidence for multifunctionality (Wells, 2006). For example, ZBP1 has been shown to repress translation of bound mRNAs (Huttelmaier et al., 2005), in addition to its role in transport and stabilization of mRNAs. This sequestration could also protect mRNAs from degradation. Taken together, these and other works indicate that RBPs impact post-transcriptional regulation at multiple levels through interaction with multi-protein complexes. However, this is not to imply that the RBPs provide these functions in isolation. Work from the Flanagan lab indicates that ribosome subunits can be tethered to the cytoplasmic domains of transmembrane receptors and held in an inactive state until the receptor is activated (Tcherkezian et al., 2010).
Similar to receptor-ligand interactions, RBPs recognize mRNAs based on primary sequence or secondary structures in the mRNA ligands. However, RBPs appear to be much more promiscuous in their interactions with these RNA ligands, since a single RBP can bind to multiple different mRNAs. This view is based largely on RNA-protein co-immunoprecipitation approaches, both traditional ‘RNA co-immunoprecipitation’ (RIP) as well as ‘cross-linking and immunoprecipitation’ (CLIP) approaches. For example, RIP assays in HEK cells identified over two hundred different mRNAs binding to IMP1, the human ortholog of ZBP1 (Jonson et al., 2007). A limitation of this traditional RIP approach is that mRNAs that are indirectly bound to the protein as part of a multi-protein RNP can also be identified and some of these may not be specific for the interactions (Darnell and Klann, 2013). Nonetheless, roles of RNA-associated proteins rather than classic RBPs through protein-protein interactions within RNPs may impart additional post-transcriptional regulatory functions. Addition of steps to query direct mRNA-protein interactions in brain lysates was used for HuD; this decreased the complexity of targets and allowed for prediction of binding sites, but still resulted in hundreds of different mRNA ligands for HuD (Bolognani et al., 2009). The CLIP approach queries mRNA-protein interactions directly in cells and tissues prior to lysis by cross linking the protein to its mRNA target. Despite the increase in specificity with the CLIP approach, these assays have still pointed to individual RBPs interacting with multiple mRNAs. For example, over 800 target mRNAs were detected from mouse brain for the fragile X mental retardation protein (FMRP) (Darnell et al., 2011). Interestingly, RBP target analyses in Saccharomyces cerevisiae indicated that many of the mRNA cohorts that individual RBPs associate with mRNAs encoding functionally related proteins (Hogan et al., 2008). For the FMRP targets noted above, ninety of those mRNAs encode for components of the pre-synaptic proteome (Darnell et al., 2011). It will be important to determine if axonal mRNA transport cohorts have evolved such that individual RBPs transport mRNAs encoding proteins with shared or complementary functions in the axons. As noted above, multiple RBPs have been isolated with individual mRNAs, so functional cohorts may also be established by the specific combination of bound RBPs or RNA-associated proteins.
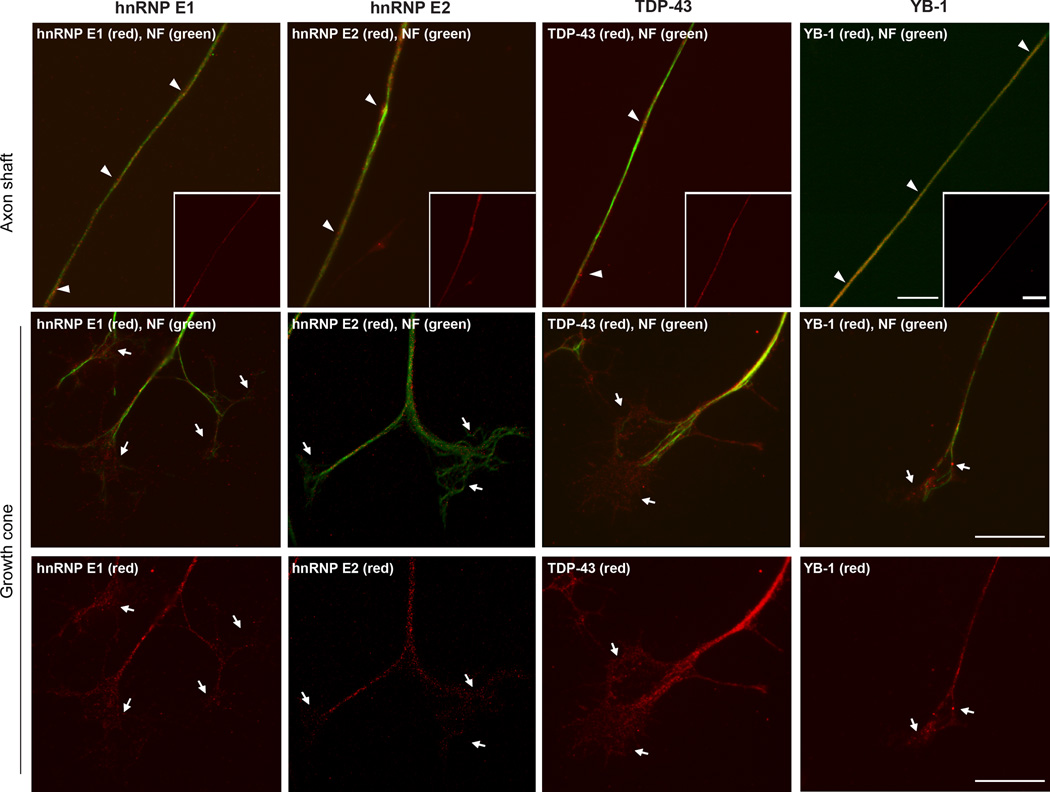
Several studies have shown the importance of RBPs in neural development and their involvement in neurite growth, as well as roles in synaptic plasticity (see (Agnes and Perron, 2004; Liu-Yesucevitz et al., 2011) and references within). Table 1 summarizes RBPs that have been demonstrated in axons from varying studies. Thus far, only a few of these have been shown to directly interact with mRNAs in the axonal compartment and most studies of these interactions have been limited to single mRNAs. Based on sheer numbers of mRNAs localizing into axons, additional RBPs must localize into the axonal compartment. Many proteins have RNA binding activities but their localization has not been thoroughly tested. Consistent with this, we previously showed that the La/SSB RBP localizes into axons (van Niekerk et al., 2007). La/SSB is an ‘RNA chaperone’ protein that is enriched in the nucleus, but has distinct mRNA-targeted functions in the cytoplasm (Wolin and Cedervall, 2002). We have used immunostaining to determine if additional RBPs that have been linked to subcellular post-transcriptional regulation in other systems might localize to axons of adult rat sensory neurons in culture. We see axonal localization of hnRNP E1, hnRNP E2, transactive response DNA-binding protein 43 (TDP-43), and Y-box binding protein 1 (YB-1), with each showing a granular localization pattern in the DRG axons similar to the appearance of transported mRNAs (Figure 1). hnRNP E1 and E2, which are also called poly-C binding proteins (PCBP) 1 and 2, respectively, are members of an hnRNP family that contains three KH domains (Leffers et al., 1995; Makeyev et al., 1999). E1 and E2 colocalize with RNA transport granules in oligodendrocytes, with E1 suggested to play a role in translational suppression (Kosturko et al., 2006). Multiple post-transcriptional roles have been attributed to TDP-43. Although much of these are considered as nuclear events (Buratti and Baralle, 2001; Buratti et al., 2001; Wang et al., 2004), TDP-43 also localizes to dendrites of hippocampal neurons (Wang et al., 2008) and to axons of spinal motor neurons (Fallini et al., 2012). Mutations in the human gene encoding TDP-43 (TARDBP) can cause amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Neumann et al., 2006; Sreedharan et al., 2008). YB-1, a member of a family of well-conserved multifunctional DNA/RNA binding protein family, plays a role in brain development and appears to function as a translational suppressor (Kohno et al., 2003; Lu et al., 2005). YB-1 also binds to Purα, a protein that has been linked to mRNA transport into dendrites (White et al., 2009).
Table 1. Axonal RNA-associated proteins and their targeted mRNAs.
Summary of RBPs and RNA-associated proteins (cumulatively termed RNA-associated proteins) identified in axons of cultured neurons and/or in vivo through localization studies is shown. These associations could be for transport, translation, or other post-transcriptional effects in the axons. Additional proteins with RNA binding activities have been identified from proteomics of growth cone and axonal preparations (Nozumi et al., 2009; Estrada-Bernal et al., 2012), although these have largely not been validated at the individual protein level. For the mRNA targets included in the third column, those in standard text have been validated by colocalization, co-immunoprecipitation, RBP knockdowns, or a combination of approaches. The RNA targets outlined in italicized text are cross-referenced from CLIP/RIP studies for the mRNA targets and axonal mRNA studies, so these are potential targets (i.e., not fully validated).
| RNA-Associated Protein |
Evidence for Axonal Localization | Axonal mRNA Targets |
|---|---|---|
| Copb1 | Cultured sensory neurons (Bi et al., 2007) | κ-opioid receptor (Bi et al., 2007) |
| CPEB1 | Cultured hippocampal neurons, optic nerve, cultured retinal ganglion cells, spinal cord commissural axons (Brittis et al., 2002; Kundel et al., 2009; Lin et al., 2009) | β-Catenin, EphA1 (Brittis et al., 2002; Kundel et al., 2009) |
| FMRP | Sciatic nerve, dorsal root, and thalamocortical axons (Hengst et al., 2006; Price et al., 2006; Murashov et al., 2007; Christie et al., 2009; Akins et al., 2012) | MAP1B (Zhang et al., 2001; Antar et al., 2005) |
| Grb7 | Neuronal-like P19 cells (Tsai et al., 2007) | κ-opioid receptor (Tsai et al., 2007) |
| Hermes/RBPMS | Xenopus retinal ganglion cell axons (Hörnberg et al., 2013) | |
| hnRNP R | Cultured spinal motor neurons (Glinka et al., 2010) | β-Actin (Glinka et al., 2010; Rossoll et al., 2003) |
| hnRNP Q1 | Cultured hippocampal neurons (Xing et al., 2012) | RhoA (Xing et al., 2012) |
| hnRNP A1* | Cultured sensory neurons (Willis et al., 2007) | |
| HuD | Cultured spinal motor neurons (Fallini et al., 2011) | GAP-43, Tau (Atlas et al., 2007; Yoo et al., 2013) |
| La/SSB | Cultured sensory neurons (van Niekerk et al., 2007) | RPL37 (Crosio et al., 2000; van Niekerk et al., 2007) |
| SMN | Cultured spinal motor neurons, NSC34 neuron-like cells, and cortical neurons (Zhang et al., 2003; Setola et al., 2007; Akten et al., 2011; Fallini et al., 2011) and zebrafish motor axons in vivo (Carrel et al., 2006) | β-Actin (Rossoll et al., 2003) CPG15/Nrn1, Cav2.2 (Jablonka et al., 2007; Akten et al., 2011) |
| Stau2 | Sciatic nerves and dorsal root axons (Price et al., 2006) | β-actin, Cofilin 1, Calreticulin, eIF2B2,GAP-43, MAP1B, RhoA, RPL21, RPS11, SepW1 (Willis et al., 2007; Maher-Laporte and DesGroseillers, 2010; Kar et al., 2013) |
| TDP-43 | Cultured spinal motor neurons (Fallini et al., 2012) | β-actin, MAP1B, RPL37, Tau (Willis et al., 2007; Sephton et al., 2011) |
| ZBP1 | Cultured forebrain neurons, hippocampal neurons, sensory neurons (Zhang etal., 2001; Donnelly et al., 2011), adult myelinated axons (Sotelo-Silveira et al., 2008) | β-Actin, GAP-43 (Zhang et al., 2001; Yoo et al., 2013) |
hnRNP A1 was identified as an axonal mRNA and validated at the protein level
Figure 1. Axonal localization of RNA Binding Proteins.
Representative fluorescent micrographs for cultures of adult DRG neurons that were immunostained with antibodies to the indicated RNA binding proteins (red) and neurofilament (green) are shown. The top row shows axon shaft (arrow heads) with red/green channels merged in the main panel and red channel only in the inset panel. Middle and bottom rows show growth cones (arrows) as red/green merged channels in the middle row and red only channels merged in the bottom row as indicated. hnRNP E1, hnRNP E2, TDP-43 and YB-1 all show localization to the axons with granular profiles. TDP-43 and YB-1 appear somewhat more concentrated in the growth cones compared with the 2 hnRNPs [scale bar = 10 µm].
hnRNP E1 and E2 have been shown to bind to mRNA encoding the medium molecular weight neurofilament (NF-M) (Thyagarajan and Szaro, 2008), and the low molecular weight neurofilament (NF-L) mRNA appears to be stabilized by binding to TDP-43 (Strong et al., 2007). Both NF-L and NF-M mRNAs localize into axons of cultured sensory neurons (Gumy et al., 2011). Work from Sotelo-Silveira et al. (2000) suggested that NF-L and NF-M mRNAs could localize into sciatic nerve axons in vivo; however, this work also demonstrated NF-L mRNA in Schwann cells by a radioactive in situ hybridization (ISH) technique. As indicated above, limitations of the ISH approach used by Sotelo-Silveira et al. (2000) are now being overcome by advancing to confocal microscopy with high sensitivity fluorescent ISH coupled with simultaneous detection of axonal proteins and with viral and transgenic reporter-UTR approaches (for recent examples see Yoo et al., 2013; Walker et al. 2012; Merianda et al., 2013b; and Perry et al., 2013). It should be noted that similar to the other known axonal RBPs noted in Table 1, immunolocalizations do not tell if the axonally localizing hnRNP E1, hnRNP E2 and/or TDP-43 interact with NF-L, NF-M or other mRNAs in the axons. Regardless, these images emphasize the potential for many different RNA-associated proteins to localize into the axonal compartment. Moreover, it is intriguing to speculate that changes in local axonal mRNA dynamics may contribute to the dying back axonopathy seen in ALS given that TDP-43 localizes to axons of cultured spinal motor neurons (Fallini et al., 2012). RIP and CLIP analyses of these and other RNA-associated proteins shown in Table 1 do bring some clear overlap with mRNAs previously detected in axons by candidate screens and non-biased RNA profiling approaches (Table 1). Although the potential for functions of these RBPs in axons exists, directed studies are clearly needed to fully define the axonal RBP population and functions of each in the axonal compartment.
RNA binding proteins recognize cis-elements in mRNAs
RBPs recognize and bind to structures within the mRNAs themselves that target them for transport into subcellular domains including axons. These ‘cis-elements’ are often found in UTRs of the mRNAs. Work from several different groups now indicate that 3’UTR of some axonal mRNAs is sufficient to localize reporter mRNA into axonal processes (Aronov et al., 1999; Wu et al., 2005; Yudin et al., 2008; Vuppalanchi et al., 2010; Ben-Yaakov et al., 2012; Merianda et al., 2012; Yoo et al., 2013). The 5’UTRs can also contribute to localization. For mRNAs encoding the rodent κ-opioid receptor and Aplysia sensorin, the 5’ and 3’UTRs contribute to their localization (Bi et al., 2006; Meer et al., 2012). However, multiple roles are ascribed to these RNA elements. For κ-opioid receptor mRNA, either the 5’ or 3’UTR is sufficient for localization but the 5’UTR provides a higher translational efficiency in axons (Bi et al., 2006). For Sensorin mRNA, the 3’UTR drives export from the cell body but the 5’UTR concentrates the mRNA at synapses in sensory-motor neuron co-cultures (Meer et al., 2012). The 3’UTR of neuritin mRNA (also called CPG15) was shown to drive its localization into axons of hippocampal neurons (Akten et al., 2011). However, neuritin’s 5’UTR seems to be the driving force for axonal localization in sensory neurons, with only minimal activity from the 3’UTR (Merianda et al., 2013a). These and other studies point to the importance of UTR structures in targeting mRNAs to subcellular domains, and emphasize the need to test multiple RNA regions for post-transcriptional activity. Protein-coding sequences of mRNAs are not without any localizing activities and some of these are driven co-translationally. Targeting mRNAs to rough endoplasmic reticulum through the signal peptide sequence has long been recognized for mRNAs encoding secreted and membrane localized proteins. Still, there are additional mRNAs targeted to the ER through translational-independent means, at least in non-neuronal systems (see (Kraut-Cohen and Gerst, 2010) and references within). Adding yet another level of complexity to mRNA targeting, the yeast ABP140 mRNA is targeted to the cell periphery through a co-translational mechanism that is able to override more conventional 3’UTR localization signals (Kilchert and Spang, 2011). It is highly likely that such mechanisms will be uncovered for axons as interest in RNA transport and localized translation grows. Additionally, there can be multiple cis-elements within a single UTR that can have distinct functions (Vuppalanchi et al., 2010).
The importance of RNA sequence/structure for RNA localization is further illustrated by works showing that alternative transcription termination sites can determine whether an mRNA localizes or not. Specifically, Importin-β1, RanBP1 and Stat3α genes can generate transcripts of differing 3’UTR length, with only the longest UTRs localizing into axons (Yudin et al., 2008; Ben-Yaakov et al., 2012; Perry et al., 2012). BDNF mRNA shows a similar differential localization of its long transcripts into dendrites, with restriction of the shorter UTR transcripts in the cell body (An et al., 2008; Chiaruttini et al., 2008). Despite that RNA segments have been linked to axonal transport of mRNAs, there has been no unifying sequence or structure identified to drive this process. Conservation of non-coding sequences between species has helped to identify candidate cis-elements in individual axonal mRNAs for analyses (Vuppalanchi et al., 2010), but this has not been a universal trait for localizing mRNAs.
The zip code element in β-actin mRNA has been the best-characterized cis-element for axonal localization to date. β-actin’s 3’UTR drives its transport axons in cultured CNS and PNS neurons (Zhang et al., 2001; Willis et al., 2007). This 3’UTR drives mRNA localization in sensory neurons in vivo, both in the peripheral and centrally projecting processes (Willis et al., 2011). ZBP1 binds to this cis-element and is essential for subcellular localization of β-actin mRNA (Ross et al., 1997). Although RIP assays demonstrated over 200 mRNAs co-precipitating with the human ZBP1 ortholog, IMP1 (Jonson et al., 2007), there does not appear to be any common RNA cis-element defining these ZBP1 targets. Consistent with this, we recently showed that axonal transport of GAP-43 mRNA requires ZBP1, yet this mRNA shows no homology to β-actin’s zip code element (Donnelly et al., 2011). Clearly, more work needs to be done to uncover common structures driving transport of different mRNAs. As we outline below, mRNAs can compete for binding to RBPs, thereby providing an inherent mechanism for modifying the localized transcriptome. However, this also presents an experimental opportunity to define cohorts of mRNAs that share transport mechanisms.
mRNAs can compete for transport into axons
The mechanisms that dictate which mRNAs and how much of a particular mRNA localizes into axons and other subcellular domains are just beginning to be tested. With functions in transport and stability of mRNAs, at least some RBPs are perfectly positioned as determinants of the subcellular mRNA population. Consistent with this, we recently showed that availability of ZBP1 in adult sensory neurons limits the axonal localization of β-actin mRNA. By introducing a GFP reporter mRNA with the axonally targeting 3’UTR of β-actin mRNA into DRG neurons, growing axons showed decreased levels of endogenous β-actin mRNA (Donnelly et al., 2011). The effects of this exogenous 3’UTR competitor extend to in vivo as well, since adult mice using expressing a GFP transgene with the 3’UTR of β-actin mRNA show depletion of endogenous β-actin mRNA from their peripheral axons (Donnelly et al., 2011). Since this transport deficit extended to other mRNAs, these data point to the possibility that ZBP1 is responsible for localizing a cohort of mRNAs into axons.
Such a ‘squelching mechanism’, where mRNAs compete for binding to limited amounts of an RBP and/or associated proteins, has been seen for other mRNAs. For example, overexpression of an exogenous reporter mRNA with the 3’UTR of Integrin α3 mRNA prevented localization of the endogenous Integrin α3 mRNA to the periphery of migrating cancer cells (Adereth et al., 2005). Similar to the decreased axonal regeneration seen in DRG neurons with the exogenous β-actin 3’UTR competitor (Donnelly et al., 2011), depleting Integrin α3 mRNA from the cell periphery attenuated cell migration (Adereth et al., 2005). Donnelly et al. (2013) very recently showed that expressing an exogenous mRNA with the 3’UTR of amphoterin effectively depletes axons of endogenous amphoterin mRNA (Donnelly et al., 2013). Though amphoterin (also called high mobility group protein box-1 [HMGB1]) is typically considered a nuclear protein, its mRNA has been shown to localize to the periphery of migrating cells (Punnonen et al., 1999; Fages et al., 2000). Amphoterin protein has been shown to induce cell migration and neurite growth (reviewed in (Huttunen and Rauvala, 2004)). However, the effects of depleting amphoterin mRNA from DRG axons are not known, since most of the amphoterin 3’UTR overexpression constructs recently used by Donnelly et al. (2013) included the open reading frames of β-actin or GAP-43 mRNA. Though a few exogenous UTR elements have now been shown to compete with their endogenous mRNA counterpart for subcellular localization, it is not clear if this is a generalized mechanism. With RNA localization elements for several other axonal mRNAs now coming to light (Wu et al., 2005; Yudin et al., 2008; Andreassi et al., 2010; Vuppalanchi et al., 2010; Ben-Yaakov et al., 2012; Perry et al., 2012; Merianda et al., 2013a and 2013b; Yoo et al., 2013), it will be possible to determine if this squelching mechanism extends to other mRNAs as a generalized mechanism to limit subcellular localization. This mechanism also raises caution for using such cis-elements in RNA transport and translation assays since localizations of both the endogenous mRNA counterpart for that element as well as other mRNAs can be competed against, with each bringing physiological consequences.
As noted above, decreasing the availability of ZBP1, either through expression of an exogenous UTR competitor from β-actin’s mRNA or deletion of one ZBP1 allele, decreases axonal levels of endogenous β-actin mRNA (Donnelly et al., 2011). These approaches also decrease axonal levels of GAP-43 mRNA in the DRG neurons, indicating that GAP-43 requires ZBP1 for its localization (Donnelly et al., 2011). We recently showed that a short segment of GAP-43’s 3’UTR is sufficient for its localization into sensory axons, but this RNA element shows no homology to the zip code element needed for β-actin’s localization (Yoo et al., 2013). However, consistent with the previous evidence for ZBP1-dependent transport of GAP-43 mRNA into axons (Donnelly et al., 2011), expressing an exogenous reporter mRNA with the 3’UTR elements of GAP-43 mRNA depletes axons of endogenous β-actin mRNA (Yoo et al., 2013). So β-actin and GAP-43 3’UTRs clearly compete for binding to limited quantities of ZBP1. Surprisingly, the RNA sequence needed for localization of GAP-43 mRNA and that competes with β-actin mRNA for localization is the adenine-uridine rich element (ARE) that has been well characterized for binding to the Elav-like protein HuD (Mobarak et al., 2000). In our hands, both HuD and ZBP1 bind to GAP-43 mRNA (Yoo et al., 2013). HuD binding to the 3’UTR of GAP-43 mRNA stabilizes the transcript, and HuD overexpression increases neurite outgrowth in vitro and in vivo (Bolognani et al., 2006; Perrone-Bizzozero et al., 2011). HuD and the human ortholog of ZBP1 have previously been shown to bind to Tau mRNA and it was suggested that a protein complex of ZBP1, HuD, and the RAS-GAP SH3 domain binding protein (G3BP1) is needed for axonal localization of Tau mRNA (Aronov et al., 1999; Atlas et al., 2004). Interestingly, HuD in complex with SMN was recently implicated in the axonal localization of neuritin/CPG15 mRNA in cultured hippocampal neurons through its 3’UTR (Akten et al., 2011). In PNS sensory neurons, neuritin/CPG15 mRNA localizes through its 5’UTR in a mechanism that is independent of ZBP1 (Merianda et al., 2013a; Donnelly et al., 2011). These studies suggest that RBPs assembled onto localizing mRNAs can show specificity for neuronal subtypes and their makeup changes with different mRNAs and potentially in different neuron types.
Expression of the RBPs needed for transport of individual mRNAs or changes in the availability of individual RBPs could effectively determine which mRNAs can localize into axons. Some mRNAs are known to be enriched in axons (Andreassi et al., 2010) and the axonal mRNA population can change with different growth conditions or stimulations (Willis et al., 2007; Taylor et al., 2009; Gumy et al., 2011). In DRG neurons, neural membrane protein 35 (NMP35) and neuritin/CPG15 mRNAs shift from cell body predominant in naïve neurons to axon predominant during regeneration (Merianda et al., 2013a and 2013b). This suggests that the RBPs needed to transport these two mRNAs are induced by injury, either at the level of their expression or activity (Figure 2A). This increase in RBP levels or activity would provide the neuron with a means to change subcellular localization of constitutively expressed mRNAs in a stimulus dependent manner. Changes in expression of a localized mRNA could similarly alter the axonal transcriptome by competing for binding to limited levels of constitutively expressed RBPs (Figure 2B). For example, GAP-43 mRNA levels increase several fold in the L4-5 DRGs after sciatic nerve injury (Van der Zee et al., 1989). With no corresponding change in β-actin mRNA levels, this shifts the overall ratio of GAP-43 mRNA to β-actin mRNA, resulting not only in more GAP-43 mRNA in axons but also in less axonal β-actin mRNA after axotomy (Yoo et al., 2013).
Figure 2. Schematic of mechanisms regulating axonal mRNA populations.
Hypothetical ‘basal’ and ‘stimulated’ conditions are noted for inducible vs. constitutively expressed RNA binding proteins (RBPi and RBPc, respectively). A, For RBPi, an increase in the levels or activity of this RBPs with stimulation results in a redistribution of target mRNAs from cell body restricted or predominant to axonal predominant. B, For RBPc, a shift in mRNA levels upon stimulation allows the green mRNA to outcompete the red mRNA for axonal localization. This both increases axonal levels of the green mRNA and decreases axonal levels of the red mRNA.
Potential for disrupted post-transcriptional processing and RBP competition in neurological diseases
Disruptions of post-transcriptional processes have been suggested to contribute to the pathogenesis and progression of some neurodegenerative diseases. Mutations in TDP-43 or fused in sarcoma/translocated in liposarcoma (FUS/TLS) proteins are causative for some familial cases of ALS and FTD (Sreedharan et al., 2008; Kwiatkowski et al., 2009; Vance et al., 2009). These two RBPs have roles in RNA processing in the nucleus and the mutant proteins localize to the cytoplasm (Lagier-Tourenne and Cleveland, 2009). Depletion of the ubiquitously expressed SMN protein, a regulator of RNA biogenesis and splicing that has also been detected in axons and linked to RNA localization (Table 1), is causative for Spinal muscular atrophy (SMA) (Gubitz et al., 2004). Mutations in prion-like domains of hnRNPA2/B1 and hnRNPA1 RBPs lead to a multisystem proteinopathy and ALS (Kim et al., 2013). Thus, alterations in RNA associated protein levels and/or functions are clear linked to neurological diseases.
There is evidence for RNA-RBP competition or ‘RBP squelching’ in genetic disorders resulting from tandem repeats in transcribed genes. Expanded CAG repeats in translated exons result in the accumulation of mutant proteins with polyglutamine repeats (reviewed in (Orr and Zoghbi, 2007)). However, the mRNAs with repeats can also be toxic through an RNA-mediated gain of function (see (Echeverria and Cooper, 2012) and references within). Once transcribed, the pre-mRNA or mRNA containing these repeats forms complex secondary structures including hairpin loops that can alter their processing, transport, translation and interaction with RNA binding proteins. For example, a tandem CTG repeat expansion in the 3’UTR of the dystrophia myotonica-protein kinase (DMPK) that causes myotonic dystrophy type 1 generates an mRNA that can ‘trap’ splicing factors such as the muscleblind-like family (MBNL) and CUG-binding protein 1 (CUGBP1) (Timchenko et al., 1996a and 1996b; Fardaei et al., 2001; Mankodi et al., 2001). These RBPs are important splicing factors for maturation of pre-mRNAs and misregulation of their activity is considered a trigger for the pathogenesis of myotonic dystrophy (Wojciechowska and Krzyzosiak, 2011; Echeverria and Cooper, 2012). CGG repeat expansions in the gene encoding FMRP (FMR1) causes Fragile X syndrome when there are over 200 repeats through epigenetic silencing (Bassell and Warren, 2008). However, FMR1 genes with 55–200 CGG repeats are transcribed and the mRNA with repeats in the 5’UTR takes on a toxic gain of function role by binding to hnRNPA2/B1, MBNL1, and Pur α (Echeverria and Cooper, 2012). This binding presumably sequesters these RBPs away from their normal cellular functions. Consistent with this, work from the Tiedge group indicates that the CGG expansion in FRMP mRNA competes with BC1 and PKMζ RNAs for interaction with hnRNP A2 and transport into dendrites (Muslimov et al., 2011). BC1 is a non-coding RNA that is hypothesized to have translational regulatory function in dendrites (Zhong et al., 2010), indicating that such RBP competition can extend to noncoding RNAs and alter what proteins are locally generated in neurons. BC1 RNA been shown to localize into goldfish Mauthner axons (Muslimov et al., 2002), and other non-coding RNAs have been detected in mammalian axons (Aschrafi et al., 2008; Natera-Naranjo et al., 2010; Kar et al., 2013). Thus, non-coding RNAs may also compete for binding to RBPs in the axonal compartment.
A hexanucleotide repeat (GGGGCC) in an intron of the C9orf72 gene, which is causative for familial ALS and FTD, was similarly shown to function as a toxic RNA by binding to Purα, ASF/SF2, and hnRNP A3 (Mori et al., 2013a; Reddy et al., 2013; Xu et al., 2013). However, there is also recent evidence that toxic peptides can be generated from C9orf72’s GGGGCC repeat as well as from other triplet repeat RNAs through non-AUG mediated translation (Mori et al., 2013b; Reddy and Pearson, 2013). Thus, transcribed repeat expansions may have multiple mechanisms underlying their pathophysiogical effects. It is intriguing to speculate that RBP competition may extend to the axonally targeted mRNAs, considering the complexities of transcripts that at least some axons appear to contain. For example, the MBNL1 RBP that FMRP mRNA’s CGG repeats binds (Echeverria and Cooper, 2012), also drives mRNA localization through binding to an ‘ACACCC’ motif in Integrin α3’s 3’UTR (Adereth et al., 2005). This is the same minimal sequence element that is needed for function of β-actin’s zip code element (Ross et al., 1997). Thus, the CGG repeat expansions in FMRP’s mRNA may affect localization of axonal mRNAs by competing for MBNL1 target mRNAs.
Conclusion and Perspectives
Local synthesis of new proteins in distal axons has been shown to contribute to developmental and regenerative axonal growth and retrograde signaling after injury (Jung et al., 2012). Intra-axonal translation contributes to growth of axons in cultured neurons, but there is also increasing evidence for functionality of locally synthesized proteins in rodent axons in vivo (Brittis et al., 2002; Donnelly et al., 2011; Ben-Yaakov et al., 2012; Walker et al., 2012). While axons can extend a few millimeters in cultured neurons, rodent PNS axons extend for several centimeters. Thus, terminal axons are at much greater distances from the cell body in vivo so locally generated proteins could provide many more functions to the axon than has been appreciated in studies focusing on growing axons. Consistent with this notion, very recent work in sympathetic neurons indicates that retrograde infection from pseudorabies virus presented at the axon terminal requires localized protein synthesis (Koyuncu et al., 2013). This, plus observations that neuronal responses to neuropathic pain inducing stimuli activate translation factors in nerve terminals (Jimenez-Diaz et al., 2008), suggests that axonally synthesized proteins may broadly affect neuronal function, both through localized effects and retrograde signaling. As noted, there is a huge gap in knowledge when it comes to matching the localized mRNAs to the molecular machineries that transport these mRNAs. Additional work will clearly be needed to fill this gap, both for determining the composition of the transport apparatus and the RBPs that are responsible for transporting the different mRNAs.
The possibility that mRNA cohorts encoding proteins with complementary functions are co-transported by the same RBP or RBP complexes is a very appealing regulatory mechanism in that this could provide a means to coordinately regulate axonal responses to extracellular environments. Increase in the levels or activity of individual RBPs or other limiting components of the transport machinery could be used to shift axonal RNA populations by increasing transport of different mRNAs into the axonal compartment as we have seen for NMP35 and neuritin/CPG15 mRNAs (Merianda et al., 2013a and 2013b) . Alternatively, the RNA competition model presented in Figure 2 above may provide a mechanism to selectively change axonal functions and responses by shifting the population of mRNAs targeted into axons and meet the physiological needs of the axon. There is some evidence for this with ZBP1 based transport, since axonally generated GAP-43 and β-actin proteins support distinct growth morphologies (Donnelly et al., 2013). The well established transcriptional induction of GAP-43 mRNA after axotomy is accompanied by a commensurate increase in axonal GAP-43 mRNA levels, but also a decrease in axonal β-actin mRNA levels (Yoo et al., 2013). Thus, competition between the endogenous β-actin and GAP-43 mRNAs could provide a selective advantage for regeneration, both by increasing local generation of GAP-43 protein to facilitate directed axon growth and by decreasing local generation of β-actin to attenuate axon branching (Donnelly et al., 2013; Yoo et al., 2013). Thereby, the expression levels of two key mRNAs can regulate neurite growth patterns by competing for binding to a single RBP. It will be interesting to determine if this competition mechanism extends to other axonal transcripts as well as to other settings of mRNA localization. If true, this could bring opportunities for broadly altering cellular functions by targeting the squelched RBPs.
Squelching of RBPs has also been reported in miRNA processing.Sellier et al. (2013) recently showed that the double stranded RNA binding protein DiGeorge syndrome critical region gene 8 (DGCR8) binds to expanded CGG repeats in FMRP mRNA. This partially sequesters both DGCR8 and Drosha microprocessor factors, so processing of micro-RNAs (miRNAs) is reduced in brain tissue from patients with Fragile X Tremor-Ataxia Syndrome (FXTAS) that results from 55–200 CGG repeats in FMR1 gene (Sellier et al., 2013). miRNAs have been demonstrated in axons and a few axonal mRNAs are known to be targets for intra-axonal actions of localized miRNAs (Aschrafi et al., 2008; Natera-Naranjo et al., 2010; Kar et al., 2013). Endogenous mRNAs have similarly been shown to compete for binding to miRNAs (Khvorova and Wolfson, 2012). So levels of mRNAs within cohorts targeted by the same miRNA can modulate miRNA effects on other mRNA targets. Thus, the transcriptional program can be utilized to regulate not only the levels of gene products transcribed but also translation or survival of other mRNAs. Of course, this model of mRNA-miRNA as well as the mRNA-RBP or -RNA-associated protein competition requires that the stoichiometry of the binding constituents and the relative affinities of the individual constituents for binding be considered in predicting any outcomes. Nonetheless, these clearly have implications for modulating the axonal transcriptome and proteome.
Acknowledgements
The authors for this work were supported in part by funds from the National Science Foundation (MCB-1020970), National Institutes of Health (R01-NS041596 and P20-RR020173), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. The authors have no conflicts of interests in this work.
References
- Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol. 2005;7:1240–1247. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnes F, Perron M. RNA-binding proteins and neural development: a matter of targets and complexes. Neuroreport. 2004;15:2567–2570. doi: 10.1097/00001756-200412030-00001. [DOI] [PubMed] [Google Scholar]
- Akins MR, Leblanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J Comp Neurol. 2012;520:3687–3706. doi: 10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D, Huang J, Merianda TT, Twiss JL, Beattie CE, Steen JA, Sahin M. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3' UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, Devita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Aronov S, Marx R, Ginzburg I. Identification of 3'UTR region implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999;12:131–145. doi: 10.1007/BF02736927. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem. 2004;89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Sapoznik S, Ginzburg I. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J Neurosci Res. 2007;85:173–183. doi: 10.1002/jnr.21099. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan S, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Blesch A, Pilpel Y, Twiss JL, Fainzilber M. Axonal Transcription Factors Signal Retrogradely In Lesioned Peripheral Nerve. EMBO J. 2012;31:1350–1363. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci USA. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Tsai NP, Lu HY, Loh HH, Wei LN. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc Natl Acad Sci USA. 2007;104:13810–13815. doi: 10.1073/pnas.0703805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2009;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Tanner DC, Merhege M, Deschenes-Furry J, Jasmin B, Perrone-Bizzozero NI. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J Neurochem. 2006;96:790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel TL, McWhorter ML, Workman E, Zhang H, Wolstencroft EC, Lorson C, Bassell GJ, Burghes AH, Beattie CE. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:2–8. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Fainzilber M, Twiss JL. Subcellular communication through RNA transport and localized protein synthesis. Traffic. 2010;11:1498–1505. doi: 10.1111/j.1600-0854.2010.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, Vuppalanchi D, McDonald M, Kim HK, Merianda TT, Gallo G, Twiss JL. Axonally Synthesized beta-Actin and GAP-43 Proteins Support Distinct Modes of Axonal Growth. J Neurosci. 2013;33:3311–3322. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Bernal A, Sanford SD, Sosa LJ, Simon GC, Hansen KC, Pfenninger KH. Functional complexity of the axonal growth cone: a proteomic analysis. PLoS One. 2012;7:e31858. doi: 10.1371/journal.pone.0031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fages C, Nolo R, Huttunen HJ, Eskelinen E, Rauvala H. Regulation of cell migration by amphoterin. J Cell Sci. 2000;113(Pt 4):611–620. doi: 10.1242/jcs.113.4.611. [DOI] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012;21:3703–3718. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell. 2009;36:1007–1017. doi: 10.1016/j.molcel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, Winkler C, Sendtner M. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19:1951–1966. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Loraine Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond LE, Rudner DZ, Kanaar R, Rio DC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörnberg H, Wollerton-van Horck F, Maurus D, Zwart M, Svoboda H, Harris WA, Holt CE. RNA-binding protein Hermes/RBPMS inversely affects synaptic density and axon arbor formation in retinal ganglion cells in vivo. J Neurosci. 2013;33:10384–10395. doi: 10.1523/JNEUROSCI.5858-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Huttunen HJ, Rauvala H. Amphoterin as an extracellular regulator of cell motility: from discovery to disease. J Intern Med. 2004;255:351–366. doi: 10.1111/j.1365-2796.2003.01301.x. [DOI] [PubMed] [Google Scholar]
- Jablonka S, Beck M, Lechner BD, Mayer C, Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J Cell Biol. 2007;179:139–149. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Diaz L, Geranton SM, Passmore GM, Leith JL, Fisher AS, Berliocchi L, Sivasubramaniam AK, Sheasby A, Lumb BM, Hunt SP. Local translation in primary afferent fibers regulates nociception. PLoS ONE. 2008;3:e1961. doi: 10.1371/journal.pone.0001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, Macgibeny MA, Gervasi NM, Gioio AE, Kaplan BB. Intra-axonal Synthesis of Eukaryotic Translation Initiation Factors Regulates Local Protein Synthesis and Axon Growth in Rat Sympathetic Neurons. J Neurosci. 2013;33:7165–7174. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Wolfson A. New competition in RNA regulation. Nat Biotechnol. 2012;30:58–59. doi: 10.1038/nbt.2092. [DOI] [PubMed] [Google Scholar]
- Kilchert C, Spang A. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. EMBO J. 2011;30:3567–3580. doi: 10.1038/emboj.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, Korza G, Lee JW, Carson JH, Barbarese E. HnRNP E1 Binds to hnRNP A2 and Inhibits Translation of A2RE mRNAs. Mol Biol Cell. 2006;17:3521–3533. doi: 10.1091/mbc.E05-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu OO, Perlman DH, Enquist LW. Efficient retrograde transport of pseudorabies virus within neurons requires local protein synthesis in axons. Cell Host Microbe. 2013;13:54–66. doi: 10.1016/j.chom.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! TIBS. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Kundel M, Jones KJ, Shin CY, Wells DG. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent beta-catenin mRNA translation in developing hippocampal neurons. J Neurosci. 2009;29:13630–13639. doi: 10.1523/JNEUROSCI.2910-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- Lin AC, Tan CL, Lin CL, Strochlic L, Huang YS, Richter JD, Holt CE. Cytoplasmic polyadenylation and cytoplasmic polyadenylation element-dependent mRNA regulation are involved in Xenopus retinal axon development. Neural Dev. 2009;4:8. doi: 10.1186/1749-8104-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher-Laporte M, DesGroseillers L. Genome wide identification of Staufen2-bound mRNAs in embryonic rat brains. BMB Rep. 2010;43:344–348. doi: 10.5483/bmbrep.2010.43.5.344. [DOI] [PubMed] [Google Scholar]
- Makeyev AV, Chkheidze AN, Liebhaber SA. A set of highly conserved RNA-binding proteins, alphaCP-1 and alphaCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J Biol Chem. 1999;274:24849–24857. doi: 10.1074/jbc.274.35.24849. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer EJ, Wang DO, Kim S, Barr I, Guo F, Martin KC. Identification of a cis-acting element that localizes mRNA to synapses. Proc Natl Acad Sci USA. 2012;109:4639–4644. doi: 10.1073/pnas.1116269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Gomes C, Yoo S, Vuppalanchi D, Twiss JL. Axonal localization of Neuritin/CPG15 mRNA in neuronal populations through distinct 5’ and 3’ UTR elements. J Neurosci. 2013a doi: 10.1523/JNEUROSCI.0962-13.2013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal Transport of Neural Membrane Protein 35 mRNA Increases Axon Growth. J Cell Sci. 2013b;126(Pt 1):90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobarak CD, Anderson KD, Morin M, Beckel-Mitchener A, Rogers SL, Furneaux H, King P, Perrone-Bizzozero NI. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol Biol Cell. 2000;11:3191–3203. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, Edbauer D, Janssens J, Kleinberger G, Cruts M, Herms J, Neumann M, Van Broeckhoven C, Arzberger T, Haass C. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013a;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013b;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Murashov AK, Chintalgattu V, Islamov RR, Lever TE, Pak ES, Sierpinski PL, Katwa LC, Van Scott MR. RNAi pathway is functional in peripheral nerve axons. FASEB J. 2007;21:656–670. doi: 10.1096/fj.06-6155com. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Titmus M, Koenig E, Tiedge H. Transport of neuronal BC1 RNA in Mauthner axons. J Neurosci. 2002;11:4293–4301. doi: 10.1523/JNEUROSCI.22-11-04293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Patel MV, Rose A, Tiedge H. Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J Cell Biol. 2011;194:441–457. doi: 10.1083/jcb.201010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Kristensen MA, Koch G, Christiansen J. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J Cell Sci. 2002;115:2087–2097. doi: 10.1242/jcs.115.10.2087. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoës M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32:4368–4376. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi M, Togano T, Takahashi-Niki K, Lu J, Honda A, Taoka M, Shinkawa T, Koga H, Takeuchi K, Isobe T, Igarashi M. Identification of functional marker proteins in the mammalian growth cone. Proc Natl Acad Sci USA. 2009;106:17211–17216. doi: 10.1073/pnas.0904092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Perrone-Bizzozero NI, Tanner DC, Mounce J, Bolognani F. Increased expression of axogenesis-related genes and mossy fibre length in dentate granule cells from adult HuD overexpressor mice. ASN Neuro. 2011;3:259–270. doi: 10.1042/AN20110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RB-T, Doron E, Iavnilovitch E, Rishal I, Dagan S, Tsoory M, Copolla G, Gomes C, McDonald M, Geschwind D, Twiss J, Yaron A, Fainzilber M. Subcellular Knockout of Importin β1 Perturbs Axonal Retrograde Signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TJ, Flores CM, Cervero F, Hargreaves KM. The RNA binding and transport proteins staufen and fragile X mental retardation protein are expressed by rat primary afferent neurons and localize to peripheral and central axons. Neuroscience. 2006;141:2107–2116. doi: 10.1016/j.neuroscience.2006.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnonen EL, Fages C, Wartiovaara J, Rauvala H. Ultrastructural localization of beta-actin and amphoterin mRNA in cultured cells: application of tyramide signal amplification and comparison of detection methods. J Histochem Cytochem. 1999;47:99–112. doi: 10.1177/002215549904700111. [DOI] [PubMed] [Google Scholar]
- Reddy K, Pearson CE. RAN translation: Fragile X in the running. Neuron. 2013a;78:405–408. doi: 10.1016/j.neuron.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The Disease-associated RGGGGCC)n Repeat from the C9orf72 Gene Forms Tract Length-dependent Uni- and Multimolecular RNA G-quadruplex Structures. J Biol Chem. 2013b;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, Tassone F, Willemsen R, Disney MD, Hagerman PJ, Todd PK, Charlet-Berguerand N. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Natl Acad Sci USA. 2007;104:1959–1964. doi: 10.1073/pnas.0610660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee M, Kidd GJ, Munro TP, Smith R. RNA trafficking and stabilization elements associate with multiple brain proteins. J Cell Sci. 2002;115(Pt 23):4661–4669. doi: 10.1242/jcs.00137. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira J, Calliari A, Kun A, Benech JC, Sanguinetti C, Chalar C, Sotelo JR. Neurofilament mRNAs are present and translated in normal and severed sciatic nerve. J Neurosci Res. 2000;62:65–74. doi: 10.1002/1097-4547(20001001)62:1<65::AID-JNR7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Cardenas M, Koenig E, Sotelo JR. Myosin Va and kinesin II motor proteins are concentrated in ribosomal domains (periaxoplasmic ribosomal plaques) of myelinated axons. J Neurobiol. 2004;60:187–196. doi: 10.1002/neu.20015. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira J, Crispino M, Puppo A, Sotelo JR, Koenig E. Myelinated axons contain beta-actin mRNA and ZBP-1 in periaxoplasmic ribosomal plaques and depend on cyclic AMP and F-actin integrity for in vitro translation. J Neurochem. 2008;104:545–557. doi: 10.1111/j.1471-4159.2007.04999.x. [DOI] [PubMed] [Google Scholar]
- Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Bio. 2013;202:53–69. doi: 10.1083/jcb.201212089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res. 2008;1189:33–42. doi: 10.1016/j.brainres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996a;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996b;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- Tsai NP, Bi J, Wei LN. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- Van der Zee CE, Nielander HB, Vos JP, Lopes da Silva S, Verhaagen J, Oestreicher AB, Schrama LH, Schotman P, Gispen WH. Expression of growth-associated protein B-50 (GAP43) in dorsal root ganglia and sciatic nerve during regenerative sprouting. J Neurosci. 1989;9:3505–3512. doi: 10.1523/JNEUROSCI.09-10-03505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci USA. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, Blesch A, Willis DE, Twiss JL. Conserved 3'-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010;285:18025–18038. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Hengst U, Kim HJ, Jeon NL, Schmidt EF, Heintz N, Milner TA, Jaffrey SR. Reprogramming axonal behavior by axon-specific viral transduction. Gene Ther. 2012;19:947–955. doi: 10.1038/gt.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, Buchmeier S, Wahle E, Hüttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG. RNA-binding proteins: a lesson in repression. J Neurosci. 2006;26:7135–7138. doi: 10.1523/JNEUROSCI.1795-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Johnson EM, Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. doi: 10.4161/cc.8.3.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, Erenstheyn M, English AW, Schanen NC, Kirn-Safran CB, Yoon SO, Bassell GJ, Twiss JL. Axonal Localization of Transgene mRNA in Mature PNS and CNS Neurons. J Neurosci. 2011;31:14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Yao X, Williams KR, Bassell GJ. Negative regulation of RhoA translation and signaling by hnRNP-Q1 affects cellular morphogenesis. Mol Biol Cell. 2012;23:1500–1509. doi: 10.1091/mbc.E11-10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Lopez de Quinto S, Matsui Y, Shevchenko A, Shevchenko A, Ephrussi A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca(2+)-dependent growth cone guidance. Nat Neurosci. 2006;10:1265–73. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yoo S, Kim H-H, Vuppalanchi D, Kim P, Donnelly CJ, Park M, Lee S-J, Merianda TT, Willis DE, Perrone-Bizzozero NI, Twiss JL. Axonal transport GAP-43 mRNA is driven by a interaction of HuD and ZBP1. J Neurochem. 2013 doi: 10.1111/jnc.12266. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Segal-Ruder Y, Vuppalanchi D, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss J, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila Fragile X-related gene regulates the MAP1b homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhong J, Chuang SC, Bianchi R, Zhao W, Paul G, Thakkar P, Liu D, Fenton AA, Wong RK, Tiedge H. Regulatory BC1 RNA and the fragile X mental retardation protein: convergent functionality in brain. PLoS One. 2010;5:e15509. doi: 10.1371/journal.pone.0015509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular Profiling Reveals Distinct and Developmentally Regulated Repertoire of Growth Cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]