Abstract
The volume, diversity and velocity of biomedical data are exponentially increasing providing petabytes of new neuroimaging and genetics data every year. At the same time, tens-of-thousands of computational algorithms are developed and reported in the literature along with thousands of software tools and services. Users demand intuitive, quick and platform-agnostic access to data, software tools, and infrastructure from millions of hardware devices. This explosion of information, scientific techniques, computational models, and technological advances leads to enormous challenges in data analysis, evidence-based biomedical inference and reproducibility of findings.
The Pipeline workflow environment provides a crowd-based distributed solution for consistent management of these heterogeneous resources. The Pipeline allows multiple (local) clients and (remote) servers to connect, exchange protocols, control the execution, monitor the states of different tools or hardware, and share complete protocols as portable XML workflows. In this paper, we demonstrate several advanced computational neuroimaging and genetics case-studies, and end-to-end pipeline solutions. These are implemented as graphical workflow protocols in the context of analyzing imaging (sMRI, fMRI, DTI), phenotypic (demographic, clinical), and genetic (SNP) data.
Keywords: aging, pipeline, neuroimaging, genetics, computation solutions, workflows, IBS, pain, Parkinson's disease, Alzheimer's disease, shape, volume, analysis, big data, visualization
Introduction
Process understanding is frequently the core research question in many biomedical, health and environmental applications. As we rarely know the exact process characteristics, we collect data (observations) which is used as proxy of the underlying physiological, physical or environmental phenomena. As such, the observed information (data) becomes the pivotal aspect of the scientific inquiry. The data variability, complexity and heterogeneity directly affect the scientific inference, accuracy of the results and reproducibility of findings.
Three data characteristics make contemporary biomedical data different, challenging and powerful. These are the data volume (size), typically in the petabyte range (1PB = 1015 bytes), data heterogeneity, including (un)formatted, ASCII/Binary, (un)structured, and the data velocity, or data derivative, which captures the change, transfer, and discovery of raw and derived data [1-3].
Table 1 illustrates the Kryder's law for exponential increase of the volume of data [4]. Using two decades of data, this law predicts that the density of information on hard drives, areal density, increases by a factor of 1,000 every 10-11 years. This storage rate increase is driven by the rapid expansion of data volume and velocity and translates into doubling of data size each 12-13 months. Both Moore's and Kryder's laws indicate similar exponential increase (of computational power and data storage, respectively) over time [5].
Table 1.
Rapid increase of the volume of neuroimaging and genetics data.
| Volume of Data MB = megabyte = 106 bytes, GB = gigabyte = 109 bytes, TB = terabyte = 1012 bytes, PB = petabyte = 1015 bytes |
D. Computational Power (CPU transistor counts) Moore's Law | Years | ||||||
|---|---|---|---|---|---|---|---|---|
| Single Cryo brain Volume 1600 cm2 | B. Neuroimaging (annually) | C. Genomics (BP/Yr) | ||||||
| A. Voxel Resolution | Gray Scale | RGB Color | 200 GB | 10 MB | 1×105 | 1985-1989 | ||
| Size | Count | 8bits | 16bits | 24bits | 1 TB | 100 MB | 1×106 | 1990-1994 |
| 1cm | 12×15×9 | 1620 | 3000 | 4860 | 50 TB | 10 GB | 5×106 | 1995-1999 |
| 1mm | 120×150×90 | 1.62 MB | 3.24 MB | 4.86 MB | 250 TB | 1TB | 1×107 | 2000-2004 |
| 100 μm | 1200×1500×900 | 1.62 GB | 3.24 GB | 4.86 GB | 1 PB | 30TB | 8×106 | 2005-2009 |
| 10 μm | 12000×15000×9000 | 1.62 TB | 3.24 TB | 4.86 TB | 5 PB | 1 PB | 1×109 | 2010-2014 |
| 1 μm | 120000×150000×90000 | 1.62 PB | 3.24 PB | 4.86 PB | 10+ PB | 20+ PB | 1×1011 | 2015-2019 (estimated) |
Legend:
A. Recent technological advances enable significant increases of the level of detail of optical imaging (e.g., cryotomographic brain images) into the micron (μm) resolution [6-8].
B. By 2012, there were 55PBs of neuroimaging data [9, 10], which may exaggerate the volume of neuroimaging data due to different publications sharing the same datasets. As of 2010, the Imaging Data Archive, a Laboratory of Neuro Imaging brain database, stored about 5×1015B=5PBs data. Recent neuroimaging studies may generate 1.5 TB of data each week [11].
C. In 2011, the size of the genetics data is estimated to be 30TBs (based on 10,000 human genomes) [12, 13]. As the total number of complete human genomes sequenced by the end of 2011 worldwide was >10,000, this figure may be orders of magnitude smaller than the real genomics data size. Furthermore, data derived from genome sequencing of other species and ‘partial genomes’ (e.g., exome capture sequencing, RNA sequencing and chromatin immunoprecipitation sequencing) is not included in this estimate. By 2015 more than a 106 human genomes will be sequenced [12]. Assuming each genome takes about 1011B (100GB) this translates into a total data volume of 1017B (100PB). Some of the sequences may be whole-genome 100X depth/coverage acquisitions, and some may be acquired at lower depth.
D. Data volume may be increasing at a faster pace compared to the well-established growth of computational power, Moore's law [14, 15].
There are thousands of software tools for acquisition, processing, storage/databasing, service, migration, mining, analysis, visualization, annotation, and data-driven process understanding. For example, the field of biomedical imaging includes hundreds of different types of image processing algorithms and filters. For each type of process, there may be dozens of concrete software products (instance implementations). More specifically, the Neuroimaging Informatics Tools and Resources Clearing House (NITRC) [16] lists over 500 openly shared neuroimaging software tools. For each openly shared tool, there may be dozens proprietary or less commonly used analogues. Similarly, in genomics and bioinformatics there are over 200 data and cloud computing service providers, and hundreds of public, private and non-profit organizations that provide thousands of stand-alone tools [17]. Resource organization, classification, discovery, traversal and utilization of these software products require flexible human and machine interfaces [18].
Another computational challenge is the proliferation of millions of hardware devices. According to Cisco [19], by the end of 2012, the number of mobile-connected devices will exceed the number of people on Earth and there will be over 10 billion mobile-connected devices in 2016; i.e., there will be more than 1.3 mobile devices per capita worldwide. These include phones, tablets, laptops, handheld gaming consoles, e-readers, in-car entertainment systems, digital cameras, and “machine-to-machine modules.” There is a clear need for bridges between these mobile devices and for efficient connections to distributed databases, clients, servers, compute-nodes, web-services, variety of interfaces.
Methods
The LONI Pipeline environment (http://Pipeline.loni.ucla.edu) [20, 21] is a graphical workflow middleware providing an interface to computational libraries, informatics resources, computational expertise and cloud services (e.g., cloud data storage, cloud computing services). The Pipeline facilitates the design, validation, execution, monitoring and sharing of advanced heterogeneous computational protocols as graphical workflows. It also mediates the tool discovery and interoperability and provides distributed computing infrastructure for en masse data processing. The Pipeline's user-friendly interface enables access to disparate data, services, hardware infrastructure, computational expertise and cloud computing services [20].
Alternative infrastructures to the Pipeline environment that also facilitate visual informatics and computational genomics include Taverna [22], Kepler [23], Triana [24], Galaxy [25], AVS [26], VisTrails [27], Bioclipse [28], KNIME [29], NyPipe [30], PSOM [31] and others. The choice of a workflow environment depends on the specific research domain, scientific application and computational need. The Pipeline environment provides some advantages over the alternative architectures. These include distributed client-server architecture, an array of scheduler grid plug-ins, external lightweight data manager, easy incorporation of new software tools and libraries, and dynamic workflow design, validation, execution, monitoring and dissemination of complete end-to-end computational solutions [32].
The main types of computational tools available in the Pipeline library include software for neuroimaging and genetics data processing and visualization. For each of these types there are 3 categories of resources – data, atomic modules, and workflows. These resources can be explored via the Pipeline Navigator (http://pipeline.loni.ucla.edu/explore/library-navigator/) and can be tested via the guest-access Pipeline Web-Start server (http://pipeline.loni.ucla.edu/PWS). Many interesting end-to-end computational workflow solutions (pipelines) are documented online (http://pipeline.loni.ucla.edu/explore/pipeline-workflows/). There are also many video tutorials, screencasts, and training materials (http://pipeline.loni.ucla.edu/learn/basic-videos/), which illustrate the basic and advanced features of the pipeline client-server architecture, and the protocols for workflow design, execution and management.
Neuroimaging Processing Tools
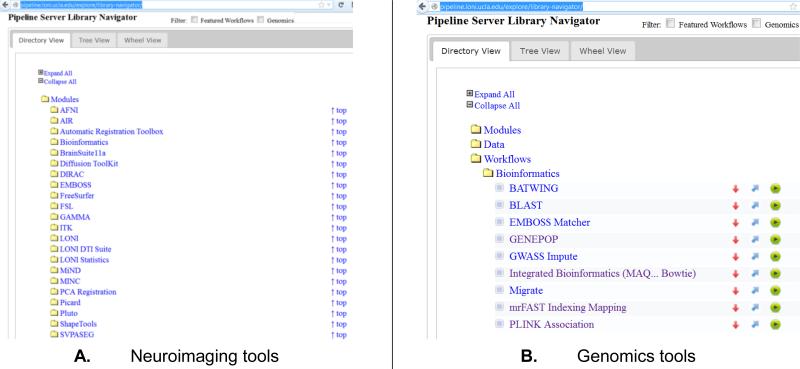
There are several hundred atomic neuroimage processing tools, from a variety of software suites available in the LONI pipeline library, Figure 1.A. These tools may be used for analysis of structural brain images (e.g., AFNI [33], ROBEX [34], MDT Atlasing [35, 36], BrainParser [37], SVPASEG [38, 39], AIR [40], FSL [41], BrainSuite [42], SSMA [43, 44], ANTS [45], ITK [46], MINC [47]), functional brain data (e.g., FLIRT [48], AFNI [33], WAIR [49], Matlab [50]), diffusion data (e.g., DTK [51], DIRAC [52], MiND [53]), statistical analyses (e.g., R [54], GAMMA [55], SOCR [56, 57], SPM [58, 59]), shape and surface modeling (e.g., sulcal analysis [60], local and global shape analyses [32], shape mapping DHM [61], FreeSurfer surface extraction, and cortical thickness [62, 63]).
Figure 1.
Examples of classes of tools available in the Pipeline computational library.
Informatics and Genomics Computational Library
The breadth of genomics tools available as pipeline modules and workflows is illustrated by the variety of sequence alignment solutions [20], Figure 1.B. Some different categories of informatics and genomics computing software tools available in the Pipeline library include: sequence alignment (Mosaik [64], MAQ [65], PERM [66], BWA/BWA-SW [67, 68], Bowtie [69], Novoalign [70], SOAPv2 [71], BLAST [72]), indexing (mrFAST/mrsFAST [73]), genome-wide association studies (GWASS [74], PLINK [75]), basic and advanced quality control (SAMTools [76], GATK [77]), CNV calling (CNV/CNVR [78, 79]), annotation (Artemis [80]), de novo assembly (Trinity [81], Velvet [82]), molecular biology (EMBOSS [83]), population genetics (GENEPOP [84]), and many others.
Backend Pipeline Servers
Pipeline web-start server (PWS) uses Java Web-Start technology enabling guest users to test the LONI Pipeline application from a web browser without the installation of either a pipeline client or a server. The PWS server provides access to all of the functions and features included in the downloadable version. PWS is accessible via an anonymous guest login or user-authentication to connect to remote Pipeline servers, e.g., http://ucla.in/GRSc8a. Several alternative Pipeline servers provide secure access-controlled connections to independent computational infrastructures. Examples include LONI Genomics Server (Genomics.loni.ucla.edu, 1TB RAM/40-core), Cranium Server (Cranium.loni.ucla.edu, 16GB RAM/core, 1,200 cores) and Medulla Server (Medulla.loni.ucla.edu, 24GB RAM/core, 4,300 slots). The Distributed Pipeline Server infrastructure (http://pipeline.loni.ucla.edu/DPS) facilities the deployment of independent disparate Pipeline services on available hardware resources, including Amazon EC2 (http://pipeline.loni.ucla.edu/products-services/pipeline-server-on-ec2/).
Big Data
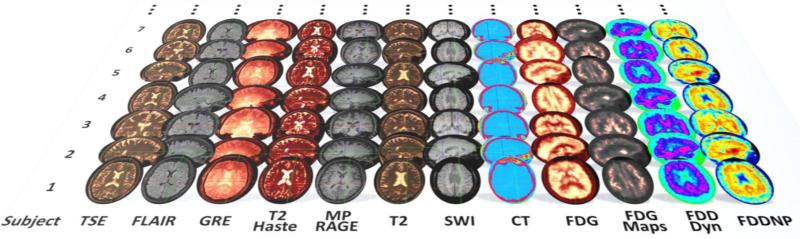
Modern protocols for imaging and genetics data collection generate enormous amounts of data. Table 2 illustrates some of the data-management, storage and processing challenges associated with common neuroimaging and genetics analysis protocols. Figure 2 shows an example of the multi-channel imaging brain data typically acquired in traumatic brain injury studies.
Table 2.
Storage and processing of Big neuroimaging and genetics data.
| Types of Studies (Number of Subjects) | Neuroimaging Depending on acquisition & analysis protocols |
C. Genetics Depending on Coverage(X) |
||
|---|---|---|---|---|
| A. Storage | B. Processing | Storage | Processing | |
| N=1 | Raw data: 10GB (e.g., 512 directional diffusion data) Derived: 100GB |
100+ GB RAM 70+ hrs CPU |
320GB (at 80X) | 2+ TB RAM 100+ hrs CPU |
| D. Cohort Studies (N~100) | 100GB – 1TB | 1TB RAM 100's hrs CPU |
3+ TB | 2+ TB RAM 100's hrs CPU |
| Multi-site population wide studies (N>1,000) | 1-10 TB | 1+ TB RAM 1000's hrs CPU |
30+ TB | 2+ TB RAM 1000's hrs CPU |
| Longitudinal (Time ≥ 2) | > 5TB | > 2 TB RAM > 5,000 hrs CPU |
--- | --- |
Legend:
A. Relative to the mouse brain, the field of view of human brain imaging data is several orders of magnitude larger [85]. Diffusion imaging of mouse brain may reach 1.9 GB (7 × 512 × 256 × 256 points with real and imaginary parts, represented as 4 bits float numbers) [86], and correspondingly diffusion spectral or high-angular resolution images may exceed 10GB per human subject and session [8]. The Global Shape Analysis pipeline workflow [32] includes about 100 processing steps and depending on the server load and the number of subjects provided as input may take 7 days to complete on the LONI Pipeline Medulla cluster (4TB RAM, 3,000 slots).
B. Many computationally intensive neuroimaging processing tools require significant hardware resources including storage, memory and CPU cycles [21].
C. In 2011, many alternative commercial DNA sequencing platforms generated whole genome sequences of size 100-600GB [87], which require days of computations on powerful grid systems. For example, our experience shows that Trinity whole-genome de novo assembly [88, 89] takes over 14 days of calculations on the LONI Pipeline Genomics server (1.4TB RAM, 40-core).
D. The infrastructure needs of cohort-based and multi-institutional studies increase linearly with the increase of the number of cases that require processing. Thus, a brain study of 1,000 subjects (e.g., Chinese Probabilistic Brain Atlas [90], vGWAS [91]) or a computational genetics study of 1,000 whole-genome sequences (e.g., prostate cancer [92], autism spectrum disorder [93]) may require Terabytes of storage and extensive infrastructure for data management, processing and interrogation. Longitudinal neuroimaging studies add another layer of complexity, as these typically require baseline as well as several (1+) follow up scans, which increases proportionately the volume of the imaging data.
Figure 2.
Traumatic brain injury (TBI) studies demonstrate the diversity of the neuroimaging data in clinical applications. Imaging modalities included in many TBI studies include: TSE: Turbo-Spin-Echo magnetic resonance imaging (MRI); FLAIR: Fluid Attenuated Inversion Recovery MRI; GRE: Gradient-Recalled-Echo (MRI); T2 Haste: Half-Fourier Acquisition Single-Shot Turbo Spin-Echo MRI; MP RAGE: Magnetization-Prepared Rapid Acquisition with Gradient Echo (MRI); T2: T2-weighted MRI; SWI: Susceptibility Weighted Imaging (MRI); CT: Computed Tomography; FDG: Fludeoxyglucose Positron Emission Tomography (PET); FDG Maps: Statistical maps of Fludeoxyglucose; FDDNP: 2-(1-{6-[(2-[F-18]fluoroethyl)(methyl)amino]-2-naphthyl}ethylidene)malononitrile PET imaging.
Applications and Results
To demonstrate the Pipeline management of heterogeneous neuroimaging, genetics, phenotypic and clinical data, and the diversity of computational data processing tools available through the Pipeline library, we have chosen three complementary applications. These include studies of imaging-based genome-wide association, hippocampal morphometry, persistent pain and irritable bowel syndrome. Each of these three applications demonstrates exemplary solutions to the resource-scalability and processing-efficiency challenges related to the data complexity (size, heterogeneity and velocity), software tools interoperability and diversity of hardware devices. Specifically, these case-studies demonstrate (1) how seemingly incongruent imaging, phenotypic and clinical data can be jointly processed and analyzed in an integrated computational workflow protocol; (2) how pipeline workflows can wrap independent software tools to make them interoperate; and (3) how these data and computational resources (tools and services) can be accessed via different client devices (e.g., desktop or laptop computers or mobile devices running different operating systems and browser configurations).
ADNI Imaging-Genetics GWAS Study
The Alzheimer's disease data used in this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2.For up-to-date information, see www.adni-info.org.
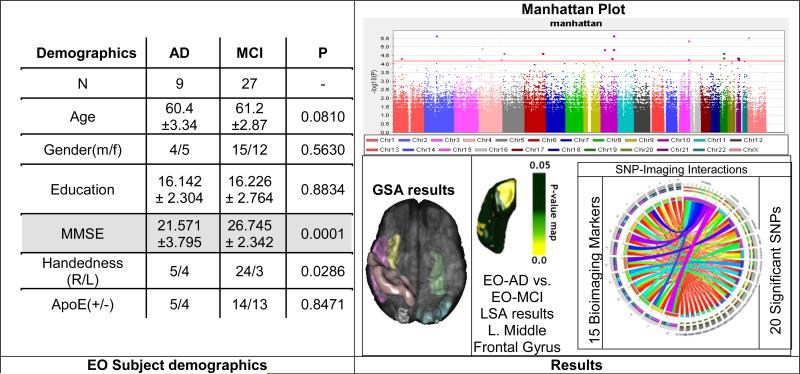
The Alzheimer's Disease Neuroimaging Initiative (ADNI) [94-96] data was screened and from 589 study participants, 188 qualified for an Alzheimer's Disease (AD) diagnosis at baseline, 401 had mild cognitive impairment (MCI). Among them, 9 were early-onset (EO) AD (Male: 4, Female: 5) and 27 were early-onset MCI (Male: 15, Female: 12). Subjects (ages 55 to 65) were divided into two groups: EO-AD and EO-MCI. Individual ADNI genotype and imaging data were downloaded and merged to form a single dataset containing genome-wide information for 36 individuals. Genetic analysis, including quality control, were performed using PLINK version 1.09. All the genetic processing was done via the LONI Pipeline environment. The 20 most significant single nucleotide polymorphisms (SNPs) were chosen by Manhattan plot and were associated with specific neuroimaging biomarkers. The structural ADNI data (1.5T MRI) was parcellated using BrainParser, and the 15 most important neuroimaging markers were extracted by the Global Shape Analysis (GSA) Pipeline workflow.
The goal of this application is to demonstrate the use of the pipeline environment for genome-wide association study (GWAS) using early-onset ADNI data including cognitive impairment measures, neuroimaging and genetics biomarkers. After standard SNP quality control [97, 98], the raw SNP data (630K SNPs) was reduced to 360K SNPs. A new pipeline workflow was designed to integrate the global shape analysis, tensor-based morphometry and SOCR multivariate regression analyses. The results of the automated pipeline workflows included significant correlations between SNPs and various neuroimaging biomarkers in the EO subjects and discriminated between EO-AD and EO-MCI cohorts, Figure 3. A connectomics diagram can be used to illustrate the strength of the associations between the 15 derived neuroimaging biomarkers and the top 20 SNP genetic markers. In this case-study, the small sample-size (N=36) has a negative effect on the (statistical) power to detect significant associations between the biomedical imaging markers (e.g., regional volume and shape metrics) and the genetic traits (SNPs/chromosomes). However, the same computational pipeline workflows can be used to analyze similarly larger cohorts (e.g., N>700), where sufficient power may be available to detect interactions between imaging and genetics effects (after Bonferonni correction for multiple testing). The imaging, genetics and clinical data used in this example were directly imported into the Pipeline workflow environment from the ADNI database using the Pipeline's IDAGet module. This pipeline workflow protocol can be designed on one client, and execution may be initiated on a user-specified pipeline server from another pipeline client, and the workflow progress, and final result inspection, may be monitored or examined on a different client device.
Figure 3.
Early Onset (EO) ADNI Imaging-Genetics GWAS Study using the pipeline environment.
Genetic Associations with Hippocampal Function and Shape
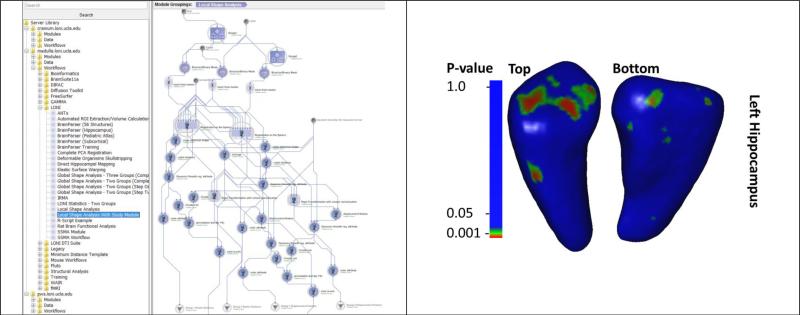
A recent study investigated the genetics effects (single-nucleotide polymorphisms, SNP, associated with FKBP5 gene regulation, rs1360780) related to attention, behavioral, and hippocampal morphometrics [99]. The FKBP5 gene regulates glucocorticoid receptor sensitivity and is associated with hypothalamic-pituitary-adrenal axis functioning and stress-related psychiatric disorders [100]. In this cross-sectional study using fMRI/MRI, African American cohort of adults (N = 103) separated into 2 groups by genotype: Group 1 included carriers of the rs1360780 T allele, associated with increased risk for posttraumatic stress disorder; Group 2 included non-carriers. The study used the local shape analysis pipeline workflow to identify attention bias toward threat (F1,90=5.19, p=0.02), and revealed alterations in the hippocampal shape for TT/TC compared with the CC genotype groups. Figure 4 shows part of the computational protocol implemented as a pipeline workflow and the exemplary result from this morphometric analysis.
Figure 4.
Example of using the pipeline environment to complete a neuroimaging genetics study of FKBP5 gene (rs1360780) association with attention, measured through behavioral response (dot probe task) and hippocampal morphometrics. The superior and inferior vies of the hippocampal surface map illustrate the vertex locations, on the mean left hippocampus, where FKBP5 carriers (group 1) and non-carriers (group 2) showed significant shape differences.
Persistent Pain and Irritable Bowel Syndrome (IBS)
A UCLA IRB approved study recruited 328 female normal controls (NC) and IBS subjects. A diagnosis of IBS was made using the ROME III symptom criteria [101, 102] based on the assessment by one of 4 gastroenterologists experienced in the diagnosis of functional bowel disease and the exclusion of organic disease. A subject's medical history and physical examination were obtained by a gastroenterologist. IBS patients with all types of predominant bowel habit were included. Subjects with a history of any chronic functional symptom or syndrome, or symptoms suggestive of disordered mood or affect, by history or by questionnaire, were excluded. In addition, potential subjects are excluded if by either history or questionnaire they a) have a serious medical condition or are taking medications which may interfere with interpretation of the brain imaging or physiological measures (other than IBS); b) have an ongoing major psychiatric diagnosis or psychotropic medication use over the past 6 months (subjects are not excluded for lifetime incidence of psychiatric disorder, or for intake of low dose tricyclic antidepressant for non-psychiatric indication); c) have a positive symptom score on the Hospital Anxiety and Depression Scale consistent with depression or anxiety d) do excessive physical exercise (i.e., marathon runners).
Brain images were obtained from all 328 subjects (107 IBS, 221 NC) using 1.5 and 3T MRI scanners [103]. We collected phenotyping data on catastrophizing (Coping Strategies Questionnaire) [104], early life trauma (Early Trauma Inventory) [105], state anxiety and depression (Hospital Anxiety and Depression Scale) [106], health status (12-Item Short-Form Health Survey) [107], trait anxiety scores (State Trait Anxiety Inventory) [108] and IBS symptom severity and duration (Bowel Symptoms Questionnaire) [109].
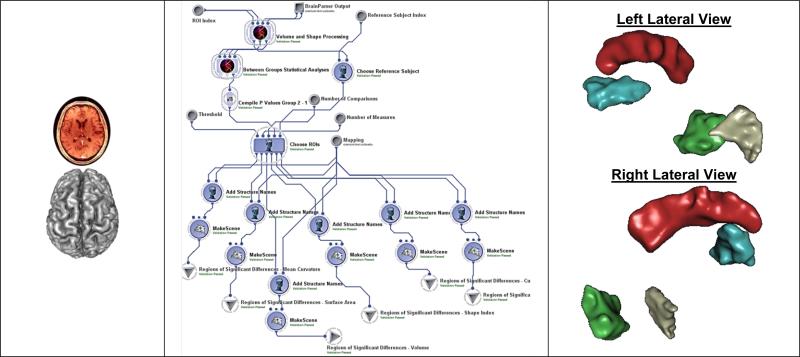
As a first step for shape-based neuroimage analysis, we reconstruct surface representation of anatomical structures of interest. Then, we analyze both cortical and subcortical structures. The cortical surfaces, including both white matter and pial surfaces, are reconstructed from T1-weighted MR images using FreeSurfer [110]. For sub-cortical structures, we applied the LONI BrainParser [37] to automatically segment the T1-weighted MR image into fifty-six regions. Using masks generated by BrainParser, accurate surface representations of the segmented regions are reconstructed with a novel algorithm we developed recently. This tool can remove segmentation artifacts without volume shrinkage and guarantees all surfaces guaranteed have the correct topology. All surfaces are represented as triangular meshes with spherical topology. The global shape analysis (GSA) pipeline workflow was used to identify regional differences between the NC and IBS subjects using the 56 regions of interest (ROIs) on 6 different volumetric and shape metrics (average mean curvature, surface area, volume, shape index, curvedness, and fractal dimension). Figure 5 shows the 3 steps in this analysis (data inputs, pipeline workflow and results of regional group differences).
Figure 5.
Analyzing IBS/NC regional differences: (Left) raw sMRI data, (Middle) GSA workflow including data processing, surface reconstruction, 3D parcellation, and statistical analysis, (Right) Statistically significant ROI between-differences rendered as 3D scenes (left cuneus is green, and right angular gyrus is gray; the red cingulate gyrus and the blue insula are shown for orientation only).
Conclusions
Although there are a number of useful software discovery and navigation frameworks [18, 111, 112], the protocols for tool interoperability continue to present significant biomedical computing challenges. There are considerable design differences between independent software suites. Furthermore, the varieties of computer programming languages for algorithm implementation, the substantial diversity of compilers and optimization strategies, and the gamut of hardware resources present additional hurdles in biomedical computing. Mediating these computational issues, coping with the enormous amounts of incongruent data, and handling a wide spectrum of devices require a paradigm shift of how we manage, process, interrogate and utilize biomedical and health related data.
The evidence is clear that we are in the front of an enormous storm of exponentially increasing wave of data, processing power and resource diversity. Multidisciplinary science efforts, technologies like Hadoop [113], OpenStack [114], Elastic Cloud Computing [115], Pipeline workflow systems [32, 116] and super high-bandwidth networking [117, 118] will be critical for riding this storm and uncovering novel biomedical knowledge. Embracing the science interactome (the multidisciplinary interactions between biomedical, computational and basic scientific areas, which often lead to new discoveries) will also be essential for establishing, maintaining and expanding the cyclical flow from Biomedical Challenges ↔ Scientific Models ↔ Data Analysis ↔ Computational Infrastructure ↔ Sustainable Education.
In this manuscript, we presented evidence of the rapid increase of the volume, diversity and velocity of biomedical data (e.g., neuroimaging and genetics [119-121]), and the growth of computational models, algorithms, software tools, services and electronic devices that manipulate these data [122-124]. There is evidence that software tool expansion always occurs within the limits of the available hardware infrastructure [125]. This close connection between the Moore's law for increase of computational power facilitates the observed expansion of new and more powerful software tools (e.g., Software as a Service (SaaS) [126], Platform-as-a-Service (PaaS) [127]). For example, in 1993, Windows NT OS 3 consisted of 5-million lines of code, which 10-years later grew 10-fold to 50-million lines in Windows, Server OS 2003 [128]. Similarly, from 2000 to 2007, the Linux Debian OS grew from 59-million to 280-million lines of code [129]. Web and mobile applications, or webapps, are software systems running on portable devices, which have significantly grown since 2005 into a multi-billion dollar business [130]. The explosion of webapp software development can be measured in terms of pure source code, usage of third-party APIs, and historical data. Studies of lines of code in specific areas indicate that over the past few decades there is an exponential increase of software development efforts [131, 132]. This advancement of the software tool capabilities in turn pushes the introduction of more efficient and omnipotent hardware devices (e.g., Infrastructure as a Service (IaaS) and Virtual Machines (VMs) [133]).
The Pipeline workflow environment is one of many solutions that provide a distributed and platform-independent management of heterogeneous resources using dispersed clients and servers, elaborate exchange protocols, and flexible mechanisms for control, execution, monitoring and sharing of complete computational protocols. We demonstrated three advanced end-to-end computational pipeline solutions for neuroimaging, genetics and computational morphometry. These solutions are implemented as graphical workflow protocols in the context of analyzing imaging (sMRI, fMRI, DTI), phenotypic (demographic, clinical), and genetic (SNP) data.
Acknowledgments
This study was partially supported by National Institutes of Health grants NIA P 50 AG16570, NIBIB EB01651, NLM LM05639, NIMH R01 MH071940, NIBIB P41EB015922, U24-RR025736, U24-RR021992, U24 GM104203, as well as National Science Foundation grants 0716055 and 1023115. The authors are also indebted to the faculty, staff and students in the Laboratory of Neuro Imaging (LONI) for their support and dedication.
Some of the data processed in this study was partly funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles.
As of September 2013, the Laboratory of Neuro Imaging (LONI) will be relocated to the University of Southern California (USC). Thus, some of the URL links, web-page references, and internet resources cited throughout this manuscript may be relocated to appropriate subdomains under http://www.loni.usc.edu. If you find broken links or defunct URLs please contact help@loni.usc.edu.
References
- 1.Foster K, Spicer M, Nathan S. IBM Infosphere Streams: Assembling Continuous Insight in the Information Revolution. International Technical Support Organization; San Jose, California: 2011. [Google Scholar]
- 2.Howe D, et al. Big data: The future of biocuration. Nature. 2008;455(7209):47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch C. Big data: How do your data grow? Nature. 2008;455(7209):28–29. doi: 10.1038/455028a. [DOI] [PubMed] [Google Scholar]
- 4.Walter C. Kryder's law. Scientific American. 2005;293(2):32–33. doi: 10.1038/scientificamerican0805-32. [DOI] [PubMed] [Google Scholar]
- 5.Sood A, et al. Predicting the Path of Technological Innovation: SAW vs. Moore, Bass, Gompertz, and Kryder. Marketing Science. 2012;31(6):964–979. [Google Scholar]
- 6.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nature methods. 2010;7(8):603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 7.Roy D, et al. 3D Cryo-Imaging: A Very High-Resolution View of the Whole Mouse. The anatomical record. 2009;292(3):342–351. doi: 10.1002/ar.20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholl I, et al. Challenges of medical image processing. Computer science-Research and development. 2011;26(1-2):5–13. [Google Scholar]
- 9.Breeze JL, Poline J-B, Kennedy DN. Data sharing and publishing in the field of neuroimaging. GigaScience. 2012;1(1):1–3. doi: 10.1186/2047-217X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mennes M, et al. Making data sharing work: The FCP/INDI experience. Neuroimage. (0) doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olabarriaga SD, Glatard T, de Boer PT. A virtual laboratory for medical image analysis. Information Technology in Biomedicine, IEEE Transactions on. 2010;14(4):979–985. doi: 10.1109/TITB.2010.2046742. [DOI] [PubMed] [Google Scholar]
- 12.Grossman R, White K. A vision for a biomedical cloud. Journal of internal medicine. 2012;271(2):122–130. doi: 10.1111/j.1365-2796.2011.02491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marusina K. Big Data Requires Big Solutions. Genetic Engineering & Biotechnology News. 2012;32(15):1, 34–40. [Google Scholar]
- 14.Fuller SH, Millett LI. Computing performance: Game over or next level? Computer. 2011;44(1):31–38. [Google Scholar]
- 15.Rupp K, Selberherr S. The economic limit to Moore's Law. Semiconductor Manufacturing, IEEE Transactions on. 2011;24(1):1–4. [Google Scholar]
- 16.Luo X.-z.J., Kennedy DN, Cohen Z. Neuroimaging informatics tools and resources clearinghouse (NITRC) resource announcement. Neuroinformatics. 2009;7(1):55–56. doi: 10.1007/s12021-008-9036-8. [DOI] [PubMed] [Google Scholar]
- 17.Eliceiri KW, et al. Biological imaging software tools. Nature methods. 2012;9(7):697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinov I, et al. iTools: A Framework for Classification, Categorization and Integration of Computational Biology Resources. PLoS ONE. 2008;3(5):e2265. doi: 10.1371/journal.pone.0002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisco Systems Inc. Cisco Visual Networking Index: Global Mobile Data Traffic Forecast Update, 2012–2017. Cisco. 2012 Available from: http://www.cisco.com/en/US/solutions/collateral/ns341/ns525/ns537/ns705/ns827/white_paper_c11-520862.pdf.
- 20.Torri F, et al. Next Generation Sequence Analysis and Computational Genomics Using Graphical Pipeline Workflows. Genes. 2012;3(3):545–575. doi: 10.3390/genes3030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinov I, et al. Applications of the Pipeline Environment for Visual Informatics and Genomics Computations. BMC Bioinformatics. 2011;12(1):304. doi: 10.1186/1471-2105-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oinn T, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20(17):3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 23.Ludäscher B, Altintas I, Berkley C, Higgins D, Jaeger E, Jones M, Lee EA, Tao J, Zhao Y. Scientific workflow management and the Kepler system. Concurrency and Computation: Practice and Experience. 2006;18(10):1039–1065. [Google Scholar]
- 24.Taylor I, Shields M, Wang I, Harrison A. Visual Grid Workflow in Triana. Journal of Grid Computing. 2006;3:153–169. [Google Scholar]
- 25.Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lord HD. Improving the application development process with modular visualization environments. SIGGRAPH Comput. Graph. 1995;29(2):10–12. [Google Scholar]
- 27.Freire J, et al. In: Managing Rapidly-Evolving Scientific Workflows, in IPAW 2006. L.M.a.I.F., editor. Springer-Verlag; Berlin Heidelberg: 2006. pp. 10–18. [Google Scholar]
- 28.Spjuth O, et al. Bioclipse: an open source workbench for chemo- and bioinformatics. BMC Bioinformatics. 2007;8(1):59. doi: 10.1186/1471-2105-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berthold MR, et al. KNIME: The Konstanz Information Miner, in Data Analysis, Machine Learning and Applications. In: Preisach C, et al., editors. Springer; Berlin Heidelberg: 2008. pp. 319–326. [Google Scholar]
- 30.Gorgolewski K, et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Frontiers in Neuroinformatics. 2011;5 doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellec P, et al. The pipeline system for Octave and Matlab (PSOM): a lightweight scripting framework and execution engine for scientific workflows. Frontiers in Neuroinformatics. 2012;6 doi: 10.3389/fninf.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinov I, et al. Neuroimaging Study Designs, Computational Analyses and Data Provenance Using the LONI Pipeline. PLoS ONE. 2010;5(9):e13070. doi: 10.1371/journal.pone.0013070. doi:10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 34.Iglesias JE, et al. Robust brain extraction across datasets and comparison with publicly available methods. Medical Imaging, IEEE Transactions on. 2011;30(9):1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, et al. Construction and validation of mean shape atlas templates for atlas-based brain image segmentation. in Information Processing in Medical Imaging. Springer; 2005. [DOI] [PubMed] [Google Scholar]
- 36.Tang Y, et al. The construction of a Chinese MRI brain atlas: A morphometric comparison study between Chinese and Caucasian cohorts. Neuroimage. 2010;51(1):33–41. doi: 10.1016/j.neuroimage.2010.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu Z, et al. Brain Anatomical Structure Segmentation by Hybrid Discriminative/Generative Models. IEEE Transactions on Medical Imaging. 2008;27(4):495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tohka J, et al. Genetic Algorithms for Finite Mixture Model Based Voxel Classification in Neuroimaging. Medical Imaging, IEEE Transactions on. 2007;26(5):696–711. doi: 10.1109/TMI.2007.895453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tohka J, et al. Brain MRI tissue classification based on local Markov random fields. Magnetic Resonance Imaging. 2010;28(4):557–573. doi: 10.1016/j.mri.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods RP, Dapretto M, Sicotte NL, Toga AW, Mazziotta JC. Creation and use of a Talairach-compatible atlas for accurate, automated, nonlinear intersubject registration, and analysis of functional imaging data. Hum Brain Mapp. 1999;8(2-3):73–9. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<73::AID-HBM1>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Supplement 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 42.Shattuck D, Leahy R. BrainSuite: An Automated Cortical Surface Identification Tool, in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2000. Lecture Notes in Computer Science. 2000:50–61. [Google Scholar]
- 43.Leung KTK. Principal Ranking Meta-Algorithms. UNIVERSITY OF CALIFORNIA; LOS ANGELES: 2011. [Google Scholar]
- 44.Leung K, et al. SSDBM 2008. Springer-Verlag; 2008. IRMA: an Image Registration Meta-Algorithm - evaluating Alternative Algorithms with Multiple Metrics. [Google Scholar]
- 45.Avants BB, Tustison N, Song G. Advanced Normalization Tools (ANTS). Insight J. 2009 [Google Scholar]
- 46.Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community. Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on. 2006.
- 47.Evans A. Neuropsychopharmacology: The Fifth Generation of Progress: American College of Neuropsychopharmacology. Nature Publishing; London: 2002. Automated 3D analysis of large brain MRI databases. pp. 301–313. [Google Scholar]
- 48.Smith SM, et al. Variability in fMRI: A re-examination of inter-session differences. Human Brain Mapping. 2005;24(3):248–257. doi: 10.1002/hbm.20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinov ID, et al. Quantitative comparison and analysis of brain image registration using frequency-adaptive wavelet shrinkage. Ieee Transactions on Information Technology in Biomedicine. 2002;6(1):73–85. doi: 10.1109/4233.992165. [DOI] [PubMed] [Google Scholar]
- 50.Hanselman D, Littlefield BC. Mastering MATLAB 5: A comprehensive tutorial and reference. Prentice Hall PTR; 1997. [Google Scholar]
- 51.Wang R, et al. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. Proc Intl Soc Mag Reson Med. 2007 [Google Scholar]
- 52.Patel V, et al. Mesh-based spherical deconvolution: A flexible approach to reconstruction of nonnegative fiber orientation distributions. Neuroimage. 2010;51(3):1071–1081. doi: 10.1016/j.neuroimage.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel V, et al. LONI MiND: Metadata in NIfTI for DWI. Neuroimage. 2010;51(2):665–676. doi: 10.1016/j.neuroimage.2010.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of computational and graphical statistics. 1996;5(3):299–314. [Google Scholar]
- 55.Chen R, Herskovits EH. Graphical-model-based morphometric analysis. Medical Imaging, IEEE Transactions on. 2005;24(10):1237–1248. doi: 10.1109/TMI.2005.854305. [DOI] [PubMed] [Google Scholar]
- 56.Che A, Cui J, Dinov I. SOCR Analyses: Implementation and Demonstration of a New Graphical Statistics Educational Toolkit. JSS. 2009;30(3):1–19. doi: 10.18637/jss.v030.i03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dinov I. Statistics Online Computational Resource. Journal of Statistical Software. 2006;16(1):1–16. doi: 10.18637/jss.v016.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu D, et al. Unified SPM–ICA for fMRI analysis. Neuroimage. 2005;25(3):746–755. doi: 10.1016/j.neuroimage.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 59.Friston KJ, et al. Statistical Parametric Mapping: The Analysis of Functional Brain Images: The Analysis of Functional Brain Images. Academic Press; 2011. [Google Scholar]
- 60.Joshi SH, et al. Diffeomorphic Sulcal Shape Analysis on the Cortex. Medical Imaging, IEEE Transactions on. 2012;PP(99):1–1. doi: 10.1109/TMI.2012.2186975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y, Thompson PM, Dinov ID, Osher S, Toga AW. Direct cortical mapping via solving partial differential equations on implicit surfaces. Medical Image Analysis. 2007;11(3):207–23. doi: 10.1016/j.media.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fennema-Notestine C, et al. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Human Brain Mapping. 2006;27(2):99–113. doi: 10.1002/hbm.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97(20):11050–5. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith DR, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Research. 2008;18(10):1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Research. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y, Souaiaia T, Chen T. PerM: efficient mapping of short sequencing reads with periodic full sensitive spaced seeds. Bioinformatics. 2009;25(19):2514–2521. doi: 10.1093/bioinformatics/btp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Briefings in bioinformatics. 2010;11(5):473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 72.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Research. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nature methods. 2010;7(8):576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 75.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, et al. The Sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 81.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Briefings in bioinformatics. 2002;3(1):87. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 84.Raymond M, Rousset F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. Journal of Heredity. 1995;86(3):248–249. [Google Scholar]
- 85.Sultan F, Braitenberg V. Shapes and sizes of different mammalian cerebella. A study in quantitative comparative neuroanatomy. Journal für Hirnforschung. 1993;34(1):79. [PubMed] [Google Scholar]
- 86.Jiang Y, Johnson GA. Microscopic diffusion tensor imaging of the mouse brain. Neuroimage. 2010;50(2):465–471. doi: 10.1016/j.neuroimage.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glenn TC. Field guide to next-generation DNA sequencers. Molecular Ecology Resources. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 88.Zhang W, et al. A Practical Comparison of De Novo Genome Assembly Software Tools for Next-Generation Sequencing Technologies. PLoS ONE. 2011;6(3):e17915. doi: 10.1371/journal.pone.0017915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Research. 2010;20(2):265. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xing W, et al. Probabilistic MRI Brain Anatomical Atlases Based on 1,000 Chinese Subjects. PLoS ONE. 2013;8(1):e50939. doi: 10.1371/journal.pone.0050939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hibar DP, et al. Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N= 1345 young and elderly subjects. Brain Imaging and Behavior. 2012:1–14. doi: 10.1007/s11682-012-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gudmundsson J, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nature Genetics. 2012 doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buxbaum JD, et al. The autism sequencing consortium: Large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76(6):1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jack CR, et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mueller SG, et al. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimer's and Dementia: The Journal of the Alzheimer's Association. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen L, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53(3):1051. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hibar DP, et al. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011;56(4):1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rimol LM, et al. Sex-dependent association of common variants of microcephaly genes with brain structure. Proceedings of the National Academy of Sciences. 2010;107(1):384–388. doi: 10.1073/pnas.0908454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fani N, G.D.T.E.B., et al. Fkbp5 and attention bias for threat: Associations with hippocampal function and shape. JAMA Psychiatry. 2013:1–9. doi: 10.1001/2013.jamapsychiatry.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34:S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 101.Sperber AD, et al. A comparative reappraisal of the Rome II and Rome III diagnostic criteria: are we getting closer to the ’true’ prevalence of irritable bowel syndrome? European journal of gastroenterology & hepatology. 2007;19(6):441. doi: 10.1097/MEG.0b013e32801140e2. [DOI] [PubMed] [Google Scholar]
- 102.Drossman D, Dumitrascu D. Rome III: New standard for functional gastrointestinal disorders. Journal of gastrointestinal and liver diseases: JGLD. 2006;15(3):237. [PubMed] [Google Scholar]
- 103.Jiang Z, et al. Sex-Related Differences of Cortical Thickness in Patients with Chronic Abdominal Pain. 2013 doi: 10.1371/journal.pone.0073932. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geisser ME, Robinson ME, Henson CD. The Coping Strategies Questionnaire and chronic pain adjustment: A conceptual and empirical reanalysis. The Clinical journal of pain. 1994 doi: 10.1097/00002508-199406000-00003. [DOI] [PubMed] [Google Scholar]
- 105.Bremner JD, Vermetten E, Mazure CM. Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: the Early Trauma Inventory. Depression and Anxiety. 2000;12(1):1–12. doi: 10.1002/1520-6394(2000)12:1<1::AID-DA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 106.Zigmond AS, Snaith R. The hospital anxiety and depression scale. Acta psychiatrica scandinavica. 1983;67(6):361–370. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 107.Ware JE, Jr, Kosinski M, Keller SD. A 12-Item Short-Form Health Survey: construction of scales and preliminary tests of reliability and validity. Medical care. 1996;34(3):220. doi: 10.1097/00005650-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 108.Spielberger CD. State-trait anxiety inventory. Wiley Online Library; 2005. [Google Scholar]
- 109.Talley N, et al. Initial validation of a bowel symptom questionnaire* and measurement of chronic gastrointestinal symptoms in Australians. Internal Medicine Journal. 1995;25(4):302–308. doi: 10.1111/j.1445-5994.1995.tb01894.x. [DOI] [PubMed] [Google Scholar]
- 110.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kennedy DN. The Internet Analysis Tools Registry: A Public Resource for Image Analysis. Neuroinformatics. 2006;4:263–270. doi: 10.1385/NI:4:3:263. [DOI] [PubMed] [Google Scholar]
- 112.Tenenbaum JD, et al. The Biomedical Resource Ontology (BRO) to enable resource discovery in clinical and translational research. Journal of Biomedical Informatics. 2011;44(1):137–145. doi: 10.1016/j.jbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.White T. Hadoop: The definitive guide. O'Reilly Media. 2012 [Google Scholar]
- 114.Wen X, et al. Fuzzy Systems and Knowledge Discovery (FSKD), 2012 9th International Conference on. IEEE; 2012. Comparison of open-source cloud management platforms: OpenStack and OpenNebula. [Google Scholar]
- 115.Ostermann S, et al. A performance analysis of EC2 cloud computing services for scientific computing. Cloud Computing. 2010:115–131. [Google Scholar]
- 116.Heinis T. Workflow-based services: infrastructure for scientific applications. Suedwestdeutscher Verlag fuer Hochschulschriften; 2010. [Google Scholar]
- 117.Chowdhury A, et al. World of Wireless Mobile and Multimedia Networks (WoWMoM), 2010 IEEE International Symposium on a. IEEE; 2010. Next-generation E-health communication infrastructure using converged super-broadband optical and wireless access system. [Google Scholar]
- 118.Wang W, Guo L. Computer Science and Electronics Engineering (ICCSEE), 2012 International Conference on. IEEE; 2012. The Development and Applications of Wireless Streaming Media Technology. [Google Scholar]
- 119.Novak NM, et al. EnigmaVis: online interactive visualization of genome-wide association studies of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) consortium. Twin research and human genetics: the official journal of the International Society for Twin Studies. 2012;15(3):414. doi: 10.1017/thg.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Van Essen DC, et al. The human connectome project: a data acquisition perspective. Neuroimage. 2012;62(4):2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thompson PM, et al. Genetics of the Connectome. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Toga AW, et al. The Center for Computational Biology: resources, achievements, and challenges. Journal of the American Medical Informatics Association. 2012;19(2):202–206. doi: 10.1136/amiajnl-2011-000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berger B, Peng J, Singh M. Computational solutions for omics data. Nature Reviews Genetics. 2013;14(5):333–346. doi: 10.1038/nrg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meir A, Rubinsky B. Distributed network, wireless and cloud computing enabled 3-D ultrasound; a new medical technology paradigm. PLoS ONE. 2009;4(11):e7974. doi: 10.1371/journal.pone.0007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuller SH, Millett LI. The Future of Computing Performance: Game Over or Next Level? The National Academies Press; 2011. [Google Scholar]
- 126.Hashizume K, Fernandez EB, Larrondo-Petrie MM. BioMedical Computing (BioMedCom), 2012 ASE/IEEE International Conference on. IEEE; 2012. A pattern for Software-as-a-Service in Clouds. [Google Scholar]
- 127.Truong H-L, Dustdar S. A survey on cloud-based sustainability governance systems. International Journal of Web Information Systems. 2012;8(3):278–295. [Google Scholar]
- 128.Maraia V. The Build Master: Microsoft's Software Configuration Management Best Practices. Addison-Wesley Professional; 2005. [Google Scholar]
- 129.Matellán Olivera V. Studying the evolution of libre software projects using publicly available data. 2012 Available from: https://buleria.unileon.es/handle/10612/1796.
- 130.Minelli R, Lanza M. Software Maintenance and Reengineering (CSMR), 2013 17th European Conference on. IEEE; 2013. Software Analytics for Mobile Applications--Insights & Lessons Learned. [Google Scholar]
- 131.Knobloch J. Four Decades of Computing in Subnuclear Physics-from Bubble Chamber to LHC. :2013. arXiv preprint arXiv:1302.2974. [Google Scholar]
- 132.German DM, Adams B, Hassan AE. Software Maintenance and Reengineering (CSMR), 2013 17th European Conference on. IEEE; 2013. The Evolution of the R Software Ecosystem. [Google Scholar]
- 133.Alarifi S, Wolthusen S. Network and System Security. Springer; 2013. Anomaly Detection for Ephemeral Cloud IaaS Virtual Machines; pp. 321–335. [Google Scholar]