Abstract
The field of non-coding RNA (ncRNA) has expanded over the last decade following the discoveries of several new classes of regulatory ncRNA. A growing amount of evidence now indicates that ncRNAs are involved even in the most fundamental of cellular processes. The heat shock response is no exception as ncRNAs are being identified as integral components of this process. Although this area of research is only in its infancy, this article focuses on several classes of regulatory ncRNA (i.e., miRNA, lncRNA, and circRNA), while summarizing their activities in mammalian heat shock. We also present an updated model integrating the traditional heat shock response with the activities of regulatory ncRNA. Our model expands on the mechanisms for efficient execution of the stress response, while offering a more comprehensive summary of the major regulators and responders in heat shock signaling. It is our hope that much of what is discussed herein may help researchers in integrating the fields of heat shock and ncRNA in mammals.
Keywords: ncRNA, miRNA, lncRNA, circRNA, ceRNA, HSP, HSP90, HSP70, HSF1, HSR1, 7SK, IGS, Pseudogenes, Exosome, Cell stress, Thermal stress, Hyperthermia
Introduction
Cells undergo stress in response to a variety of conditions including transient exposure to hot or cold temperatures, heavy metals, exogenous chemicals, oxidative stress, ischemia/reperfusion (I/R), salt, pH shifts, etc. Activation of the stress response results in a reprioritization of cell physiology to support survival. This process is generally associated with repression in basal transcription and translation; however, heat shock proteins (HSP) and other chaperones are the exception as they become actively transcribed and translated in response to cellular stresses (Calderwood 2005). A number of mechanisms exist for rapid execution of the stress response. In eukaryotes, heat-mediated activation of HSP gene expression is facilitated by the transcription factor HSF1 (heat shock factor 1). In unstressed cells, HSF1 is present in the cytoplasm as an inactive monomer bound to several chaperones (e.g., HSPA1A/HSP72, HSPC1/HSP90, etc.). Upon heat shock, HSF1 is released from the complex resulting in its trimerization and translocation to the nucleus. HSF1 binds to sequence motifs termed heat shock elements (HSEs) in promoters of HSP genes leading to its transactivation. HSF1 is controlled, in part, by a negative feedback loop in which HSP induction functions to re-sequester DNA-bound HSF1. Regulation of HSF1 activity and expression of HSP genes are fundamentally important for cytoprotection and cell survival following heat stress.
In association with transcriptional activation, the transcripts of HSP genes are also stabilized in response to heat stress. This phenomenon appears to be mediated, in part, though the 3′UTR (3′ untranslated region) of HSP transcripts involving decreased deadenylation and suppression of mRNA decay via ARE (AU-rich domain)-dependent mechanisms (Dellavalle et al. 1994; Moseley et al. 1993). Conversely, other non-HSP transcripts are generally destabilized in response to heat stress and/or accumulate within stress granules or p-bodies (Buchan and Parker 2009). As such, regulating mRNA stability is also a fundamental component of managing the heat shock response in stressed cells.
A resurgence of interest in the field of non-coding RNA (ncRNA) has emerged over the last decade as a result of new discoveries defining novel classes of ncRNAs with diverse biological activities. In fact, ncRNAs are continually being implicated in a vast number of cellular processes including cell growth, disease, embryogenesis, gene regulation, signal transduction, receptor activation, etc. Once defined by only the traditional housekeeping ncRNAs (i.e., rRNA, tRNA, etc.), the recent cast of regulatory ncRNAs has expanded to include such species as miRNAs (microRNAs), lncRNAs (long non-coding RNAs), and circRNAs (circular RNAs). While the activities associated with ncRNAs are approaching those of proteins, many functions still remain unknown. As such, ncRNA is a growing ‘hot spot’ for biological research even in fields thought to be well-defined. The mammalian heat shock response is no exception as ncRNAs are being identified as integral components in this process. Although this area of research is only in its infancy, this review article focuses on several classes of regulatory ncRNAs and summarizes the activities of select examples implicated in mammalian heat shock. In addition, it proposes a greater involvement for ncRNA than what is currently known and defines some putative roles in which ncRNA may regulate the heat shock response. It is our hope that much of what is discussed herein may be taken into consideration by researchers to help push the field forward.
The basics of miRNA
MicroRNAs (miRNAs) comprise a large class of small ncRNA that have emerged as key regulators of gene expression in nearly all multicellular organisms (reviewed in Kim et al. 2009; Pasquinelli 2012). Core components of the miRNA pathway are generally well-conserved with only subtle variations in the animal kingdom. In mammals, miRNAs are predominantly transcribed by RNA polymerase II (Pol II) into long primary transcripts termed pri-miRNA. Approximately one-third of known miRNAs are embedded within the introns of protein-coding genes, which are often co-transcribed with their host gene. Other miRNAs can be found in exons or processed from much larger ncRNAs. Biogenesis of most miRNAs is dependent on the sequential activity of RNase III family members Drosha and Dicer. Following transcription, Drosha excises short hairpin structures (∼60–100 nucleotides in length) from the primary transcripts in the nucleus to form premature miRNA (pre-miRNA). Pre-miRNAs are subsequently exported to the cytoplasm by the protein Exportin 5 (XPO5) and processed by Dicer into double-stranded RNAs ∼19–24 nucleotides in length. These resulting duplexes are the mature form of miRNA. Some miRNAs bypass the conventional order of biogenesis and mature independent of Drosha processing. These include mirtrons and tailed mirtrons, which release their cognate pre-miRNA through splicing and exonuclease trimming (Berezikov et al. 2007).
Canonical miRNA activity is dependent on its incorporation into the RNA-induced silencing complex (RISC). Members of the argonaute (AGO) protein family are at the core of RISC, which function to recognize mature miRNAs and process them to single-stranded forms by expelling one strand of the miRNA duplex. In mammals, AGO1 and AGO2 are the major facilitators of miRNA activity (Wang et al. 2012). Sequence of the loaded miRNA guides RISC to complementary transcripts and suppresses gene expression by blocking translation and/or degrading messenger RNA (mRNA). Most miRNAs regulate gene expression by imperfect base-pairing with sequence in the 3′UTRs of target transcripts; however, atypical target site locations have been identified in 5′UTRs and coding sequence, as well (Hafner et al. 2010). Specificity is largely dependent on the first ∼7–8 nucleotides at the 5′-end of each miRNA (termed the ‘seed’ region) in which a high degree of complementarity is essential for effective targeting. It has been estimated that miRNAs control ∼50 % of all protein coding genes in mammals (Krol et al. 2010). In fact, functional studies have indicated that miRNAs participate in the regulation of nearly every cellular process investigated thus far.
miRNAs target heat shock protein transcripts
It has been shown that miRNAs play a critical role in regulating gene networks in response to cellular stresses (reviewed in Leung and Sharp 2010). Depending on where miRNAs are embedded in stress pathways, they can function to restore homeostasis via feedback loops or enforce new gene expression programs. For instance, HSPA5/GRP78 is an essential chaperone critical for protein quality control in the endoplasmic reticulum that functions to promote cell survival in response to certain stress conditions (Tranter et al. 2010). During stroke, miR-181 levels increase in ischemic tissue potentiating injury by targeting and depleting levels of HSPA5/GRP78 (Ouyang et al. 2012). High glucose stress damages cardiomyocytes in the diabetic heart by increasing expression of miR-1 and miR-206. Both miRNAs target and repress HSPD1/HSP60, which is an important protein in the defense against diabetic myocardial injury (Shan et al. 2010). In rat ventricular cells, miR-1 also functions to promote apoptosis during oxidative stress by targeting both HSPD1/HSP60 and HSPA1A/HSP70 (Xu et al. 2007). Conversely, ischemia preconditioning in the murine heart decreases levels of miR-378* and miR-711, which aid in tissue protection by increasing levels of downstream target HSPA1A/Hsp70.3 (Tranter et al. 2011). In addition, depletion of miR-320 enhances levels of its downstream target HSPB6/HSP20 to protect cardiomyocytes from I/R-induced death (Ren et al. 2009). It has also been shown that miR-3120 plays a role in regulating constitutive levels of HSPA8/Hsc70 in neuronal cells by targeting multiple conserved sites within its 3′UTR (Scott et al. 2012). Because HSPA8/Hsc70 catalyzes the uncoating of clathrin-coated vesicles, miR-3120 overexpression prevents vesicle uncoating. Such results imply that changes in miRNA expression can interfere or supplement target HSP activity.
Transcriptional regulation of the heat shock response is orchestrated largely by HSF1. It has been disclosed that miR-378 directly targets and represses the expression of HSF1 in cardiomyocytes (Yuan et al. 2010). As such, depletion or overexpression of miR-378 may likely impact HSF1 activity and induction of downstream HSPs in the heart. Studies in human breast cancer have also implicated miR-608 as a putative regulator of HSF1. HER2-positive breast cancer requires aberrant HSF1 activity during tumorigenesis. A polymorphism in miR-608 (rs4919510) that putatively lowers its binding affinity for a target site within the HSF1 3′UTR has been shown to contribute to HER2-positive breast cancer risk (Huang et al. 2012). While speculative, the pathological mechanism is that rs4919510 weakens suppression of HSF1 mRNA by miR-608 contributing to aberrant HSF1 levels in HER2-positive breast cancer cells.
Constitutively expressed miRNAs can also function to sequester basal expression of stress response genes in unstimulated cells. This is best exemplified in the case of extracellular ligands MICA and MICB, which are readily induced by a variety of stressors including heat shock (Stern-Ginossar et al. 2008). In unstressed cells, basal expression levels are inhibited by miR-20a, miR-93, and/or miR-106b in order to maintain minimal production of each ligand. However, upon stimulation, the levels of these miRNAs do not change; rather the transcription of MICA and MICB surges and protein levels become readily detectable. By this mechanism, levels of transcript after stress saturate the concentration threshold for effective inhibition by the miRNAs resulting in protein production.
Because of their ability to stabilize oncoprotein function, HSPC/HSP90 family members have been the target of drug development for a number of malignancies. Despite the inherent importance in cancer therapeutics, little has been disclosed in regards to miRNA regulation of HSPC/HSP90 transcripts. A recent study has shown that miR-223 may function as a putative tumor suppressor by targeting HSPC4/Hsp90B1 in osteosarcoma (Li, Cai et al. 2012). Additionally, miR-101 has been identified to indirectly regulate HSPC/HSP90 isoforms by targeting co-chaperone p23 (Liu, Zou et al. 2012). Like members of the HSPC/HSP90 family, p23 has also been implicated in several cancers (e.g., childhood acute lymphoblastic leukemia), in part, by protecting HSPC/HSP90 proteins from drug inhibitors (Forafonov et al. 2008). In the interest of drug development, it would be relevant to compare or combine the effects of such miRNAs with HSPC/HSP90 inhibitors currently being evaluated in the clinic (e.g., 17-AAG).
miRNA target sites are abundant in heat shock gene transcripts
Many different algorithms exist for prediction of miRNA targets sites in gene transcripts (e.g., TargetScan, miRanda, DIANA-MicroT, RegRNA, miRTar, etc.). While all share common rules, each possess unique features or search criteria for defining putative target sites. For example, all algorithms utilize the ‘seed’ region in miRNAs as the main determinant for target site prediction; however, TargetScan considers additional features such as target site conservation across different species (Friedman et al. 2009). Differences can also arise from the source of transcript sequence. For instance, TargetScan utilizes the Ensembl database by default to define 3′UTRs, whereas miRanda retrieves sequences from the University of California Santa Cruz (UCSC) genome database (Bina 2008; Flicek et al. 2008). If the miRanda algorithm was applied to both databases, only a 65 % overlap in target prediction would be observed (Ritchie et al. 2009). Other search engines like RegRNA offer a different approach by allowing users a simple interface to perform custom scans of input sequence (Huang et al. 2006). Regardless of search method, there is always a possibility of false-positive prediction. It has been calculated that the false-positive rate can vary between 24 and 70 % depending on algorithm and search criteria (Thomson et al. 2011). In addition, some analyses will also miss genuine miRNA target sites. Each program often generates different lists of predicted targets. As such, it is generally good practice to combine results from multiple programs in order to encompass all possible target sites, as well as define overlap to maximize predictive power (Sethupathy et al. 2006).
In silico analyses of HSF1 and several major inducible HSPs (i.e., HSPA1A, HSPA6, and HSPC1) reveal numerous putative miRNA target sites in 3′UTRs. Table 1 lists example search results for select algorithms including TargetScan, miRanda, DIANA-MicroT, and RegRNA. Although the search outcomes vary between each algorithm, all queries share overlapping results. This simple observation reveals HSF1 and its major downstream regulators are not deficient of putative miRNA target sites thereby supporting a hypothesis in which miRNAs likely play a more prevalent role in regulating heat shock signaling.
Table 1.
miRNA target sites are detectable in heat shock gene transcripts
| Gene name | Alt. name(s) | 3'UTR sizea | miRNA target search algorithm | miRNA target site search resultsb | Predicted regulatory miRNAsc |
|---|---|---|---|---|---|
| HSF1 | HSTF1 | 387 nt | TargetScan | 72 | miR-3714; miR-445-3p; miR-4666-3p; miR-4668-3p; miR-548c-3p; etc. |
| miRanda | 1 | miR-431 | |||
| DIANA-MicroT-CDS | 10 | miR-654-5p; miR-541-3p; miR-615-5p; miR-3619-5p; miR-92a-2-5p; etc. | |||
| RegRNA | 84 | miR-1205; miR-1226; miR-3934; miR-1825; miR-1972; etc. | |||
| HSPA1A | HSPA1A HSP72; HSPA1; | 258 nt | TargetScan | 33 | miR-4699-3p; miR-4635; miR-127-5p; miR-527; miR-561; etc. |
| miRanda | 9 | miR-146a; miR-146b-5p; miR-183; miR-34c-5p; miR-449a; etc. | |||
| DIANA-MicroT-CDS | 8 | miR-146b-5p; miR-146a; miR-548ao-5p; miR-34a; miR-412; etc. | |||
| RegRNA | 62 | miR-127-5p; miR-146b-5p; miR-561; miR-570; miR-1264; etc. | |||
| HSPA6 | HSP70B' | 307 nt | TargetScan | 40 | miR-106a; miR-20a; miR-20b; miR-106b; miR-519d; etc. |
| miRanda | 23 | miR-376a; miR-20b; miR-17; miR-106a; miR-93; etc. | |||
| DIANA-MicroT-CDS | 10 | miR-4646-5p; miR-298; miR-5688; miR-20a; miR-106b; etc. | |||
| RegRNA | 70 | miR-103b; miR-106a; miR-298; miR-20b; miR-376a; etc | |||
| HSPC1 | HSP90; HSP90N; HSP90A; LAP2; HSPN; HSPCA; HSP90AA1 | 974 nt | TargetScan | 136 | miR-9; miR-361-5p; miR-495; miR-224; miR-370; etc. |
| miRanda | 14 | miR-96; miR-1271; miR-411; miR-148a; miR-377; etc. | |||
| DIANA-MicroT-CDS | 62 | miR-4778-3p; miR-578; miR-515-5p; miR-529e; miR-205-5p; etc. | |||
| RegRNA | 182 | miR-1224; miR-142-3p; miR-632; miR-3119; miR-518d-5p; etc. |
a3′UTR sequence of human genes was obtained through the UCSC genome browser
bAll searches were performed with default settings. Search results refer to the total number of putative miRNA regulatory sites listed following each search query
cTarget sites are defined by their cognate human miRNA. All miRNAs in bold were detected by at least one other search algorithm
One of the more interesting aspects of miRNA biology is their ability to elicit pleiotropic effects by targeting multiple transcripts. In fact, it is not uncommon for a single miRNA to regulate multiple genes within a specific signaling cascade or cellular mechanism. The heat shock signaling pathway should be considered no exception to this phenomenon. Search algorithms like miRTar allow analysis of multiple scenarios including the ability to predict recurring miRNA target sites shared among a series of selected genes (Hsu et al. 2011). Analysis of 3′UTR sequences from 36 heat shock-related genes (i.e., 17 members of the HSP70 superfamily, 11 members comprising the HSPB family, five members of the HSPC/HSP90 family, HSPD1, HSPE1, and HSF1) reveal several miRNAs with putative target sites across multiple gene transcripts (Table 2). For instance, miR-92a-2-5p has target sites predicted in eight of the 36 heat shock-related genes including HSF1 and HSPA1A. Such information supports a hypothesis that the heat shock response and its related factors may be subject to coordinate control by the same miRNAs.
Table 2.
Recurring miRNA target sites exist across multiple heat shock-related genes
| miRNAa | Number of predicted gene targets | Names of predicted heat shock-related gene targetsb |
|---|---|---|
| miR-92a-2-5p | 8 | CRYAB; HSF1; HSPA12B; HSPA1A; HSPB6; HSPB7; HYOU; TRAP1 |
| miR-4270 | 8 | CRYAA; HSPA12A; HSPA14; HSPA4; HSPB6; HSPB7; HSPB8; HYOU1 |
| miR-762 | 7 | HSPA12A; HSPA12B; HSPA14; HSPA5; HSPB6; HSPB7; HYOU1 |
| miR-680 | 7 | HSF1; HSPA12B; HSPA4; HSPB1; HSPB6; HSPB7; HYOU1 |
| miR-4268 | 7 | HSP90AA1; HSP90B1; HSPA12A; HSPA12B; HSPA14; HSPA4; TRAP1 |
| miR-3189 | 7 | HSP90AA1; HSPA12B; HSPA1A; HSPA4; HSPA5; HSPB2; HSPD1 |
| miR-3127 | 7 | HSPA12A; HSPA12B; HSPA1A; HSPB2; HSPB6; HSPB7; HYOU1 |
| miR-220c | 7 | HSPA121A; HSPA12B; HSPA14; HSPA1A; HSPA9; HSPB8; HYOU1 |
| miR-1909 | 7 | CRYAA; HSP90AA1; HSP90AB1; HSP90B1; HSPA14; HSPB1; HSPB6 |
| miR-637 | 6 | HSF1; HSPA14; HSPA1A; HSPB6; HSPB7; HYOU1 |
| miR-370 | 6 | CRYAA; HSP90AA1; HSP90AB1; HSPA12A; HSPB7; HYOU1 |
| miR-30c-1-3p | 6 | HSPA4; HSPA5; HSPB6; HSPB7; HYOU1; ODF1 |
| miR-92a-1-5p | 5 | HSF1; HSPA12B; HSPA14; HSPB6; HSPB8 |
| miR-761 | 5 | HSP90AB1; HSPA12A; HSPA1A; HSPB1; HSPB7 |
| miR-298 | 5 | HSPA6; HSPA9; HSPB8; HSPH1; HYOU1 |
amiRNAs correspond to human sequence
b3′UTRs from 36 heat shock-related genes were scanned using the miRTar search engine with default settings in order to identify common regulatory miRNAs shared by the different gene transcripts
Heat shock-responsive miRNAs
While alterations in miRNA expression have been recorded in several organisms following exposure to various stressors, minimal data has been generated analyzing miRNA expression following heat stress in mammals. Microarray analyses have shown that recurring heat exposure in rats alters miRNA expression patterns in the small intestine resulting in the upregulation and downregulation of 18 and 11 miRNAs, respectively (Yu et al. 2011) (Table 3). Integrated bioinformatics analyses also revealed that the changes in miRNA expression are inversely correlated with alterations in mRNA levels of putative downstream targets to imply heat-induced changes in the transcriptome are, in part, mediated by miRNA. In mice, thermal preconditioning increases the expression of miR-1, miR-21, and miR-24 in the heart to generate a cardioprotective phenotype resistant to I/R injury (Yin et al. 2008) (Table 3). Interestingly, direct injection of the miRNAs was able to substitute for heat shock and inhibit cardiac infraction (Yin et al. 2008; Yin et al. 2009). In vitro analyses in human cell lines have also shown that miR-125b, miR-154, and miR-382 were activated in response to mild hyperthermia (Oshlag et al. 2013) (Table 3).
Table 3.
Reported miRNAs deregulated by heat stress
| Citation | Organism | Tissue/cell type | Heat stress | Deregulated miRNAsa |
|---|---|---|---|---|
| Yu et al. 2011 | Sprague–Dawley rats | Small intestine tissue | 40 °C for 2 h over 10 consecutive days | UP: miR-34b, -137, -154, -672, -219-5p, -375, -7a, -34a, -30a*, -500, -185, -203, -200a, -140*, let-7d, -125a-5p, -27b, -210 |
| DOWN: miR-322, -142-5p, -434, -204, -142-3p, -193, -31, -150, -148b-3p, -223, -23a | ||||
| Yin et al. 2008 | ICR mice | Heart tissue | 42 °C for 15 min with 2 h recovery at RT | UP: miR-1, -21, -24 |
| DOWN: not reported/evaluated | ||||
| Oshlag et al. 2013 | Human | Hela and JAR cell lines | 39.5 °C for 1.5 to 8 h | UP: miR-125b, -154, -382 |
| DOWN: not reported/evaluated | ||||
| Wilmink et al. 2010 | Human | HDF cells in culture | 44 °C for 40 min with 4 h recovery at RT | UP: miR-125b, -452, -133b, -192, -382, -378, -101, -424, -22 |
| DOWN: miR-138, -7, -376a, -31, -222, -33a, -29b, -606, let-7c, let-7d, - 218, -196a, -204, -196b, -154, -1298, -18a, -487b |
ICR imprinting control region, RT room temperature, UP upregulated miRNAs, DOWN downregulated miRNAs
amiRNA sequences correspond to cognate species
One of the more compelling studies identified 27 annotated miRNAs differentially regulated by heat shock in cultured normal adult human dermal fibroblast (HDF) cells (Wilmink et al. 2010) (Table 3). Comparative analyses with other miRNA expression profiles from different stressors (i.e., folate deficiency, arsenic exposure, radiation, hypoxia, and cigarette smoke) identified a group of inducible miRNAs (miR-125b, miR-222, miR-22, and let-7c) conserved by most stress signals. As such, it is possible to envision that some miRNAs may be universal responders analogous to minimal stress proteins. Conversely, other inducible miRNAs were unique to only heat stress (e.g., miR-452, miR-382, and miR-378) implying certain miRNAs may be stress-type dependent.
miRNA expression patterns following heat shock are likely context dependent in regards to cell type, as well. For instance, miR-1 has been reported to be muscle specific with important pathophysiological functions in the heart (Yang et al. 2007). As such, miR-1 may not be induced and/or participate in cytoprotection following heat shock in other tissues. While these examples indicate miRNA levels do change in response to heat exposure, it is unclear if the mechanisms regulating expression are at the level of transcription and/or involve HSF1. Like the core set of inducible HSPs (e.g., HSPA1A, HSPA6, etc.), it would be interesting to determine if there is a key miRNA signature indicative of heat shock.
Heat shock may also manipulate miRNA levels by affecting their biogenesis. In support, it has been shown in mammalian cells that thermal stress increases Dicer levels, which may influence miRNA maturation (Oshlag et al. 2013). Additionally, HSPC1/Hsp90 has been shown to be an important regulatory of RISC by functioning to mediate loading of miRNA into the complex (Iwasaki et al. 2010). As such, heat-induced changes in HSP expression may directly alter RISC activity/assembly.
The short on long non-coding RNAs
Less than 2 % of the mammalian genome actually codes for proteins, yet ∼70–90 % is transcribed in some context as long non-coding RNAs (lncRNAs). As such, lncRNAs are proposed to be the largest transcript class in the mammalian transcriptome (Derrien et al. 2012). Originally thought to be only genomic “dark matter” manifesting as transcriptional noise, lncRNAs have now been shown to possess biological function participating in a variety of processes including gene regulation, signal transduction, development, etc. LncRNAs are generally defined as transcripts longer than 100–200 nucleotides in length in order to distinguish them from smaller RNA species such as miRNAs, piRNAs (piwi-interacting RNAs), siRNAs (small interfering RNAs), and small nucleolar RNAs (snoRNAs); although, size should be considered a rather subjective limit when defining lncRNAs (reviewed in Wapinski and Chang 2011). Some subclasses include asRNAs (antisense RNAs), lincRNAs (long intergenic non-coding RNAs), paRNAs (promoter-associated RNAs), eRNAs (enhancer RNAs), etc. While the term has only been recently defined, some ncRNAs discovered in years past are now appropriately referred to as lncRNAs (e.g., H19, Xist, etc.).
LncRNAs can function in cis and/or in trans. Cis-acting lncRNAs are restricted to their site of synthesis and act on one or more proximal genes transcribed at the same locus. Their expression is often coordinated with that of neighboring genes. Such lncRNAs can behave as structural components of chromatin and offer sites of interaction for modifiers, remodelers, or other regulatory proteins (Rodriguez-Campos and Azorin 2007). For instance, an antisense lncRNA transcribed within the INK4/ARF locus referred to as ANRIL recruits chromatin modifiers PRC1 and PRC2 to promote heterochromatin formation and silence gene expression (Kotake et al. 2011; Yap et al. 2010; Yu et al. 2008). Alternatively, cis-acting lncRNAs can displace bound factors. LncRNAs transcribed in CpG islands (CGIs) have been proposed to inhibit binding of DNMT3B and prevent local DNA methylation at active genes (Ginno et al. 2012).
In contrast, trans-acting lncRNAs diffuse from their site of synthesis and act on many genes located at distant loci on same or different chromosomes. As such, these types of lncRNAs can act within large genic networks. For example, steroid receptor RNA activator (SRA) is an lncRNA transcribed in humans from chromosome 5q31.3 that elicits its pleotropic effects on gene expression by functioning as a molecular subunit and essential co-activator for a variety of nuclear hormone receptors (i.e., estrogen, androgen, progesterone, etc.) (Lanz et al. 1999). LncRNAs can also regulate gene expression networks in trans by acting as molecular decoys for endogenous miRNAs. These lncRNAs possess multiple miRNA target sites, which function as competitive inhibitors for other transcripts. For instance, HULC is an lncRNA transcribed from chromosome 6p24.3 that appears to function as a molecular decoy by sequestering the activity of miR-372 in liver cancer cells (Wang et al. 2010). Any transcribed RNA (e.g., lncRNA, pseudogene, mRNA, etc.) that influences the levels of another transcript by competing for the same pool of miRNAs are referred to as competing endogenous RNAs (ceRNAs). Collectively, trans-acting lncRNAs have been described to function as signals, guides, decoys (i.e., ceRNAs), or scaffolds for chromatin or other cellular machinery to regulate such processes as gene expression, protein activity, and/or subcellular localization.
lncRNA is an auxiliary factor of HSF1 activation
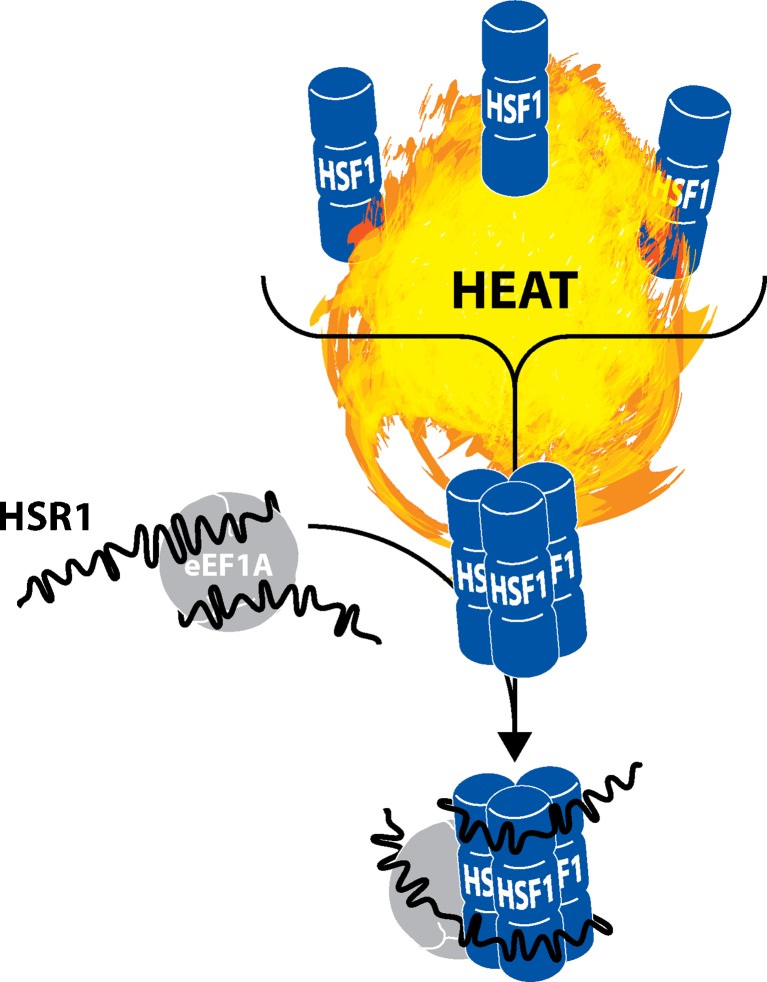
The hallmark of the heat shock response is the rapid and robust induction of cytoprotective genes driven predominantly by the transcription factor HSF1. Under normal conditions, HSF1 is present in the cell cytoplasm as an inactive monomer. Upon heat stress, HSF1 localizes to the nucleus as a homotrimer and binds to HSE stimulating the transcription of stress response genes. HSF1 is largely controlled by bound chaperone proteins (i.e., HSPA1A/HSP70, HSPC1/HSP90, p23, etc.), which disassociate upon heat shock making it possible for HSF1 to trimerize. In human cells, another key determinant in this process is an lncRNA termed HSR1 (heat shock RNA-1) (Shamovsky and Nudler 2009). In vitro, HSR1 forms a complex with eEF1A (eukaryotic elongation factor 1A), which is required for HSF1 trimerization and its subsequent DNA binding activity (Fig. 1). As such, HSR1 depletion hinders induction of stress response genes and dampens cell survival to prolonged heat shock (Shamovsky et al. 2006). In this regards, HSR1 functions as an auxiliary factor required for the trimerization and subsequent activation of HSF1.
Fig. 1.
HSF1 trimerization in response to heat shock requires lncRNA HSR1. In unstressed cells, HSF1 is predominantly monomeric. Upon heat shock, chaperone proteins dissociate from HSF1 freeing monomers to form functional homotrimers. HSR1 is an lncRNA ∼2 kb in length containing a long poly(A) tail. It is constitutively present in cells, but following heat shock HSR1 undergoes conformational changes and associates with protein eEF1A. Though typically involved in translational regulation, eEF1A acquires an apparent non-canonical function upon binding HSR1. Both eEF1A and HSR1 associate with HSF1 bringing about its trimerization. Within this mechanism, HSR1 behaves much like a molecular thermosensor that effectively regulates HSF1 mobilization
lncRNA mediates general transcription repression by heat stress
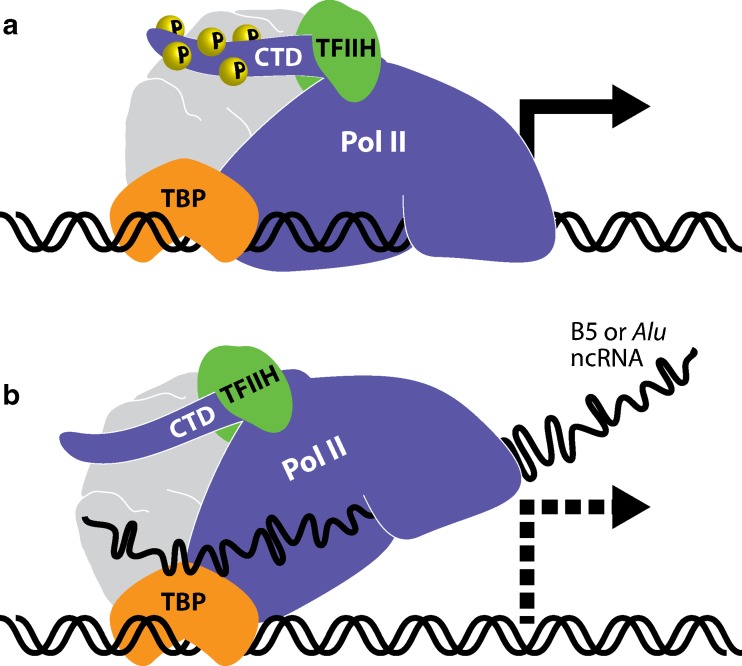
Upon heat shock, basal transcription is sequestered in eukaryotic cells at the same time HSF1 is activated to drive transcription of heat shock-specific genes. One mechanism by which heat shock suppresses gene expression is through the upregulation of inhibitory ncRNA that function in trans to block general RNA polymerase II (Pol II) activity. Two ncRNA species including B2 RNA in mice and Alu RNA in humans are transcribed by RNA polymerase III (Pol III) from short interspersed elements (SINEs). During normal cellular growth, these RNA species accumulate at relatively low levels; however, their abundance transiently increases by as much as 40-fold under certain conditions of stress (i.e., heat shock) (Li et al. 1999; Liu et al. 1995). Although the kinetics of induction resembles that of heat shock proteins, upregulation of B2 and Alu RNAs is achieved by a separate stress-activated transcriptional mechanism. There is no shared sequence homology between B2 and Alu RNAs, but rather their biological functions are considered to be very similar. Following stress, B2 or Alu RNAs directly bind to the active site within Pol II, disrupting its interaction with promoter DNA and interfering with phosphorylation of the carboxy terminal domain (CTD; Fig. 2) (Yakovchuk et al. 2009, 2011). As such, both B2 and Alu RNAs function in trans as direct repressors of general Pol II activity contributing to the global downregulation of basal gene expression following heat stress.
Fig. 2.
Suppression of basal transcription following heat shock is mediated by lncRNA. a Under normal conditions, RNA polymerase II (Poll II) associates with gene promoters through the help of accessory factors such as TATA-binding protein (TBP) to drive basal transcription of house-keeping genes. Phosphorylation (P) of heptapeptide repeats at serine-5 within the carboxyl terminal domain (CTD) of Pol II is facilitated by TFIIH to signify transcription initiation. b LncRNAs B5 in mice (∼178 nts) or Alu in humans (∼300 nts) are upregulated in response to heat stress. Both RNA species function in trans as repressors of basal transcription by binding to the active site of Pol II, preventing proper association with DNA, and suppressing CTD phosphorylation. TFIIH does not appear to disassociate from the complex, rather its kinase activity is suppressed following lncRNA interaction with Pol II
lncRNA regulates transcriptional elongation of stress response genes
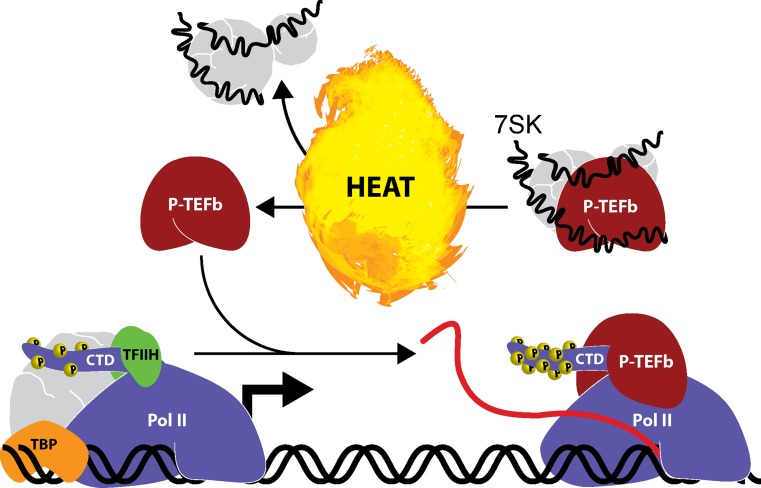
7SK is an abundant nuclear ncRNA widely conserved in the Metazoan kingdom. In fact, it has been shown to be one of the most abundant nuclear transcripts in human cells (Wassarman and Steitz 1991). It is transcribed by Pol III and plays a major role in regulating transcription. Under normal conditions, Pol II briefly pauses following release from gene promoters. In order for transcription to continue, Pol II is stimulated by the kinase activity of P-TEFb (positive transcription elongation factor b) resulting in CTD hyperphosphorylation and transcription elongation. 7SK functions in trans as an essential structural component and repressor of the P-TEFb complex. In mammalian cells, a fraction of P-TEFb is sequestered in an inactive state by 7SK and several other factors (e.g., HEXIM1, etc.). In fact, it has been estimated that more than 50 % of its total cellular levels are bound to 7SK (Nguyen et al. 2001). P-TEFb is subsequently activated upon release from the repressor complex as a result of conformational changes in 7SK (Peterlin et al. 2012; Yang et al. 2001). Under conditions of cellular stress, nearly all P-TEFb is ejected from 7SK causing a substantial increase in active P-TEFb levels. Although a direct role of 7SK in the heat shock response has not yet been examined, it seems likely as P-TEFb is known to rapidly localize to the promoters of stress response genes following heat shock (Fig. 3) (Lis et al. 2000). As such, prompt mobilization of P-TEFb following heat stress supports efficient transcription of heat shock response genes.
Fig. 3.
Transcription elongation of heat shock genes is regulated by lncRNA. RNA polymerase II (Pol II) pauses following transcription initiation at gene promoters. P-TEFb promotes transcription elongation by phosphorylating (P) heptapeptide repeats at serine-2 within the carboxyl terminal domain (CTD) of Pol II. 7SK is a nuclear lncRNA (∼331 nts) that interacts with a subpopulation of P-TEFb repressing its activity and sequestering its ability to interact with transcription complexes. Induction of the stress response (e.g., heat shock) causes P-TEFb to disassociate from 7SK allowing it to interact with Pol II and promote transcription elongation of susceptible genes including heat shock response factors. By this mechanism, 7SK functions in trans as a scaffold component of P-TEFb and regulator of gene transcription in response to stress signals
lncRNA immobilizes proteins to the nucleolus in response to heat stress
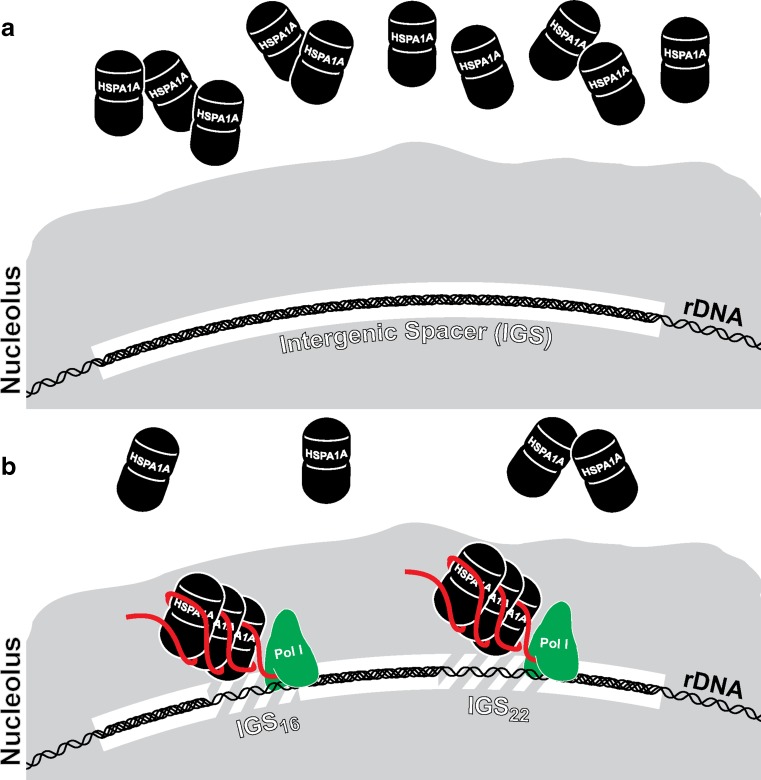
The nucleolus coordinates several vital cellular processes by sequestering or releasing proteins in response to specific physiological signals. Upon cellular stress, nucleolar detention of proteins becomes an important step in maintaining cellular homeostasis (Boulon et al. 2010). This process is mediated, in part, by a group of inducible lncRNAs transcribed by RNA polymerase I (Pol I) from intergenic spacers (IGS) located between ribosomal genes (Audas et al. 2012). Different stress signals induce subsets of IGS transcripts, which function to sequester various proteins. In response to heat stress, IGS transcription occurs at two loci positioned 16 kb (IGS16) and 22 kb (IGS22) downstream of the rRNA (ribosomal RNA) transcription start site (TSS) in human cells. The resulting lncRNAs function in cis to recruit and immobilize HSPA1A/HSP72, as well as other proteins (e.g., RNF8), to the rDNA (ribosomal DNA) cassette in the nucleolus (Fig. 4). Furthermore, silencing of specific IGS lncRNAs prevents enrichment of nucleolar proteins at targeted loci (Audas et al. 2012). Nucleolar translocation of HSPA1A/HSP72 is known to be a key event during the heat response reported to accelerate nucleolar recovery and protect cellular integrity (Kotoglou et al. 2009; Pelham 1984; Welch and Feramisco 1984). As such, transcription of lncRNAs in the IGS region and their biological activity is essential for facilitating some of the cytoprotective effects associated with heat stress.
Fig. 4.
Nucleolar sequestration of HSPA1A is facilitated by lncRNA. a Transcription of ribosomal DNA (rDNA) occurs in the nucleolus of mammalian cells. Between each ribosomal gene is a region of repetitive sequence long believed to be devoid of transcriptional activity referred to as the intergenic spacer (IGS). HSPA1A/HSP72 is a prototypical inducible protein activated in response to heat stress. b One hallmark of the heat shock response is nucleolar recruitment of HSPA1A/HSP72 in addition to its overexpression. Within the IGS region, heat stress activates transcription at specific loci located 16 kb (IGS16) and 22 kb (IGS22) downstream of the rRNA TSS. Other stress stimuli trigger transcription at different IGS loci. The resulting lncRNAs (∼300 nts) function in cis to capture a subfraction of HSPA1A/HSP72 at the sites of transcription forcing its sequestration in the nucleolus. RNA polymerase I (Pol I) is responsible for transcription within the IGS region; however, it is unclear if the lncRNAs remain tethered to chromatin as nascent transcripts (as shown) or bound by other factors
Pseudogenes as lncRNAs
Long regarded as a genomic graveyard, pseudogenes might actually possess biological function in context as lncRNAs. For instance, expression of the pseudogene PTENP1 sequesters the same miRNAs that normally repress translation of the tumor suppressor PTEN thereby increasing its protein levels (Poliseno et al. 2010). As such, pseudogenes may actually be a vast repository for regulatory lncRNAs, in part, by functioning as molecular decoys for their ancestral genes. The HSPA/HSP70 superfamily has roughly 30 pseudogenes in the human genome, while HSPC1/HSP90 is estimated to have 12 pseudogenes including HSP90B3P; a known lncRNA (Brocchieri et al. 2008). Each one of these pseudogene transcripts contains numerous putative miRNA target sites, which may function in sequestering miRNAs that would otherwise repress HSP levels or other target transcripts.
The significance of circular RNAs
Discovered more than 20 years ago, circular RNA (circRNA) was an enigmatic class of ncRNA thought to exist only in low abundance as a result of occasional errors in splicing (Nigro et al. 1991). However, it is now known that circRNAs are a prevalent form of ncRNA in the mammalian transcriptome (Jeck et al. 2013). Unlike linear RNA with exposed termini (e.g., mRNA, lncRNA, etc.), cirRNAs are formed when the 5′ and 3′ end of exons within the same transcript are covalently linked together to form a circular molecule. Because circRNAs are predominantly comprised of only exonic sequence, their biogenesis appears to represent a novel form of alternative splicing (Salzman et al. 2012). In some instances, circRNAs are >10 times more abundant than their cognate linear transcript of the same gene (Jeck et al. 2013). Like lncRNAs, circRNAs have been shown to function as a type of ceRNA capable of sequestering miRNA activity. For example, a circRNA termed ciRS-7/CRD1as contains more than 70 conserved miRNA target sites and acts as a molecular “sponge” for miR-7 (Hansen et al. 2013; Memczak et al. 2013). However, circRNAs are inherently more stable than linear molecules as their unique structure provides protection against exonuclease activity (Hansen et al. 2013; Memczak et al. 2013).
Similar to lncRNAs, it is plausible circRNAs have additional roles in the cell other than strict ceRNA activity. For instance, circRNAs may function as structural components or allosteric regulators for RNA-binding proteins. CircRNAs may also possess non-folding chaperone-like function acting to store, sort, or localize bound factors such as miRNA. Studies have shown that numerous circRNAs are expressed under a variety conditions across different tissues and developmental stages (Hansen et al. 2013; Memczak et al. 2013). As such, it is reasonable to speculate that circRNAs function in the stress response, as well. CircRNAs may act as molecular decoys for stress-induced miRNAs, stressors may elevate their expression levels, or known stress response genes may generate cognate circRNAs. These ncRNA species represent a novel topic that should be explored within the field of heat shock and stress response.
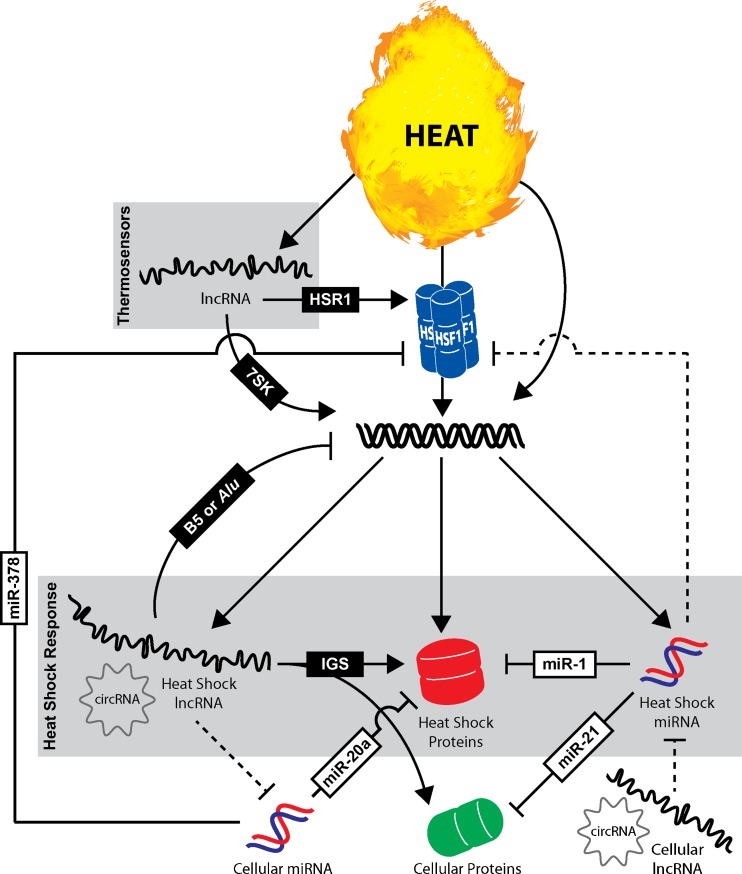
An integrated model for the heat shock response pathway
The conventional heat shock response pathway is characterized by the rapid trimerization/activation of HSF1 and subsequent transcriptional upregulation of cytoprotective genes such as HSPs (e.g., HSPA1A, HSPA6, etc.). While it is becoming increasingly clear that ncRNAs are integral components of a variety of cellular processes, they are frequently overlooked in context to the heat shock response pathway in mammals. As such, it is our hope that introduction of an updated model integrating the traditional heat shock response with the activities of regulatory ncRNA (i.e., miRNA, lncRNA, and circRNA) may offer a more comprehensive summary highlighting the major regulators and responders of heat shock signaling (Fig. 5). In this model, lncRNAs in the form of molecular thermosensors also function as primary responders to heat stress. Thermosensors undergo conformational changes upon heat stress leading to alterations in activity and/or complex composition. For instance, heat shock activates HSR1 to assist in HSF1 trimerization and triggers 7SK to release P-TEFb reserves for efficient transcription of heat shock response genes (Peterlin et al. 2012; Shamovsky et al. 2006). Although there is some debate over the evolutionary origin of HSR1, its function in vitro is preserved and still exemplifies the role of lncRNAs as key regulators of thermal stress (Kim et al. 2010).
Fig. 5.
ncRNAs function as integral components of the heat shock response in mammals. The heat shock response is typically characterized by the rapid trimerization/activation of HSF1 and subsequent transcriptional induction of heat shock proteins (e.g., HSPA1A, HSPA6, etc.). ncRNAs including lncRNAs and miRNAs are often overlooked regulators of the heat shock response pathway. This model illustrates the diverse functional roles that ncRNAs play or likely play in heat stress. Select examples of lncRNA (black boxes) and miRNA (white boxes) involved in the heat shock response are delineated by solid lines. Dashed lines define putative functional interactions of ncRNA not yet examined/reported
In addition to HSPs, it is important to recognize that HSF1 also likely regulates the expression of ncRNAs including lncRNAs, circRNAs, and/or miRNAs. By this mechanism, transcriptional activation would mirror that of other heat shock response genes in which HSF1 binds HSE sites in gene promoters to drive transcription of ncRNAs. HSF1-independent mechanisms also regulate gene transcription including the expression of several regulatory lncRNAs. For instance, B5 in mice and Alu in humans are transcribed in response to heat stress and function in trans to repress basal transcription of housekeeping genes (Yakovchuk et al. 2009, 2011). In addition, lncRNAs transcribed within the IGS region of rDNA function to immobilize HSPA1A and other cellular proteins (e.g., RNF8) to the nucleolus during heat shock (Audas et al. 2012).
Depending on downstream targets, inducible miRNAs may provide an additional feedback mechanism for controlling HSP levels and/or restoring homeostasis. For example, miR-1 is activated in the heart upon thermal stress, which has independently been shown to target and suppress HSPA1A expression (Yin et al. 2008; Xu et al. 2007). Other inducible miRNAs may enforce the stress program by targeting other cellular proteins. In heart tissue, heat shock activates expression of miR-21, which contributes to the cytoprotection phenotype by reducing the levels of proapoptotic genes like BCL-10 (Yin et al. 2008). Other cellular miRNAs constitutively present in unstressed cells may quell basal expression of stress response genes and/or control HSF1 levels in order to suppress unnecessary stimulation or accumulation of stress factors. This is best exemplified by miR-20a, which has been shown to silence basal expression of stress-induced proteins MICA and MICB in unstressed cells (Stern-Ginossar et al. 2008). Additionally, it has been disclosed that miR-378 targets HSF1, which may function to titrate intracellular HSF1 levels (Yuan et al. 2010).
Based on their defined activities and general abundance, lncRNAs and/or circRNAs may have additional functions as ceRNAs in the heat shock response. Inducible lncRNAs or circRNAs could putatively contribute to the rapid induction of stress response genes by sequestering miRNA activity. Other cellular lncRNA or circRNAs present in unstressed cells may function to titrate basal activity of heat shock-inducible miRNAs. By this mechanism, heat stress would increase miRNA levels and exceed the regulatory thresholds established by cellular concentrations of ceRNAs.
Beyond intracellular interactions
Members of the HSPA/HSP70 and HSPC/HSP90 family are known to be secreted into circulation. One mechanism by which they enter the extracellular environment is through packaging of small membrane vesicles (∼30–100 nm in diameter) called exosomes (Lv et al. 2012). Exosomes are secreted by numerous cell types and function as an intercellular delivery system of internalized molecules. In additions to HSPs, ncRNAs (e.g., miRNA, lncRNA, etc.) are also found within exosomes, although miRNAs comprise up to ∼75 % of all mappable reads following sequencing experiments (Huang et al. 2013). Exosomal miRNAs can exist as mature duplexes or associated with AGO proteins as part of RISC. Given that intracellular cross-talk is prevalent between miRNAs and HSPs, it is possible that such interactions occur in context of the extracellular environment, as well. For instance, HSPC1/Hsp90 has been shown to be required for miRNA loading and RISC assembly in the cell (Iwasaki et al. 2010). Exosomal HSPs may also play a role in maintaining miRNA integrity in the exosome by stabilizing RISC and/or mature duplexes, as well as facilitating loading of exosomal miRNA upon intercellular delivery. Additionally, both HSPs and miRNAs have been implicated in the activation of toll-like receptors in the tumor microenvironment via non-canonical mechanisms (Fabbri et al. 2012; Tamura et al. 2012). Given their common destination, one could hypothesize they work as co-ligands in the extracellular environment to regulate immune-tumor communication.
The field of ncRNA is a growing ‘hot spot’ for biological research as it represents a novel source of discovery for most any cellular pathway. The heat shock response should be treated as no exception. While examples of regulatory ncRNAs in the mammalian heat shock response have been highlighted in this article, many more remain undiscovered. We propose ncRNAs have a greater involvement than what is currently known and have defined several putative roles in which ncRNA may function in regulating heat stress. It is our hope that this article may serve as a stepping stone for researchers to pursue projects interrogating the roles of ncRNA in the mammalian heat shock response.
Footnotes
R.F. Place and E.J. Noonan contributed equally to this manuscript.
Contributor Information
Robert F. Place, Email: Place.Robert@gmail.com
Emily J. Noonan, Email: Emily.Noonan@gmail.com
References
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45(2):147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina M. The genome browser at UCSC for locating genes, and much more! Mol Biotechnol. 2008;38(3):269–275. doi: 10.1007/s12033-007-9019-2. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40(2):216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L, Conway de Macario E, Macario AJ (2008) hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol 8:19. doi: 10.1186/1471-2148-8-19 [DOI] [PMC free article] [PubMed]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36(6):932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK. Regulatory interfaces between the stress protein response and other gene expression programs in the cell. Methods. 2005;35(2):139–148. doi: 10.1016/j.ymeth.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Dellavalle RP, Petersen R, Lindquist S. Preferential deadenylation of Hsp70 mRNA plays a key role in regulating Hsp70 expression in Drosophila melanogaster. Mol Cell Biol. 1994;14(6):3646–3659. doi: 10.1128/mcb.14.6.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S (2008) Ensembl 2008. Nucleic Acids Res 36 (Database issue):D707–714. doi: 10.1093/nar/gkm988 [DOI] [PMC free article] [PubMed]
- Forafonov F, Toogun OA, Grad I, Suslova E, Freeman BC, Picard D. p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity. Mol Cell Biol. 2008;28(10):3446–3456. doi: 10.1128/MCB.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Hsu JB, Chiu CM, Hsu SD, Huang WY, Chien CH, Lee TY, Huang HD (2011) miRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics 12:300. doi: 10.1186/1471-2105-12-300 [DOI] [PMC free article] [PubMed]
- Huang AJ, Yu KD, Li J, Fan L, Shao ZM. Polymorphism rs4919510:C>G in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PLoS One. 2012;7(5):e35252. doi: 10.1371/journal.pone.0035252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Chien CH, Jen KH, Huang HD (2006) RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res 34 (Web Server issue):W429–434. doi: 10.1093/nar/gkl333 [DOI] [PMC free article] [PubMed]
- Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39(2):292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Lee Y, Hahn Y. Evidence for bacterial origin of heat shock RNA-1. RNA. 2010;16(2):274–279. doi: 10.1261/rna.1879610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoglou P, Kalaitzakis A, Vezyraki P, Tzavaras T, Michalis LK, Dantzer F, Jung JU, Angelidis C. Hsp70 translocates to the nuclei and nucleoli, binds to XRCC1 and PARP-1, and protects HeLa cells from single-strand DNA breaks. Cell Stress Chaperones. 2009;14(4):391–406. doi: 10.1007/s12192-008-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40(2):205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Cai M, Fu D, Chen K, Sun M, Cai Z, Cheng B (2012) Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell Physiol Biochem 30(6):1481–1490. doi:10.1159/000343336 [DOI] [PubMed]
- Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239(2):367–372. doi: 10.1016/S0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14(7):792–803. [PMC free article] [PubMed] [Google Scholar]
- Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23(10):1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zou L, Zhu L, Zhang H, Du C, Li Z, Gao C, Zhao X, Bao S, Zheng H (2012) miRNA mediated up-regulation of cochaperone p23 acts as an anti-apoptotic factor in childhood acute lymphoblastic leukemia. Leuk Res 36(9):1098–1104. doi:10.1016/j.leukres.2012.05.003 [DOI] [PubMed]
- Lv LH, Wan YL, Lin Y, Zhang W, Yang M, Li GL, Lin HM, Shang CZ, Chen YJ, Min J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J Biol Chem. 2012;287(19):15874–15885. doi: 10.1074/jbc.M112.340588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Moseley PL, Wallen ES, McCafferty JD, Flanagan S, Kern JA. Heat stress regulates the human 70-kDa heat-shock gene through the 3'-untranslated region. Am J Physiol. 1993;264(6 Pt 1):L533–L537. doi: 10.1152/ajplung.1993.264.6.L533. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- Oshlag JZ, Devasthanam AS, Tomasi TB. Mild hyperthermia enhances the expression and induces oscillations in the Dicer protein. Int J Hyperthermia. 2013;29(1):51–61. doi: 10.3109/02656736.2012.753471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Xu LJ, Xiong XX, White RE, Sun X, Giffard RG. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012;45(1):555–563. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Pelham HR. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984;3(13):3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3(1):92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119(17):2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6(6):397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007;2(11):e1182. doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott H, Howarth J, Lee YB, Wong LF, Bantounas I, Phylactou L, Verkade P, Uney JB. MiR-3120 is a mirror microRNA that targets heat shock cognate protein 70 and auxilin messenger RNAs and regulates clathrin vesicle uncoating. J Biol Chem. 2012;287(18):14726–14733. doi: 10.1074/jbc.M111.326041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3(11):881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440(7083):556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. Isolation and characterization of the heat shock RNA 1. Methods Mol Biol. 2009;540:265–279. doi: 10.1007/978-1-59745-558-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, Fu YH, Liu XY, Li YX, Zhang YY, Lin SG, Yu XY. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584(16):3592–3600. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9(9):1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Torigoe T, Kutomi G, Hirata K, Sato N. New paradigm for intrinsic function of heat shock proteins as endogenous ligands in inflammation and innate immunity. Curr Mol Med. 2012;12(9):1198–1206. doi: 10.2174/156652412803306710. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39(16):6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, Haar L, Liu Y, Ren X, Jones WK. Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem. 2011;286(34):29828–29837. doi: 10.1074/jbc.M111.221796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter M, Ren X, Forde T, Wilhide ME, Chen J, Sartor MA, Medvedovic M, Jones WK. NF-kappaB driven cardioprotective gene programs; Hsp70.3 and cardioprotection after late ischemic preconditioning. J Mol Cell Cardiol. 2010;49(4):664–672. doi: 10.1016/j.yjmcc.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26(7):693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11(7):3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WJ, Feramisco JR. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J Biol Chem. 1984;259(7):4501–4513. [PubMed] [Google Scholar]
- Wilmink GJ, Roth CL, Ibey BL, Ketchum N, Bernhard J, Cerna CZ, Roach WP. Identification of microRNAs associated with hyperthermia-induced cellular stress response. Cell Stress Chaperones. 2010;15(6):1027–1038. doi: 10.1007/s12192-010-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106(14):5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcription. 2011;2(1):45–49. doi: 10.4161/trns.2.1.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Salloum FN, Kukreja RC. A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res. 2009;104(5):572–575. doi: 10.1161/CIRCRESAHA.108.193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia–reperfusion in mice. FEBS Lett. 2008;582(30):4137–4142. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liu F, Yin P, Zhu X, Cheng G, Wang N, Lu A, Luan W, Zhang N, Li J, Guo K, Yin Y, Wang H, Xu J. Integrating miRNA and mRNA expression profiles in response to heat stress-induced injury in rat small intestine. Funct Integr Genomics. 2011;11(2):203–213. doi: 10.1007/s10142-010-0198-8. [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ma H, Gong H, Zhou N, Liang Y, Niu Y, Zou Y. MicroRNA-378, a novel regulator of heat shock transcription factor-1, involves development of cardiac hypertrophy. Beijing: World Congress Of Cardiology; 2010. [Google Scholar]