Abstract
BACKGROUND
Few genetic risk factors have been uncovered that contribute specifically to the racial disparity in prostate cancer (CaP) observed in African Americans (AA). With the advent of Ancestry Informative Marker (AIM) single nucleotide polymorphism (SNP) panels and powerful genetic strategies such as Mapping by Admixture Linkage Disequilibrium (MALD) it is possible to discover genes that underlie ethnic variation in disease risk.
METHODS
1130 AA CaP cases enrolled in the North Carolina-Louisiana Prostate Cancer Project (PCaP) were genotyped using a 1,509 AIM SNP panel. MALD was performed using ADMIXMAP to test for linkage between CaP risk and ancestry estimates at each AIM SNP.
RESULTS
The largest increase of African ancestry was observed at marker rs12543473 (P=0.0011), located on chromosome 8q24.21, and the greatest excess of European ancestry was observed at marker rs10768140 (P=0.0004) at chromosome 11p13.
CONCLUSIONS
The study confirmed the 8q24 risk loci and identified a novel genomic region on 11p13 that is associated with CaP risk. These findings should be replicated in larger AA populations and combined with fine mapping data to further refine the novel 11p13 CaP risk loci.
Keywords: ancestry informative markers, prostate cancer, African American, mapping by admixture linkage disequilibrium, MALD, SNP
INTRODUCTION
Prostate cancer (CaP) is the most common cancer among men in developed countries [1]. Despite widespread screening and improved treatment, CaP remains a major public health problem and African-American men have among the highest worldwide incidence and mortality rates [2].
CaP is a multifactorial disease with both genetic and environmental components. Quantitative estimates from twin studies indicate that 42% of CaP cases may have a heritable component [3], which is stronger than for any other type of cancer in humans [4-7]. The results from both family studies and genome-wide association studies (GWAS) indicate that many genetic loci contribute to disease risk with varying levels of penetrance [8]. The racial differences in disease prevalence suggest that some of the CaP susceptibility variants may have different risk allele frequencies in different populations (e.g., African vs. European) [9]. Recently admixed populations, such as African Americans, may have an increased probability of inheriting a chromosomal segment that contains causal variants from the ethnic population with higher disease prevalence (African). A number of CaP genetic loci have emerged from genome-wide association studies (GWAS) [10], but fewer genetic risk factors have been uncovered that contribute specifically to the racial disparity observed in African Americans (AA) [11,12].
New approaches to disease mapping in admixed populations [13] that compliment GWAS (association analyses of cases and controls within the same ancestral group) have gained attention [13,14]. The recent availability of Ancestry Informative Marker (AIM) single nucleotide polymorphism (SNP) panels (SNPs whose allele frequency vary widely by ancestral group – e.g., African, European) and new analytic tools have made possible a powerful genetic strategy known as Mapping by Admixture Linkage Disequilibrium (MALD) . MALD facilitates the discovery of genes that underlie ethnic variation in disease risk and capitalizes on the long-range linkage disequilibrium (LD) that creates large ancestral haplotype blocks, which characterize genomes of recently admixed populations, such as AAs. The average size of ancestry blocks in AA populations is much longer than the haplotype blocks of un-admixed human populations. Thus MALD is more efficient than GWAS since MALD requires 200-500-fold fewer SNPs to canvas the genome, but retains the high statistical power of association studies. Additionally control subjects are not required, because MALD compares the proportion of ancestry at each locus to the average genome-wide ancestry using affected persons only (cases). Comparisons of admixture mapping and association analysis in family, patient and population-based studies have demonstrated that each test can provide unique information and that ancestry can be more informative than direct association analysis when the causal variant has large allele frequency differences between ancestral populations [15,16].
Only two studies have reported on admixture mapping of CaP among AA [11,12]. Freedman et al. [11] in 2006 capitalized on developing AIMs panels and evaluated 1,597 CaO cases from 7 different studies, while Bock et al. [12] in 2009 evaluated 482 cases from two independent studies. While both studies confirmed the 8q24 locus, Bock et al. [12] reported several other ancestry-specific susceptibility loci. Herein we use an established AIM panel to conduct admixture analysis on a large well-characterized AA CaP cohort; AAs enrolled in the population-based North Carolina-Louisiana Prostate Cancer Study (PCaP).
SUBJECTS AND METHODS
Study population
PCaP is a population-based, case-only study designed to investigate social, individual, and tumor-level causes of racial differences in prostate cancer that has been described [17]. PCaP recruited 1130 black/ AA CaP cases between 40 and79 years of age with newly diagnosed, histologically-confirmed, adenocarcinoma of the prostate from North Carolina (NC) and Louisiana (LA). Medical records were abstracted for information related to a CaP diagnosis which included total serum prostate-specific antigen (PSA) (defined as the PSA value closest and within 1 year prior to CaP diagnosis date), tumor stage at diagnosis (stage number was derived from stage as reported in the medical record such that 1=T1a, T1c T1(not otherwise specified (NOS)), T1C; 2=T2(NOS), T2a, T2b, T2 a or b, T2c; 3=T3/4 (NOS), T3b, T3a, 3A and 4=T4) and grade (Gleason score= sum of primary and secondary Gleason grade) for aggressiveness classification. Men were classified based on clinical Gleason score, clinical stage, and PSA at diagnosis as: (1) highly aggressive (Gleason score ≥ 8, or PSA > 20 ng/ml, or Gleason score = 7, and stage cT3–cT4); (2) non-aggressive (Gleason score < 7 and stage cT1–cT2, and PSA < 10 ng/ml); or (3) intermediate aggressive (all other cases). DNA was available on 997 AA who are the subjects of this study. Age at diagnosis was derived from self-reported date of birth and date of diagnostic biopsy as indicated in the medical record and rounded to the nearest full year. Informed consent was obtained from all research subjects prior to blood and questionnaire collection and study protocols were approved by participating institution Internal Review Boards.
Genotyping
DNA was extracted from blood samples (n=811) or buccal cells (n=44) by the University of North Carolina at Chapel Hill (UNC-Chapel Hill) Biospecimen Processing Facility, or from lymphocytes immortalized by the UNC-Chapel Hill Tissue Culture Facility (n=142). Genotyping was performed by the NIH Center for Inherited Disease Research (CIDR) using the Illumina AA Admixture Panel. The SNP panel contains 1,509 ancestry-informative SNPs with averaged allele frequency difference of 0.74 between HapMap CEU and HapMap YRI population. The averaged chromosome distance between SNPs is 1,950kb. Twenty sample duplicates and 11 HapMap trios with representation of CEU and YRI ancestral groups were analyzed.
Statistical Analysis
An affected-only MALD analysis was performed in PCaP AA CaP case genotype data using ADMIXMAP software [18] to compare observed versus expected ancestry allele copies across the genome conditional on a priori ancestral allele frequencies. ADMIXMAP fits a Bayesian probability model with computationally intensive Markov chain Monte Carlo parameter estimation algorithm.
The affected-only test requires strong prior information on allele frequencies for any specified ancestry [19]. Priors, were calculated from allele frequency based on HapMap west African Yoruban (YRI) and CEPH Europeans from Utah (CEU) genotypes (http://hapmap.ncbi.nlm.nih.gov/), and ADMIXMAP software was used on genotype data obtained using the same panel of SNPs from 608 AAs who were part of a separate study to estimate ancestry-specific allele frequencies for each SNP with the HapMap allele frequency prior. The model-fit evaluation statistics - the deviance information criterion (DIC) for affected-only tests on PCaP data were 2.16×106 and 2.15×106, respectively for HapMap prior and AA data estimated allele frequencies prior. These results indicated AA estimated allele frequency priors produced a better model fit, and thus the estimated allele frequencies were used as priors in the final PCaP data analysis. For each research subject, the affected-only method used the score test to compare ancestry estimates at each locus to the corresponding ancestry estimates of the whole genome. The score for each locus was calculated by averaging the posterior probability of linkage with ancestry across samples. The test statistic (Z-score) was the score divided by the square root of the observed information, and thus had a standard normal distribution under the null hypothesis. A positive Z-score indicated excess African ancestry allele copies at a locus while a negative Z-score indicated excess European ancestry allele copies at a locus. Similar to Bock et al[12] and Schwartz et al[20], we reported peak regions with absolute value of Z-score greater than 3.0 (P-value ~ 0.0025).
ADMIXMAP software also can evaluate residual LD among SNPs that are not accounted for by the admixture model. SNPs with genotype data highly correlated with adjacent SNPs (score test for residual LD, P<1×10−5) were removed from final analysis. Furthermore, ADMIXMAP was used to adjust for the excess of African ancestry on the X chromosome due to historical sex asymmetry with a prior of exp(3) for the population-level odds ratio female/male in founders [21]. Once this adjustment was made, the Z scores for the X chromosome were interpreted in concert with the MALD results for autosomes. The recently built AA genetic map [22] for SNP map positions was used in the analysis to increase the precision of admixture estimates.
RESULTS
Allele calling was conducted using Illumina’s Genotyping Module version 1.0.10 in GenomeStudio 1.0.2.20706. The genotype intensity cluster plots were inspected visually for each SNP. Genotypes with an Illumina GenCall (GC) score below 0.25 were assigned as missing. Seven PCaP research subjects were excluded because of failed genotyping due to poor sample quality. In total, 990 research subjects with high quality genotype data were available for analysis, which included 447 AA from NC and 543 AA from LA. A total of 58 SNPs (3.8%) were excluded due to poor clustering pattern or parent-parent-child (P-P-C) heritability errors identified using HapMap trios. In addition, 13 SNPs were excluded by the Hardy-Weinberg equilibrium test or residual LD test P-value <1×10−5. Genotypes of 1438 SNPs for 990 AA CaP cases remained after all exclusions. The overall research subject genotyping call rate was 99.96%. The reproducibility rate was 100% based on duplicates and the overall P-P-C heritability was 99.97% based on HapMap trios. Table 1 describes the characteristics of these eligible cases by study site. The mean estimated individual proportion of African ancestry for PCaP research subjects was slightly higher in North Carolina (82%) than Louisiana (79.3%). The distributions of other clinical characteristics were similar between states.
Table 1.
Clinical characteristics of PCaP AA research subjects in data analysis
| NC | LA | |
|---|---|---|
| No. of research subjects | 447 | 543 |
| Age at diagnosis | ||
| Median | 60 | 63 |
| Range | 41-79 | 43-79 |
| Mean African ancestry (%) | 82.0 | 79.3 |
| Median PSA ng/ml | 6 | 6 |
| PSA levels N (%) | ||
| < 4 ng/ml | 40 (8.9) | 65 (12) |
| 4-10 ng/ml | 276 (61.7) | 305 (54.2) |
| 10-20 ng/ml | 74 (16.7) | 83 15.3) |
| > 20 ng/ml | 52 (11.6) | 56 (10.3) |
| Unknown | 5(1.1) | 34 (6.2) |
| Gleason Score, N (%) | ||
| ≤6 | 237 (53.0) | 309 (56.9) |
| 7 | 155 (34.7) | 157 (28.9) |
| 8-10 | 55 (12.3) | 76 (14.0) |
| Unknown | 0 (0) | 1 (0.2) |
| Pathologic stage, N (%) | ||
| T1 | 274 (61.3) | 266 (49.0) |
| T2 | 162 (36.3) | 246 (45.3) |
| T3-T4 | 9 (2.0) | 8 (1.5) |
| Unknown | 2 (0.4) | 23 (4.2) |
| Aggressiveness, N (%) | ||
| Low | 203 (45.4) | 244 (45.0) |
| Intermediate | 157 (35.1) | 159 (29.3) |
| High | 87 (19.5) | 110 (20.2) |
| Unknown | 0 (0) | 30 (5.5) |
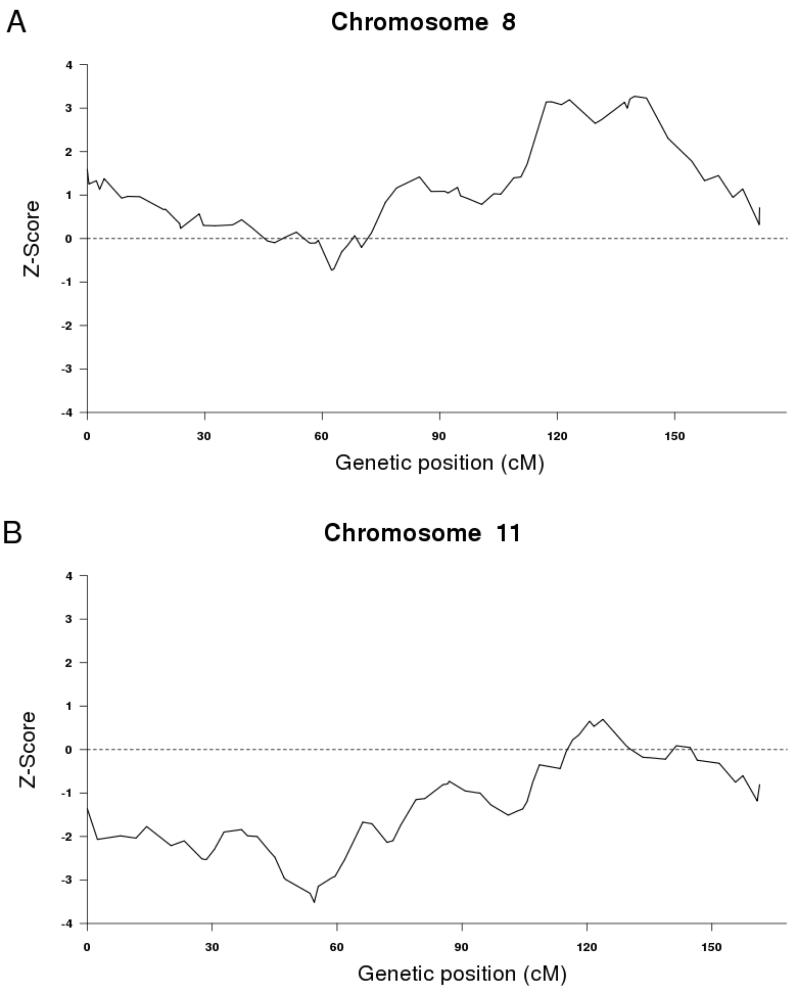
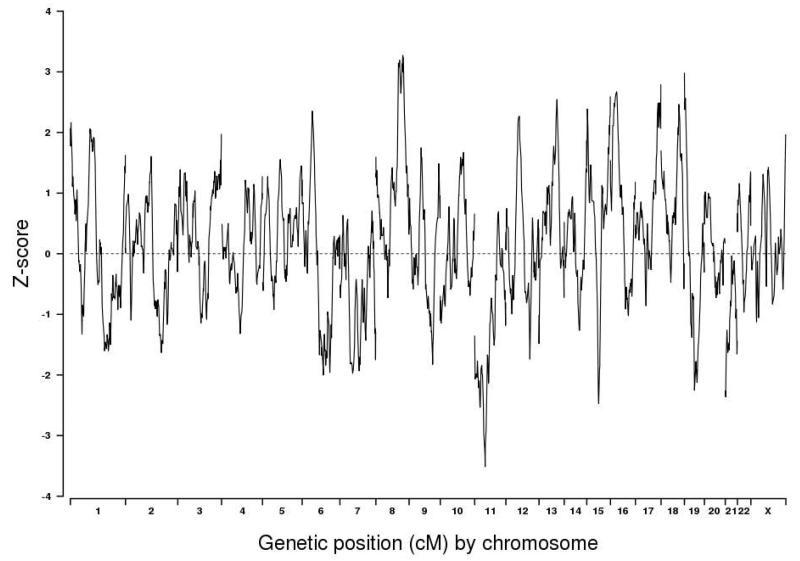
The distribution of Z-scores resulting from ADMIXMAP is shown in Figure 1. Details about the two SNP markers at local peaks with |Z|>3.0 (P-value ~ 0.0025) are summarized in Table 2. The most significant increase in African ancestry among AA PCaP cases are observed on chromosome 8q24.21, while the most significant increase in European ancestry are observed on chromosome 11p13 (Table 2). A more detailed examination of chromosomal regions with |Z|>3.0 is shown in Figures 2A and 2B. Z-scores for all AIM SNPs in the MALD analysis are provided in Supplemental Table 1.
Figure 1.
Genome-wide admixture mapping results in 990 AA men with CaP.
Table 2.
Summary of SNPs at regional ancestry association peaks with |Z| >3.0
| Locus | Location | Position | Entire sample |
Age ≤ 60 |
Age > 60 |
|||
|---|---|---|---|---|---|---|---|---|
| Z | P | Z | P | Z | P | |||
| rs12543473 | 8q24.21 | 127850622 | 3.267 | 0.0011 | 2.437 | 0.0148 | 2.317 | 0.0205 |
| rs10768140 | 11p13 | 35467483 | −3.515 | 0.0004 | −2.063 | 0.0391 | −2.857 | 0.0043 |
Figure 2.
Detailed local chromosomal regions with |Z|>3.0 in PCaP MALD analysis.
Whether reported GWAS SNPs [23-33] coincided with admixture results was investigated by mapping the chromosomal position of these SNPs onto the admixture mapping results. Only 5 of the 37 GWAS regions coincided with MALD peaks of |Z|>2 (Supplemental Table 2, Supplemental Figures 1 and 2).
DISCUSSION
Results from PCaP identified a significant increase in African ancestry on chromosome 8q24.21, which confirmed two admixture mapping analyses in AA men with prostate cancer [11,12]. In the admixture mapping analysis reported by Freedman et al. in 2006 of 1,597 AA with CaP from 7 studies, a 3.8 Mb interval on the 8q24 chromosome alone emerged as a region with the greatest increased African ancestry related to CaP using ANCESTRYMAP (LOD score = 4.1 in all cases) [11,34]. Freedman et al. in 2006 [11] genotyped 15 additional SNPs among 294 additional cases to better localize the admixture peak. rs12543473 (at physical location 127,850,622 in NCBI Genome Build 35) was among these additional SNPs, which was the SNP on our AIM panel that yielded the most significant African ancestry signal. In the additional analysis by Freedman et al., this SNP displayed the second highest case-only LOD score of 3.97, second only to its neighboring SNP, rs780321 at map position 127,152,877 with LOD score 4.01. A smaller admixture mapping study by Bock et al. [12] in 2009 of 482 AA cases and 261 controls using ADMIXMAP and the Illumina AIMs panel, similar to our study, reported finding the strongest 8q24 association (though non-significant) with rs4367565 at map position131,606,887-131,607,387 (NCBI Genome Build 35) on chromosome 8 (in both a case-control and case-only analysis). When comparing our results to the more dense SNP evaluation of the 8q24 genomic region (124-132 megabases (Mb) by Freedman et al [11]. our SNP signal is located directly within the same 3.8 Mb peak at 127 Mb. In contrast the signal observed by Bock et al., determined using a much smaller sample group, is in the tail of the peak delineated by Freedmen et al at 131 Mb with low probability density [11,12]. Our results present a somewhat bimodal distribution at 8q24, which likely reflects lack of refinement (6-7 cM) of the MALD map rather than a significant observation. In addition to excess African ancestry on 8q24, Bock et al. reported a region on chromosome 5q35 (rs7729084 and rs12474977, case-only analysis). As indicated by Bock et al., this region on chromosome 5 also demonstrated higher African ancestry in our analysis (rs7729084, Z score=1.29), though the finding did not reach statistical significance.
Several regions of the genome with European ancestry have been found to increase CaP risk in admixture mapping of AAs. Bock et al. reported an association with 7q31 (rs2141360), and although a higher proportion of European ancestry was found at this locus, the association did not reach statistical significance (rs2141360 Z-score = −1.23). The most significant increase in European ancestry was found at a novel locus on chromosome 11p13 with Z score well below −3 (rs10768140); this region has not been reported in admixture mapping studies of AA CaP cases to-date [11,12]. This SNP is located within the intron of peptidase domain-containing protein associated with muscle regeneration 1 (PAMR1) that may play a role in regeneration of skeletal muscle and is down-regulated in muscle cell lines from patients with Duchenne muscular dystrophy (http://www.ncbi.nlm.nih.gov/). Given the function of this gene, the AIM marker probably represents a marker for an associated CaP locus within the ancestral region rather than a functional variant. No small nuclear or microRNAs are reported in a region (1 Mb) surrounding this SNP (UCSC Genome Browser Build 37 [35-41]. Several other confirmed genes lie within a 1 Mb region surrounding rs10768140 (35 MB – 36 MB on chromosome 11). CD44, a cell-surface glycoprotein involved in cell-cell interaction including cell migration is located telomeric to PAMR1 (http://genome.ucsc.edu/). CD44 interacts with ligands, such as matrix metalloproteinases, and participates in tumor metastasis (UCSC Genome Browser) in cancers including prostate [42]. Additionally, CD44 is a stem cell marker for breast and CaP [43,44]. Similar to the fine mapping study by Freedman et al. that refined an 8 Mb region to a peak of 3.8 Mb [11], further fine mapping in this region is needed to identify the specific signal location. While genes within a 1 Mb region surrounding our AIM signal were reported on, genes in more distant locations could be relevant, especially because the average ancestry block size observed in African Americans is 17-20 centimorgans (cM) [34,45-49] or approximately 17-20 Mb on the physical genetic map. Bock et al. did not report excess European ancestry on chromosome 11, however it is unclear if chromosome 11 was evaluated since the genome-wide admixture plot did not delineate several autosomes including chromosomes 11, 15, 18 and 20 and supporting documentation was not provided detailing the Z scores for all genotyped SNPs. Even though the same Illumina AIM panel was used by Bock et al., there may have been a lack of coverage across specific genomic regions including chromosome 11 after genotypes were removed that had poor call rates (N=188).
Random associations may occur within MALD and may be dominated by the signal from a single SNP, producing a narrow peak that is often near the tail of a chromosome. The 11p13 peak was relatively wide and did not include SNPs from the tail of the chromosome; this peak retained a |Z| score greater than 3 even after removal of the SNP with the strongest association (|Z|=3.26 2 for adjacent SNP rs2553779 and |Z|=3.255 for adjacent SNP rs6484807).
Both 8q24, demonstrating excess African ancestry, and 11p13, demonstrating excess European ancestry, may harbor CaP susceptibility alleles that contribute to the increased CaP risk of AAs. The effect size of the 8q24 loci is large and should be detected in both GWAS and ancestry analysis and harbor alleles that contribute to both ancestry-specific and general CaP case association. The lack of identification of the 11p13 region among European GWAS analyses may be explained on several levels. First, the effect size may be less than that of 8q24 and the allele frequency differences much larger, which makes this locus easier to identify by ancestry mapping and more difficult by GWAS because of the penalties incurred in GWAS for multiple tests [14]. In addition, GWAS is not robust to allelic heterogeneity, a likely feature of CaP, while admixture mapping is. If different risk variants at the same locus within the 11p13 region are responsible for CaP risk, this region may be more readily detected by MALD.
A small but well-established AIM panel was used to infer African and European ancestral segments. New analytic methods that leverage both MALD and GWAS in either joint analyses or 2-stage design are emerging [14,50-53]. These newer designs will capitalize on the efficiency of MALD by reducing statistical penalties in the discovery phase, while providing refinement of the genetic susceptibility loci by direct fine mapping conducted in the second phase.
In conclusion, this study confirms the 8q24 risk loci and identifies a novel genomic region on 11p13 that is associated with CaP risk in AAs. These MALD findings should be replicated in larger AA populations and combined with fine mapping data to further refine this novel CaP risk loci.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff, advisory committees and participants for their important contributions. The Prostate Cancer Project (PCaP) is carried out as a collaborative study supported by the Department of Defense contract DAMD 17-03-2-0052. We would like to acknowledge the UNC BioSpecimen Facility and the LSUHSC Pathology Lab for our DNA extractions, blood processing, storage, and sample disbursement (https://genome.unc.edu/bsp). This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES049033)
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Ries L, Melbert D, Krapcho M, Mariotto A, Miller B, Feuer E, Clegg L, Horner M, Howlader N, Eisner M, Reichman M, Edwards B. SEER Cancer Statistics Review, 1975-2004. National Cancer Institute; Bethesda, MD: 2006. [Google Scholar]
- 3.Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci U S A. 1992;89(8):3367–3371. doi: 10.1073/pnas.89.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England journal of medicine. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Page WF, Braun MM, Partin AW, Caporaso N, Walsh P. Heredity and prostate cancer: a study of World War II veteran twins. The Prostate. 1997;33(4):240–245. doi: 10.1002/(sici)1097-0045(19971201)33:4<240::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 6.Ahlbom A, Lichtenstein P, Malmstrom H, Feychting M, Hemminki K, Pedersen NL. Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst. 1997;89(4):287–293. doi: 10.1093/jnci/89.4.287. [DOI] [PubMed] [Google Scholar]
- 7.Gronberg H, Damber L, Damber JE. Studies of genetic factors in prostate cancer in a twin population. The Journal of urology. 1994;152:1484–1487. doi: 10.1016/s0022-5347(17)32452-7. 5 Pt 1. discussion 1487-1489. [DOI] [PubMed] [Google Scholar]
- 8.Ostrander EA, Johannesson B. Prostate cancer susceptibility loci: finding the genes. Advances in experimental medicine and biology. 2008;617:179–190. doi: 10.1007/978-0-387-69080-3_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halder I, Shriver MD. Measuring and using admixture to study the genetics of complex diseases. Human genomics. 2003;1(1):52–62. doi: 10.1186/1479-7364-1-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Wang B, Han C. Meta-analysis of genome-wide and replication association studies on prostate cancer. The Prostate. 2011;71(2):209–224. doi: 10.1002/pros.21235. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bock CH, Schwartz AG, Ruterbusch JJ, Levin AM, Neslund-Dudas C, Land SJ, Wenzlaff AS, Reich D, McKeigue P, Chen W, Heath EI, Powell IJ, Kittles RA, Rybicki BA. Results from a prostate cancer admixture mapping study in African-American men. Human genetics. 2009;126(5):637–642. doi: 10.1007/s00439-009-0712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nature reviews Genetics. 2011;12(8):523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin H, Zhu X. Power comparison of admixture mapping and direct association analysis in genome-wide association studies. Genetic epidemiology. 2012;36(3):235–243. doi: 10.1002/gepi.21616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Siegmund DO, Johnson NA, Romieu I, London SJ. Joint testing of genotype and ancestry association in admixed families. Genetic epidemiology. 2010;34(8):783–791. doi: 10.1002/gepi.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, Tayo B, Adeyemo A, Sun YV, Li Y, Morrison A, Newton-Cheh C, Liu K, Ganesh SK, Kutlar A, Vasan RS, Dreisbach A, Wyatt S, Polak J, Palmas W, Musani S, Taylor H, Fabsitz R, Townsend RR, Dries D, Glessner J, Chiang CW, Mosley T, Kardia S, Curb D, Hirschhorn JN, Rotimi C, Reiner A, Eaton C, Rotter JI, Cooper RS, Redline S, Chakravarti A, Levy D. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Human molecular genetics. 2011;20(11):2285–2295. doi: 10.1093/hmg/ddr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder JC, Bensen JT, Su LJ, Mishel M, Ivanova A, Smith GJ, Godley PA, Fontham ET, Mohler JL. The North Carolina-Louisiana Prostate Cancer Project (PCaP): methods and design of a multidisciplinary population-based cohort study of racial differences in prostate cancer outcomes. The Prostate. 2006;66(11):1162–1176. doi: 10.1002/pros.20449. [DOI] [PubMed] [Google Scholar]
- 18.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64(Pt 2):171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 19.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74(5):965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz AG, Wenzlaff AS, Bock CH, Ruterbusch JJ, Chen W, Cote ML, Artis AS, Van Dyke AL, Land SJ, Harris CC, Pine SR, Spitz MR, Amos CI, Levin AM, McKeigue PM. Admixture mapping of lung cancer in 1812 African-Americans. Carcinogenesis. 2011;32(3):312–317. doi: 10.1093/carcin/bgq252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybicki BA, Levin AM, McKeigue P, Datta I, Gray-McGuire C, Colombo M, Reich D, Burke RR, Iannuzzi MC. A genome-wide admixture scan for ancestry-linked genes predisposing to sarcoidosis in African-Americans. Genes and immunity. 2011;12(2):67–77. doi: 10.1038/gene.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, Aldrich MC, Ambrosone CB, Amos C, Bandera EV, Berndt SI, Bernstein L, Blot WJ, Bock CH, Boerwinkle E, Cai Q, Caporaso N, Casey G, Cupples LA, Deming SL, Diver WR, Divers J, Fornage M, Gillanders EM, Glessner J, Harris CC, Hu JJ, Ingles SA, Isaacs W, John EM, Kao WHL, Keating B, Kittles RA, Kolonel LN, Larkin E, Le Marchand L, McNeill LH, Millikan RC, Murphy A, Musani S, Neslund-Dudas C, Nyante S, Papanicolaou GJ, Press MF, Psaty BM, Reiner AP, Rich SS, Rodriguez-Gil JL, Rotter JI, Rybicki BA, Schwartz AG, Signorello LB, Spitz M, Strom SS, Thun MJ, Tucker MA, Wang Z, Wiencke JK, Witte JS, Wrensch M, Wu X, Yamamura Y, Zanetti KA, Zheng W, Ziegler RG, Zhu X, Redline S, Hirschhorn JN, Henderson BE, Taylor HA, Jr., Price AL, Hakonarson H, Chanock SJ, Haiman CA, Wilson JG, Reich D, Myers SR. The landscape of recombination in African Americans. Nature. 2011;476(7359):170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nature genetics. 2006;38(6):652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 24.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nature genetics. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 25.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nature genetics. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 26.Witte JS. Multiple prostate cancer risk variants on 8q24. Nature genetics. 2007;39(5):579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 27.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics. 2007;39(5):638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Balter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Gronberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99(24):1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 29.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nature genetics. 2008;40(3):281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nature genetics. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 31.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O'Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Collaborators UKGPCS. British Association of Urological Surgeons' Section of O. Collaborators UKPS. Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nature genetics. 2008;40(3):316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 32.Xu Z, Bensen JT, Smith GJ, Mohler JL, Taylor JA. GWAS SNP Replication among African American and European American men in the North Carolina-Louisiana prostate cancer project (PCaP) The Prostate. 2011;71(8):881–891. doi: 10.1002/pros.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Wang B, Han C. Meta-analysis of genome-wide and replication association studies on prostate cancer. The Prostate. 2011;71(2):209–224. doi: 10.1002/pros.21235. [DOI] [PubMed] [Google Scholar]
- 34.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. American journal of human genetics. 2004;74(5):979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic acids research. 2004;32(Database issue):D23–26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic acids research. 2009;37(Database issue):D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, Bateman A. Rfam: Wikipedia, clans and the "decimal" release. Nucleic acids research. 2011;39(Database issue):D141–145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22(9):1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt C, Apweiler R, Jesus Martin M, O'Onovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrera Casanova E, Bely B, Bingley M, Bower L, Bursteinas B, Mun Chan W, Chavali G, Da Silva A, Dimmer E, Eberhardt R, Fazzini F, Fedotov A, Garavelli J, Castro LG, Gardner M, Hieta R, Huntley R, Jacobsen J, Legge D, Liu W, Luo J, Orchard S, Patient S, Pichler K, Poggioli D, Pontikos N, Pundir S, Rosanoff S, Sawford T, Sehra H, Turner E, Wardell T, Watkins X, Corbett M, Donnelly M, van Rensburg P, Goujon M, McWilliam H, Lopez R, Xenarios I, Bougueleret L, Bridge A, Poux S, Redaschi N, Argoud-Puy G, Auchincloss A, Axelsen K, Baratin D, Blatter M-C, Boeckmann B, Bolleman J, Bollondi L, Boutet E, Braconi Quintaje S, Breuza L, deCastro E, Cerutti L, Coudert E, Cuche B, Cusin I, Doche M, Dornevil D, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gehant S, Ferro S, Gasteiger E, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, James J, Jimenez S, Jungo F, Kappler T, Keller G, Lara V, Lemercier P, Lieberherr D, Martin X, Masson P, Moinat M, Morgat A, Paesano S, Pedruzzi I, Pilbout S, Pozzato M, Pruess M, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stanley E, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey A-L, Wu CH, Arighi CN, Arminski L, Barker WC, Chen C, Chen Y, Dubey P, Huang H, Kukreja A, Laiho K, Mazumder R, McGarvey P, Natale DA, Natarajan TG, Roberts NV, Suzek BE, Vinayaka CR, Wang Q, Wang Y, Yeh L-S, Zhang J. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic acids research. 2012;40(Database issue):D71–75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25(12):1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 43.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 45.Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet. 2004;75(5):771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. American journal of human genetics. 1998;63(6):1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale J-L, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O'Brien SJ, Reich D. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74(5):1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H, Coram M, Wang P, Zhu X, Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am J Hum Genet. 2006;79(1):1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X, Zhang S, Tang H, Cooper R. A classical likelihood based approach for admixture mapping using EM algorithm. Human genetics. 2006;120(3):431–445. doi: 10.1007/s00439-006-0224-z. [DOI] [PubMed] [Google Scholar]
- 50.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, Farlow DN, Folsom AR, Fornage M, Forrester T, Fox E, Haiman CA, Hartiala J, Harris TB, Hazen SL, Heckbert SR, Henderson BE, Hirschhorn JN, Keating BJ, Kritchevsky SB, Larkin E, Li M, Rudock ME, McKenzie CA, Meigs JB, Meng YA, Mosley TH, Newman AB, Newton-Cheh CH, Paltoo DN, Papanicolaou GJ, Patterson N, Post WS, Psaty BM, Qasim AN, Qu L, Rader DJ, Redline S, Reilly MP, Reiner AP, Rich SS, Rotter JI, Liu Y, Shrader P, Siscovick DS, Tang WHW, Taylor HA, Tracy RP, Vasan RS, Waters KM, Wilks R, Wilson JG, Fabsitz RR, Gabriel SB, Kathiresan S, Boerwinkle E. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS genetics. 2011;7(2):e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin H, Morris N, Kang SJ, Li M, Tayo B, Lyon H, Hirschhorn J, Cooper RS, Zhu X. Interrogating local population structure for fine mapping in genome-wide association studies. Bioinformatics (Oxford, England) 2010;26(23):2961–2968. doi: 10.1093/bioinformatics/btq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Zhu X, Qin H, Cooper RS, Ewens WJ, Li C, Li M. Adjustment for local ancestry in genetic association analysis of admixed populations. Bioinformatics (Oxford, England) 2011;27(5):670–677. doi: 10.1093/bioinformatics/btq709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasaniuc B, Zaitlen N, Lettre G, Chen GK, Tandon A, Kao WHL, Ruczinski I, Fornage M, Siscovick DS, Zhu X, Larkin E, Lange LA, Cupples LA, Yang Q, Akylbekova EL, Musani SK, Divers J, Mychaleckyj J, Li M, Papanicolaou GJ, Millikan RC, Ambrosone CB, John EM, Bernstein L, Zheng W, Hu JJ, Ziegler RG, Nyante SJ, Bandera EV, Ingles SA, Press MF, Chanock SJ, Deming SL, Rodriguez-Gil, Palmer CD, Buxbaum S, Ekunwe L, Hirschhorn JN, Henderson BE, Myers S, Haiman CA, Reich D, Patterson N, Wilson JG, Price AL. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS genetics. 2011;7(4):e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.