Abstract
Development of predictive in vitro assays for early toxicity evaluation is extremely important for improving the drug development process and reducing drug attrition rates during clinical development. High-content imaging-based in vitro toxicity assays are emerging as efficient tools for safety and efficacy testing to improve drug development efficiency. In this report we have used an induced pluripotent stem cell (iPSC)–derived hepatocyte cell model having a primary tissue-like phenotype, unlimited availability, and the potential to compare cells from different individuals. We examined a number of assays and phenotypic markers and developed automated screening methods for assessing multiparameter readouts of general and mechanism-specific hepatotoxicity. Endpoints assessed were cell viability, nuclear shape, average and integrated cell area, mitochondrial membrane potential, phospholipid accumulation, cytoskeleton integrity, and apoptosis. We assayed compounds with known mechanisms of toxicity and also evaluated a diverse hepatotoxicity library of 240 compounds. We conclude that high-content automated screening assays using iPSC-derived hepatocytes are feasible, provide information about mechanisms of toxicity, and can facilitate the safety assessment of drugs and chemicals.
Introduction
The pharmaceutical industry continues to face challenges associated with attrition of drug candidates late in the development pipeline, or drugs being withdrawn after approval.1 Drug safety is one of the primary concerns of drug development and liver toxicity is among the top organs for adverse drug reactions. In fact, drug-induced liver injury has been associated with over a third of acute liver failures in the United States and more than 1,000 drugs are considered potentially toxic to the liver.2,3 Drug-induced liver injury is one of the major causes of drug candidate failure in preclinical and clinical testing4 and is also the most frequently cited reason for withdrawal of approved drugs.5,6
While in vivo animal studies remain the standard for toxicity testing, they are time consuming and costly, and more importantly, are rather poor predictors of human toxicity.7,8 There is also growing momentum to move away from animal models for toxicity testing and toward cell-based in vitro assays.9,10 Predictive in vitro cell-based assays should reduce or obviate the dependence on animal models, streamline the assessment of candidate molecules and their metabolites, and facilitate evaluation of the mechanisms of toxicity, kinetics, and dose-responses. In addition, cell-based toxicity models will reduce per compound assay cost and thereby enable deployment earlier in the development process and allow focus on larger number of compounds.
A number of in vitro models have been established for hepatotoxicity testing.11,12 For example, precision-cut liver slices13,14 contain all cell types of the liver in their natural architecture and have xenobiotic metabolism capacity. This model, however, is arguably not well suited for high-throughput studies. Immortalized cell lines, such as HepG2, and more recently HepaRG cell line,15 are also widely employed. Cultures of primary (e.g., freshly isolated or cryopreserved) human, rodent, or canine hepatocytes have also been widely used for in vitro testing.16 However, high inter-individual variability, limited availability, high cost, and changes in cell morphology and liver-specific functions during long-term culture are significant limitations.
Human induced pluripotent stem cell (iPSC)–derived hepatocytes show great promise with respect to having a primary tissue-like phenotype, consistent and unlimited availability, and the potential to establish genotype-specific cells from different individuals.17 iPSC-derived tissue specific cells provide relevant human biology in vitro and are increasingly being studied for their potential to accurately predict drug-induced toxicity.18–21 As a result, iPSC-derived cell models are being adopted by the pharmaceutical industry for preclinical toxicity studies.22,23 To realize the full potential of iPSC-derived cell models, it is necessary to develop predictive in vitro assays that can be performed in a high-throughput manner. To that end, we have developed several assays for measuring general and mechanism-specific hepatotoxicity that are well-suited for automated analysis.
High-content imaging-based in vitro toxicity assays show promise for safety and efficacy testing as they can be performed using high-throughput systems for simultaneous screening of many compounds.24 High-content imaging has been used with primary hepatocytes15 and immortalized cell lines.16,25,26 In these studies, hepatotoxicity was evaluated using morphological and biochemical readouts, including cell count, nuclear shape, mitochondria potential (MP), Ca2+ content, and cell permeability.
Given the promise of both iPSC-derived hepatocytes and high-content screening, we developed imaging and analysis methods that provide tools for characterization of multiple toxicity phenotypes using live cells. Specifically, we characterized a number of toxicity assays and phenotypic read-outs, including characterization of cell shape, cell adhesion and spreading, nuclear condensation, accumulation of lipids, cytoskeleton integrity, in addition to short-term and long-term changes of MP. To improve assay workflow, we have optimized certain protocols that can be used as one step staining, reducing assay time and minimizing cell disturbance. In addition, by taking advantage of high-content image acquisition systems with large field of view cameras and improved image analysis software, we demonstrate that the analysis results can be reported in real-time. Finally, we have tested a commercially available library of compounds that have been shown to be hepatotoxic. The results illustrate that this method has significant promise for compound screening and early safety evaluation in the drug development process.
Materials and Methods
Cell Model
The cell model used for all assays were iCell® hepatocytes (Cellular Dynamics International [CDI], Madison WI), which are human iPSC-derived hepatocytes. Cells were received fresh and processed according to the protocol provided by CDI. Briefly, live cells were disaggregated by trypsinization and purified over a density gradient and then plated onto collagen-coated plates of the appropriate format using the plating and maintenance media outlined in the protocol from CDI. Characterization of the cells is shown in Figure 1 and included morphological and histochemical staining analyses and albumin production as measured by ELISA using Albumin ELISA Quantitaion Set, #E80-129 (Bethyl Laboratories, Montgomery, TX) read at 48 h in culture. Morphology characteristic of hepatocytes was displayed upon plating and images captured at 96 h postplating were observed to contain bi-nucleation and bile canalicular formation. PAS staining (#P5463; Sigma-Aldrich, St. Louis MO) was positive and indicative of glycogen storage activity. Oil red (#O0625; Sigma-Aldrich) and BODIPY staining showed evidence of lipid uptake. Albumin staining was done using anti-albumin antibodies (Cedar Lane) and secondary antibody donkey anti-mouse-AlexaFluor488 (Invitrogen, Carlsbad, CA). All images shown were taken with a 10× objective.
Fig. 1.
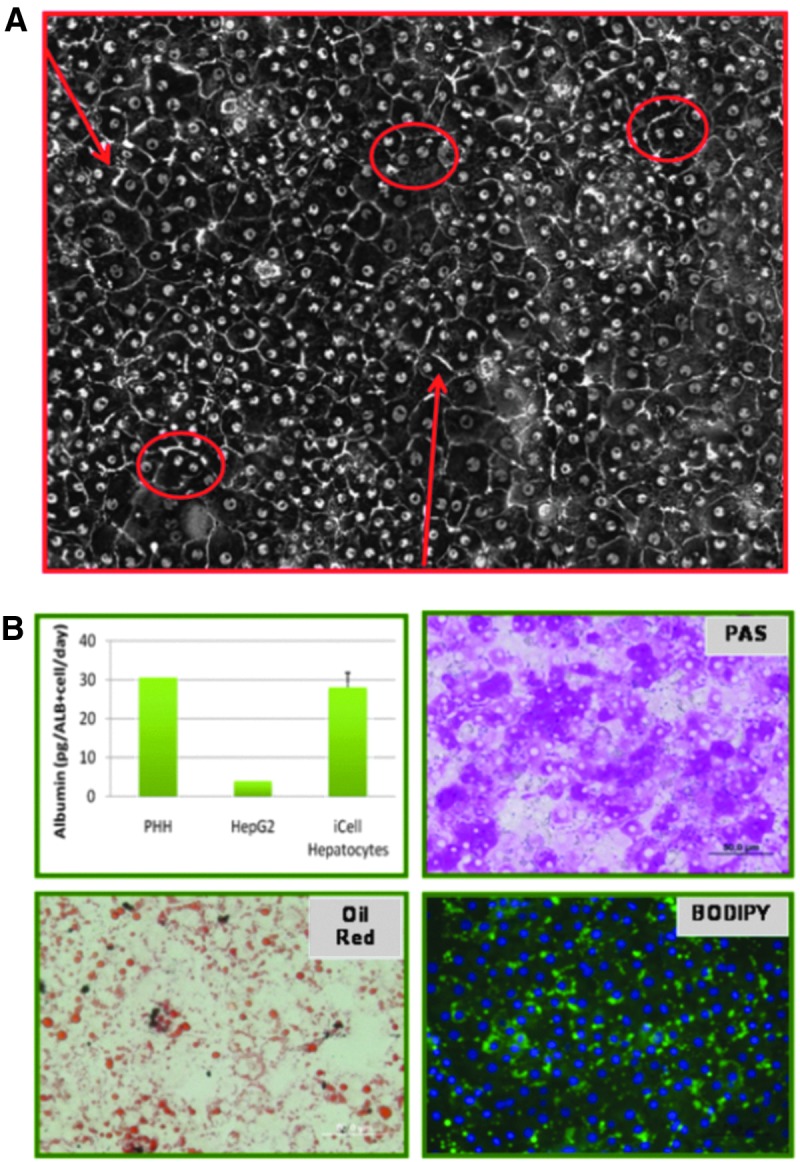
Morphological and functional characterization of iCell hepatocytes. (A) Transmitted light image of iCell hepatocytes after 4 days in culture. Binucleation is indicated by circles and bile canaliculi by arrows. (B) Indication of levels of albumin production (pg), glycogen storage (periodic acid Schiff PAS staining), and lipid accumulation (Oil Red and BODYPY staining).
Cell Culture and Compound Treatment
Cells were plated at a density of 60,000 cells/well (96-well plate) or 15,000 cells/well (384-well plate) on black clear bottom collagen-coated plates (BD Biosciences, San Jose, CA) and incubated at 37°C and 5% CO2 for 2 days in media containing glucose (media D) prior to compound treatment. We have used cell plating densities recommended by CDI protocol. In addition, we have found that using lower plating density (12,000 cells/well, 384-well plate) resulted in sub-confluent cell monolayer, while higher densities (18,000 cells/well) resulted in over-confluent cell culture. Cells were observed to exhibit a uniform confluent monolayer with cobblestone-like morphology (Fig. 1A). The media was then switched to a glucose-free, galactose-containing solution for 24 h (all media formulations provided by CDI). Dilutions of individual compounds were prepared from 100 mM dimethyl sulfoxide (DMSO) stock solutions. Final DMSO concentration in the media was 0.1%. Compounds were tested in quadruplicate in a dilution series by half-log concentration increments over a range of 10 nM–1,000 μM. Compounds from the hepatotoxicity library were prepared from 10 mM DMSO stocks and tested at five concentrations ranging from 1 to 100 μM. For the multiparameter toxicity assay, cells were treated with compounds for a 72 h period at 37°C and 5% CO2. For the measurement of MP by the addition of JC-10 dye, the cells were treated for 60 min. For the autophagy and phospholipidosis assays, the cells were incubated at 37°C and 5% CO2 for 24 or 48 h.
Assessing Multiple Parameters for Hepatotoxicity Using Fluorescent Probes
Several fluorescent probes were simultaneously used to monitor cells in culture. Calcein AM (Invitrogen) was used as a marker of cell viability, cytoskeleton and cell area (total cell count). Hoechst 33258 (Invitrogen) was used to evaluate nuclear shape. MitoTracker Orange CM-H2TMROS (Invitrogen) served as a marker of the MP. For live cell imaging assays, cell media was removed and cells incubated with a mixture of the following reagents in phosphate-buffered saline (PBS): 1 μM Calcein AM, 2 μg/mL Hoechst 33258, and 200 nM of MitoTracker Orange for 30 min at which point the staining solution was replaced with PBS containing 0.1% of fetal bovine serum (FBS; Gibco, Invitrogen) and images were acquired using the ImageXpress® Micro XL system (Molecular Devices, LLC, Sunnyvale, CA) with a 10× or 4× objective. No additional washes were necessary to obtain high quality images by this protocol. During acquisition, plates were controlled at 37°C. We found that stained plates are stable for at least 90 min after staining. The number of viable cells in control samples and staining intensities were very similar at 30 min and 120 min poststaining, while after 3 h those parameters had decreased >30%.
For the fixed cell imaging protocol, the cell culture medium was removed and cells were incubated first with the mix of Hoechst 33258 (2 μg/mL) and MitoTracker Orange (200 nM) for 30 min, then the cells were fixed using formaldehyde (4% final concentration; Sigma-Aldrich) in PBS for 15 min at the room temperature. Fixed cells were then washed once with PBS, once with permeabilization solution and permeabilized for 1 h using 0.02% saponin (Sigma-Aldrich) plus 2% FBS. After permeabilization the cells were stained with AlexaFluor-488 (AF488)-conjugated phalloidin (Invitrogen) for 2 h at room temperature. Note that this step may be combined with the permeabilization step if desired. Cells were washed twice with PBS before images were acquired. It should also be noted that fixing the cells and phalloidin staining could be done after the live cell imaging protocol with Calcein AM, if the subsequent evaluation of additional markers or cell storage is desired.
MP Assay
In addition to the multiparameter staining protocol, a MP assay was also used to further elaborate the toxicity assessment. In contrast to the 72 h incubation during the general toxicity assay described above, the MP assay measures rapid (30–90 min) effects of compounds on MP. Cells were treated with compounds for 60 min and mitochondria membrane potential was monitored by the addition of the mitochondria active dye JC-10 (AAT Bioquest, Sunnyvale, CA) following the recommended protocol and appropriate concentrations of included reagents. Mitochondrial toxicity assays were performed on separate sets of plates and typically combined with the nuclear (Hoechst) stain. As the JC-10 staining protocol requires 15–30 min incubation with reagents, in the typical experiment, a 6× solution of JC-10 reagent was directly added to the cell culture media without removal of the compound. The plates were incubated at 37°C and 5% CO2 for 30 min (total time for compound treatment is 60 min), and then images were acquired using ImageXpress Micro XL system at 37°C.
Autophagy and Phospholipidosis Assays
A selected set of compounds were tested for their ability to induce autophagy and phospholipidosis. Autophagy was assessed by the Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, Farmingdale, NY) after treatment of cells with compounds for 24 h. Phospholipidosis was measured using the HCS LipidTOX Phospholipidosis and Steatosis Detection Kit (Invitrogen) for neutral lipids and phospholipids after treatment of cells with compound for 48 h. Separate assay plates were used for both autophagy and phospholipidosis assays. The staining protocol followed the product recommended reagent dilutions and buffers.
Image Acquisition and Analysis
Images were acquired using the ImageXpress Micro XL system using 20×/0.45NA ELWD Plan Fluo, 10×/0.3NA Plan Fluor, or 4×/0.2NA Plan Apo objectives. The light source was a solid state white light engine with emission from 380 to 680 nm. A fluorescein isothiocyanate (FITC) filter cube (ex 482/35, em 536/40) (center wavelength [nm]/bandpass width [nm]) was used for Calcein AM, Cyto-ID, and LipidTOX staining of neutral lipids. A tetramethyl rhodamine isothiocyanate (TRITC) filter cube (ex 543/22, em 593/40) was used for MitoTracker Orange and LipidTOX staining of phospholipids. A 4′,6-diamidino-2-phenylindole (DAPI) filter cube (ex 377/50, em 447/60) was used for Hoechst nuclear stain. Typically, one image per well was taken for 384-well plates and two to four images per well were taken for 96-well plates when using 10× or 20× objectives. Some assays were done using a 4× objective, in this case one image per well was taken. The number of images was determined by a balance of number of cells per field of view and acquisition time. Multiparameter toxicity assay can be done with either 10× or 4× objectives. A 10× objective provides higher resolution of cells and subcellular structures while allowing one to capture relatively large numbers of cells (2,000–3,000) in a single image (27% of the 384-well area and ∼7% of 96-well area captured in one image). When using a 4× objective, the image captures an entire 384-well (>10,000 cells) with sufficient resolution to detect cells and nuclei. However, detection of nuclear size or cell size is less accurate than with 10× magnification. Other assays described here (MP, autophagy, and phospholipidosis) require at least 10× magnification. 20× magnification provides even better resolution but typically requires taking multiple sites per well and adds to acquisition time.
Typical exposure times ranged from 10 ms to 100 ms. All image analysis was done using MetaXpress® 4 or MetaXpress 5 software (Molecular Devices, LLC) and various image processing application modules including Multi-Wavelength Cell Scoring, Count Nuclei, and Granularity. In addition, the Custom Module Editor (CME) in MetaXpress 5 software was used to combine the above algorithms into one analysis module and characterize additional features important for analysis of phenotypic changes associated with toxicity. The CME algorithm for the general hepatotoxicity assay involved the following steps: (1) identification of nuclei and determination of nuclear area; (2) segmentation of cells and determination of cell area; (3) measurement of Calcein AM intensity in cell area; (4) measurement of MitoTracker Orange intensity in cell area; (5) determination of nuclear brightness; (6) classification of nuclear condensation; and (7) measurement of output parameters.
Images for the MP assay images were acquired using the TRITC and FITC channels for JC-10 staining along with a DAPI channel if Hoechst staining was included. For the phospholipidosis assay, the TRITC and FITC channels were used for phospholipids and neutral lipids, respectively. Autophagy was detected using the FITC channel. Typically one image per well was taken for 384-well plates using 10× objective for these assays. Images were acquired with 20× objective when higher resolution and improved analysis of small features such as granules was desired. Images were analyzed using MetaXpress 4 or MetaXpress 5 software and the Granularity application module with or without single cell segmentation based on optional nuclear staining. A similar image acquisition and analysis setup was used for the phospholipidosis and autophagy assays.
The statistical analysis included calculation of a Z′-factor coefficient.27 As used here, it represents the separation of positive and negative control wells on a given plate. Typically, 8–16 control wells each for positive and negative are included on each plate, and Z′=1−[(3×σpos+3×σneg)/|(Mpos−Mneg)|], where σ=standard deviation and M=average value. In certain cases, only four control wells each were used for controls; these are noted in the text.
Compound Library Screening
The Screen-Well™ Hepatotoxicity Library (Enzo Life Sciences) contains 240 compounds including anti-cancer, anti-inflammatory, neuroleptics, antibiotics, and other classes of compounds. Major toxicity categories include steatosis, cholestatic effects, mitochondrial toxicity, CYP450 inactivation, toxic metabolites, Mallory body formation, and elevation of liver enzymes. In addition to hepatotoxic compounds, the library contains 30 control compounds that have no reported liver toxicity. Two wells were blank. Therefore, the library contains 208 hepatotoxic compounds and 30 negative (“hepato-safe”) compounds. The compounds were dissolved in DMSO at 10 mM and aliquoted into deep-well plates.
Cells were treated with library compounds for 72 h in a 384 well format plate in duplicate. In addition, each plate contained 48 vehicle only wells treated with the corresponding DMSO concentration. Hit selection criteria were based on a decrease or increase of each measured response parameter being greater than three standard deviations (SDs) from average DMSO control well response. At least two parameters were required to respond with a change greater than three SDs for a compound to be classified as a hit. Based on the toxic compounds identified by these combined assay measures, the assay sensitivity was calculated as the percentage of positive compounds out of the total number of compounds expected to be positive (208 hepatotoxic compounds). Assay specificity was calculated as percentage of negative compounds out of the total number of compounds expected to be negative (30 safe compounds). The predictive value of the assay was calculated as the mean value between sensitivity and specificity.
Concentration-Response Curve Fitting, Derivation of Benchmark Concentrations and Ranking of Compounds
Normalized (to corresponding DMSO-containing wells) data for each chemical and endpoint were used to fit a concentration-response quantitative logistic function and to visualize the concentration–response relationship, using the R software version 2.15.2. Variability of the DMSO-only wells was used to derive the benchmark concentration (BMC),28 which involves modeling to obtain dose levels corresponding to specific response levels near the low end of the observable range of the data. An accepted approach for continuous data is to define a point-of-departure (i.e., benchmark) response as a change in the mean equal to one control SD from the control mean. An interactive Toxicological Prioritization Index (ToxPi™) application29 was used to integrate evidence across endpoints and to visualize the relative prioritized ranks of compounds.
Results
Characterization of Human iPSC-Derived Hepatocytes
Human iPSC can be developed from adult tissue via noninvasive methods, expanded indefinitely, and differentiated into multiple cell types.30,31 The iCell hepatocytes used in the studies were provided as a fully differentiated and highly pure population of cells that formed an adherent cell monolayer with a cobblestone morphology similar to primary human hepatocytes (Fig. 1A). Purity was measured by counterstaining for iPSC markers; typically less than 5% of the population is found positive for iPSC markers (data not shown32). The cells have round nuclei, distinct nucleoli, and a high cytoplasm to nuclear ratio. In addition, as indicated in Figure 1A, some of cells show bi-nucleation (circles), and visible evidence of bile canaliculi (arrows). iCell hepatocytes express multiple hepatocyte markers, including albumin, alpha-1-antitrypsin, and HNF4α protein (not shown). In addition, iCell hepatocytes secrete albumin at a level similar to primary human hepatocytes and greater than HepG2 cells, exhibit glycogen storage, lipid accumulation, and tight junction formation (Fig. 1B). We have confirmed that incubation of cells in galactose media does not perturb their phenotype or ability to secret albumin. Such characterization was done by immunostaining followed by imaging with a 10× objective (data not shown).
Multiparameter Assays for Hepatotoxicity
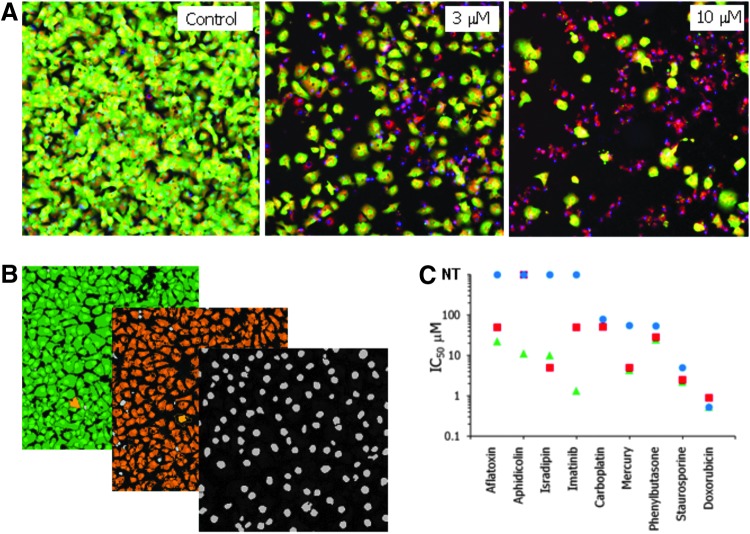
The primary goal of this study was to evaluate and optimize several assays to test for general and mechanism-specific hepatotoxicity. We posit that a mixture of three dyes provides phenotypic readouts indicative of general cellular toxicity that occurs through various pathogenetic mechanisms. The phenotypic readouts include cell viability assessment and characterization of cell area (by Calcein AM), MP (by MitoTracker Orange), and total cell count along with nuclear characterization by area and fluorescence intensity (Hoechst stain). Advantages of this method, especially for screening environments, include simplicity, cost, and ease of workflow. Cell staining is performed by adding a mix of dyes in one step, without fixing cells or repeated washes. This procedure minimizes automation steps required and reduces cell losses by washes that may lift off loosely attached cells and impact assay quality. Representative composite images of cells treated with amitriptyline from this assay are shown in Figure 2A.
Fig. 2.
Live cell imaging toxicity assessment. (A) iCell hepatocytes treated with indicated concentrations of amitryptiline for 72 h, then stained with Calcein AM, Hoechst 33258, and MitoTracker Orange. Images were taken using a 10× objective and analyzed using Multi-Wavelength Cell Scoring module. (B) Image processing by Multi-Wavelength Cell Scoring application module. Masks shown define Calcein AM staining (green), MitoTracker staining (orange), and Hoechst 33258 staining (grey). (C) Comparison of IC50s obtained by using total cells (nuclear count, blue circles), Calcein AM–positive cells (green triangles), or MitoTracker Orange–positive cells (red squares). NT, no significant toxicity detected from the four-parameter curve fit to the data.
For a more in-depth investigation into cytotoxic mechanisms elicited by the compounds assayed, several analyses were run in parallel to fully characterize phenotypic changes detected. Images were first segmented into individual cells by nuclear staining and then standard cell segmentation algorithms were used to associate specific image areas with each cell (Fig. 2B). Cell segmentation based on nuclei worked very efficiently for confluent cells and allowed the scoring of cells as positive or negative for additional markers to define the total cell number (nuclear count), number of viable cells (Calcein AM–positive), and cells with intact mitochondria (MitoTracker Orange–positive). In addition to cell scoring and count, this analysis provided additional parameters, which could be quantified specifically for each cell or averaged over the entire population in the image, including average and total cell area positive for Calcein AM or MitoTracker, average and total fluorescent intensities for Calcein AM or MitoTracker orange, and average and total nuclear areas and fluorescence intensities. The nuclear area and intensity was used to determine the presence of nuclear condensation as evidenced by increased nuclear brightness and decreased nuclear area (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/adt), which may indicate apoptosis. These readouts can be used either selectively or in combination for characterizing toxicity effects and determining concentration-response.
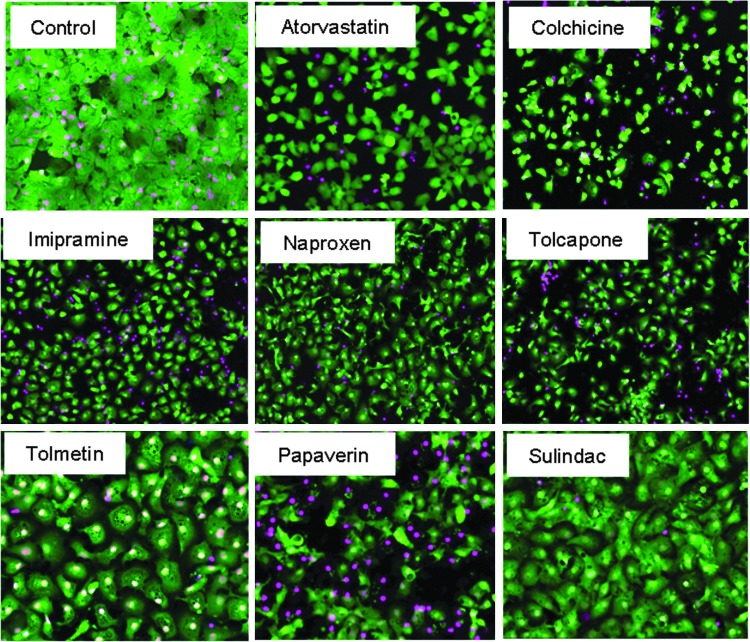
A comparison was made between characterizing toxicity by use of total cell count via nuclear stain, live cell count via Calcein AM, and count of cells with intact mitochondria via MitoTracker Orange. Detailed results of average, SD, Z′-factor coefficient, and assay window for ten different output parameters calculated for nine compounds are given in Supplementary Table S1. The Z′-factor coefficients for the most robust output parameters (“number of cells positive for Calcein AM,” “positive W2”) were typically >0.5. For other parameters, Z′-factor coefficients were dependent on individual compounds and specific compound effect: for example, “all nuclei mean area” was significantly decreased and Z′-factor coefficients higher for staurosporine (0.46) and aflatoxins (0.7), consistent with nuclear condensation phenotype. Z′-factor coefficients were >0.5 for “positive W2 mean stain area” for mitomycin C and phenylbutasone, indicating decreased cell spreading (Supplementary Tables S1 and S2). The multi-dye assay IC50 values were evaluated for nine representative hepatotoxic compounds using the three methods (Fig. 2C). The IC50s determined by “count of cells positive for Calcein AM” were generally lower than those determined by “total cell count” for several compounds, for example, aflatoxin, imatinib, and aphidicolin. This can be partially attributed to the fact that, often, dead or apoptotic cells are still present in an image and contribute to the nuclear count (Fig. 2A). For other compounds, IC50s determined by the different readouts were found to be closely correlated (e.g., for doxorubicin). Several other parameters were defined to assist the phenotypic characterization of cells. While a decrease in cell number is a very strong indicator for cytotoxicity, more subtle changes in cell appearance and morphology may be good early indicators of cell stress yet are more difficult to quantify. We aimed to detect and quantify change in appearance as an indicator of cell stress. Figure 3A shows representative images of treated cells that exhibited changes in average cell area and/or total cell area. Cells treated with atorvastatin, imipramine, naproxen, or tolcapone did not exhibit a dramatic decrease in cell number, yet showed a significant decrease in cell area as a result of rounding or loss of cell–cell contact. Specifically, cell count in the case of atorvastatin was decreased by only 25%, while total area of viable cells was decreased by 50%. In other examples (e.g., tomeltin or suldinac), an increase in average cell size was observed as a result of vacuolization or swelling. In the case of papaverin treatment, a decrease in average cell size was observed as a result of reduced cell spreading. Such changes can be quantified using the total area covered with viable cells or average individual cell area (Supplementary Fig. S2A). Significantly, changes in these parameters appear earlier, and can be a more sensitive indicator of toxicity than live cell count, since cell rounding up or vacuolization usually appears once the cell has committed, but still prior to cell death. Therefore, in addition to “total cell count,” “total” or “integrated fluorescent intensity by Calcein AM” and “total positive cell area” can be used for dose-dependent characterization of compound effects and determining IC50 values.
Fig. 3.
Phenotypic characterization of hepatotoxicity by quantitation of the average and total stained cell areas. Images of iCell hepatocytes treated with 100 μM of indicated compounds for 72 h, then stained with Calcein AM and Hoechst 33258. Examples presented show impact of different compounds on total positive cell area and/or average positive cell area. Images were acquired using a 10× objective.
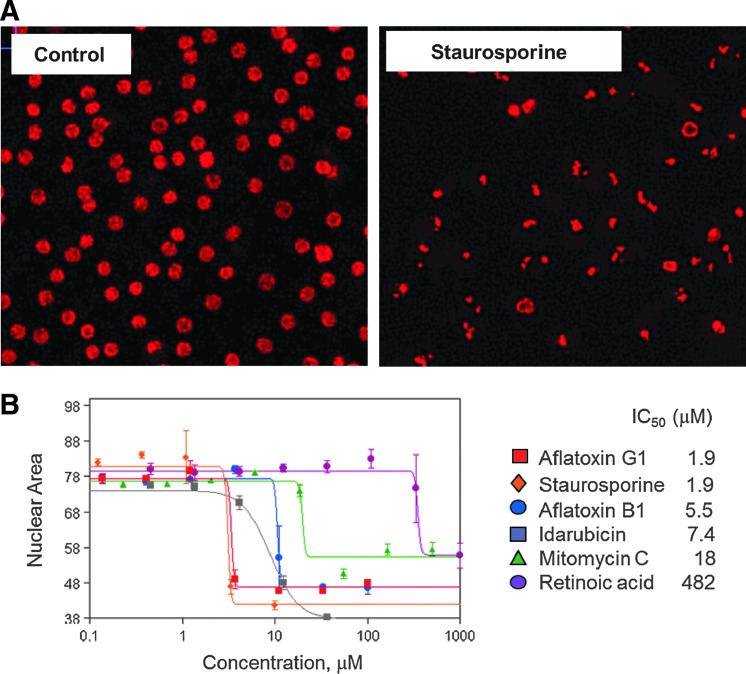
Nuclear characterization is another important parameter to assess cell death, allowing detection of the phenomenon of nuclear condensation, usually a hallmark of apoptosis.33 Nuclear condensation can be characterized by increased nuclear fluorescent intensity and decrease of nuclear area.25 We have used the nuclear characterization measures of average nuclear intensity and nuclear area as additional read-outs for multiparametric toxicity assessment (see Supplementary Fig. S2B and Supplementary Table S1). Images showing this effect for cells treated with staurosporine and resulting concentration response curves are shown in Figure 4. This effect was concentration-dependent, as evident from Figure 4B. Caspase staining (CellEvent Caspase 3/7; Invitrogen) done in parallel with the assay for staurosporine and anisomycin confirmed the presence of apoptosis for those compounds (data not shown). Nuclear characterization can provide additional sensitivity for certain compounds and does not add appreciable time to the image analysis process.
Fig. 4.
Phenotypic characterization of toxicity by nuclear shape and intensity. (A) Images of iCell hepatocytes treated with 10 mM of staurosporine, stained with Hoechst 33258. Images were acquired using a 10× objective. (B) Concentration-dependent responses for several compounds using “average nuclear area” as a readout. The IC50s for the tested compounds were aflatoxin G1 (red squares), 1.9 μM; staurosporine (orange diamonds), 1.9 μM; aflatoxin B1 (blue circles), 5.5 μM; idarubicin (grey squares), 7.4 μM, mitomycin C (green triangles), 18 μM; and retinoic acid (purple circles), 482 μM.
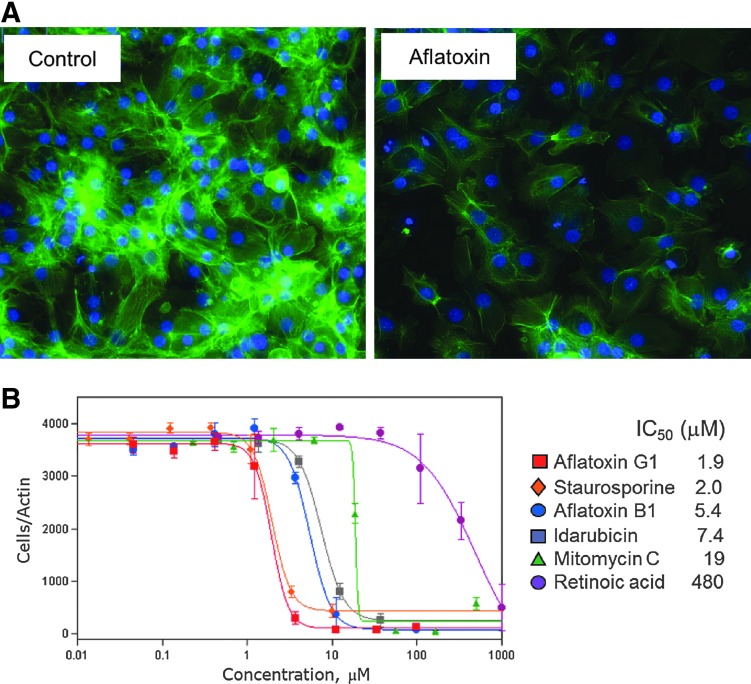
In addition to the assays described above, we evaluated an assessment of cytoskeleton integrity by phalloidin staining. Figure 5A shows intact cells and cells treated with aflatoxin B1. The cytoskeleton integrity assay can be combined with Hoechst nuclear stain and, if desired, with MitoTracker Orange if the latter is done prior to cell fixing. Detailed results of average, SD, Z′-factor coefficient, and assay window for eight different output parameters calculated for five compounds are given in Supplementary Table S2. The output parameter with highest Z′-factor coefficients and assay windows was “count of cells positive for actin staining.” This parameter was used to measure concentration response of six known hepatotoxic compounds (Fig. 5B). The cytoskeleton integrity assay had an excellent assay window and low variance leading to good 4-parameter fits of the concentration–response curves for most of the compounds tested. This assay could be used as a supporting alternative method for hepatotoxicity measured by Calcein AM. However, it does require the additional steps of fixing, permeabilizing, and washing the cells. A distinct advantage is that the assay plates can be stored, providing a fixed time point for readout if that is desired in a workflow.
Fig. 5.
Cytoskeleton integrity assessment by actin staining. (A) iCell hepatocytes treated with 30 mM of aflatoxin B1 for 72 h, then fixed and stained with AlexaFluor-488-Phalloidin and Hoechst 33258. (B) Concentration-dependent responses for several compounds using “number of actin-positive cells” as a readout. The IC50s for the tested compounds were aflatoxin G1 (red squares), 1.9 μM; staurosporine (orange diamonds), 2.0 μM; aflatoxin B1 (blue circles), 5.4 μM; idarubicin (grey squares), 7.4 μM; mitomycin C (green triangles), 19 μM; and retinoic acid (purple circles), 480 μM.
In addition, we have evaluated effect of timing of compound exposure on cell viability and other markers. Cells were treated with a set of hepatotoxic compounds (amiodarone, tamoxifen, pimozide, mitomycin C, idarubicin, and haloperidol) for 1, 3, or 7 days, on parallel plates. We have noticed that while some compounds (e.g., idarubicin) had comparable effects in all time points, other compounds (e.g., haloperidol) had time-dependent incremental effect (Supplementary Fig. S1C). IC50s were apparently smaller for 7 days exposure in comparison to 1 day exposure (Supplementary Table S3).
Mechanism-Specific Assays for Hepatotoxicity
In addition to general cellular toxicity assessment, high-content analysis can address other mechanism-specific types of toxicity, and there are many reagents available for such assays. Specifically, we have evaluated the mitochondrial dye JC-10, Cyto-ID dye, and LipidTOX reagents to determine the effects of compounds on MP, autophagy, and phospholipidosis/steatosis, respectively.
MP assay
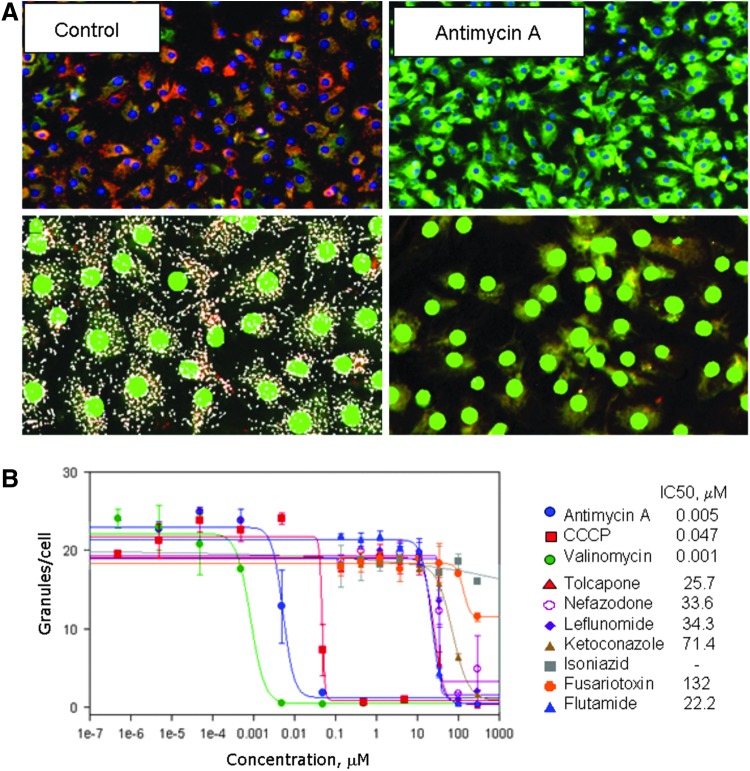
Mitochondrial depolarization is an early signal for hypoxic damage or oxidative stress. In addition to the 72 h compound treatment assay, which can show mitochondrial damage as a result of cell death caused by various mechanisms, we performed a short-term (60 min) compound treatment followed by JC-10 staining in combination with a nuclear stain. This assay allows examination of the immediate direct effects of compounds on MP. Figure 6 shows images of control cells and cells treated with an inhibitor of oxidative phosphorylation, antimycin A, and a dramatic difference in the content of intact mitochondria (orange stain) is observed. We used the MetaXpress 5 software Granularity application module for the analysis of mitochondrial damage using the nuclear stain to define and segment individual cells. The module finds granules (mitochondria) and allows defining such characteristic parameters as total number of granules, total granule area, number of granules per cell, and average intensity of the granules. Detailed results of average, SD, Z′-factor coefficient, and assay window for five different output parameters calculated for two compounds are given in Supplementary Table S4. We found that “average number of granules per cell,” “average area of granules per cell,” and “total number of granules per image” all gave a 10× or greater assay window. Since this assay can be used in real time, we characterized the compound response at several time points: 60 min, 90 min and 2 h. We found that a 60 min time point gave the most robust results, while 90, 120, or 180 min time points resulted in decreased signal intensity. A time course study was done to show effect of compound incubation time on assay results (Supplementary Tables S5 and S6). We characterized the assay using a panel of known poisons of oxidative phosphorylation including antimycin A, CCCP, and valinomycin. The Z′-factor coefficient for the assay was 0.68. We also tested various compounds whose expected mechanism of hepatotoxicity is mitochondrial damage (tolcapone, flutamide, nefasodone, leflunomide, ketoconasole, isoniazid, and fusariotoxin). Compounds were tested over a concentration range from 1 nM to 100 μM. The resulting concentration–response curves for these compounds and measured IC50 values are shown in Figure 6B.
Fig. 6.
JC-10 assay for mitochondrial potential (MP). (A) iCell hepatocytes treated with indicated compounds for 60 min. Top: Cells stained with Hoechst 33258 (nuclei), and JC-10 (MP); bottom: overlay of Granularity analysis result masks. “Total” or “average number of granules per cell,” “total granule area,” or “total granule intensity” were used as output parameters. (B) Concentration-dependent responses of “average number of granules per cell” for several compounds and corresponding IC50 values (in μM): antimycin A (blue circles), 0.005; CCCP (red squares), 0.047; valinomycin (green squares), 0.001; tolcapone (red triangles), 25.7; nefazodone (white circles), 33.6; leflunomide (purple diamonds), 34.3; ketoconazole (brown triangles), 71.4; isoniazid (grey squares), n/d; fusariotoxin (orange circles), 132; and flutamide (blue triangles), 22.2.
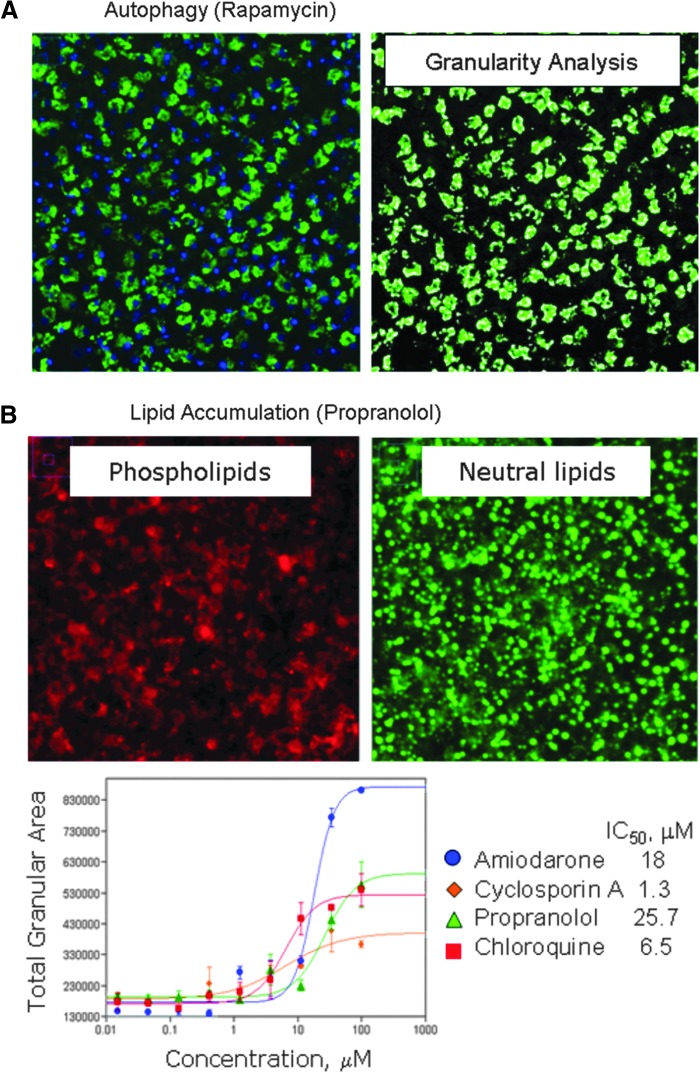
Autophagy is a process of selective degradation of intracellular targets such as mis-folded proteins and damaged organelles. This process is important for maintaining homeostatic function of the cell, and it may also function as a survival mechanism to maintain viability during periods of stress and a number of drugs have been shown to increase or decrease autophagy. We have evaluated the performance of the autophagy assay for high-content analysis. iPSC-derived hepatocytes were treated with compounds for 24 h, then stained with Cyto-ID dye plus a nuclear stain and images analyzed using MetaXpress 5 software and the Granularity application module. The total granule area was used as an output parameter. Detailed results of average, SD, Z′-factor coefficient, and assay window for three compounds are given in Supplementary Table S7. Representative images and granularity analysis results from the autophagy assay for rapamycin are shown in Figure 7A. We measured the dose response for several representative compounds (chloroquine, verapamil, and rapamycin) and determined their IC50 values in this assay. The measured fold increases of autophagy staining were 4.7 for chloroquine, 2.1 for verapamil, and 1.4 for rapamycin, and IC50s were determined to be 1.3, 33.5, and 3.1 μM respectively.
Fig. 7.
(A) Autophagy detection assay. Image of iCell hepatocytes treated with rapamycin for 24 h and stained with Cyto-ID autophagy detection kit. Right: Analysis with Granularity module. (B) Phospholipidosis, lipid accumulation assay. Images of iCell hepatocytes treated with propranolol for 48 h. Phospholipidosis and steatosis detected using LipidTOX reagent showing phospholipids and neutral lipids as indicated. Concentration-dependent responses for phospholipidosis using “total granular area” output and corresponding IC50 values (in μM) for several compounds: amiodarone (blue circles), 18; cyclosporin A (orange diamonds), 1.3; propranolol (green triangles), 25.7; and chloroquine (red squares), 6.5.
Phospholipidosis, or excessive accumulation of phosphilipids in tissues, is an important concern for drug development, since many cationic amphiphilic compounds can accumulate within cells. This drug trapping is followed by a gradual accumulation of drug-phospholipid complexes (myeloid bodies) in tissues. Compounds including antidepressants, antianginal, antimalarial, and cholesterol-lowering drugs are reported to cause drug-induced phospholipidosis. Drug-induced phospholipidosis represents a concern as a safety risk assessment; therefore, we have tested this assay for utility in hepatotoxicity screening. We used the LipidTOX reagent to evaluate the effects of several compounds known to cause phospholipidosis. iCell hepatocytes were treated with compound for 48 h, at which time phospholipidosis and steatosis were detected using the LipidTOX reagent. A significant amount of neutral lipids and phospholipid granules were observed using the phospholipidosis assay, an example of which is shown for the case of proproanolol treatment in Figure 7B. Images were analyzed using the Granularity application module of MetaXpress 5 software with the total granule area used as the output parameter. Detailed results of average, SD, Z′-factor coefficient, and assay window for three compounds are given in Supplementary Table S8. It was found that untreated hepatocytes had significant background levels of neutral lipids and phospholipids, which limited the capability of the assay. We were only able to detect modest increases of total lipids or phospholipids in response to compound treatment for total granule area or intensity in comparison to control (see line graph in Fig. 7B). The measured fold increases of phospholipidosis were 2.4–3.7× for amiodarone, 2× for cyclosporine, 1.4× for tamoxifen, and 4.5× for propranolol.
Multiparameter Hepatotoxicity Assay
A combination of the general toxicity assessment by the multi-dye assay (Fig. 2) and mechanism specific JC-10 assay (Fig. 6) were chosen to perform a Multiparameter Hepatotoxicity assay and screen a small compound library. A customized analysis was setup for the first assay using the MetaXpress 5 software CME, which allows the image analysis to be tailored and optimized for multiparametric outputs from toxicity indicators. This is important because it is necessary to characterize the previously described multiple phenotypic changes that occur during hepatotoxicity to provide insight into the specific mechanism of toxicity. The simultaneous analysis of parameters provides a simple and efficient way to increase the amount and quality of information provided by the assay.
The CME algorithm involved number of steps described in the Materials and Methods section. Representative images and masks derived from the analysis for untreated and amiodarone-treated cells are shown in Supplementary Figure S1A. Shown are examples of cell images and analysis masks representing cell area (spreading) and nuclear characterization including the results for Live Cell Area and Apoptotic Cells. Multiple phenotypic changes are easier to visualize using compound profiles across multiple parameters. Figure 8 presents the profiles from several hepatotoxic compounds from different groups (cardiac drugs, anticancer drugs, antibiotics, etc). Each compound has its specific signature across the different readouts. Importantly, the multiparameter analysis allows for comparing compound profiles and grouping similar profiles together. In addition, it may also provide insight into the mechanism of action.
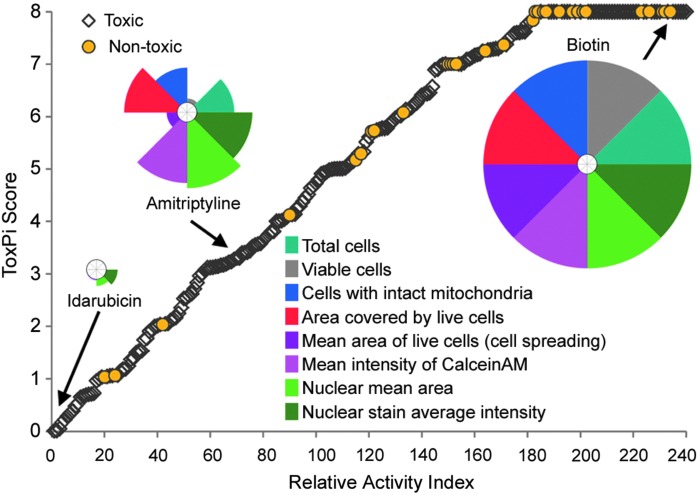
Fig. 8.
Multiparametric hepatotoxicity analysis. ToxPi scores were used to create a relative ranking of 240 screened compounds using multiparametric data (total cells, viable cells, cells with intact mitochondria, area covered by live cells, mean area of live cells [cell spreading], mean intensity of Calcein AM, nuclear mean area, and nuclear stain average intensity). Yellow circles, nontoxic drugs; clear diamonds, hepatotoxic drugs. Representative ToxPi-es for three compounds are shown to visualize the relative contribution of each parameter (represented in colors of each slice) to the overall score. Note: Nontoxic compounds are expected to align with a higher relative activity index value.
Compound Library Screening Results
The Screen-Well Hepatotoxicity library was used for validation of the multiparametric hepatotoxicity (MPH) assays with iCell hepatocytes. As mentioned previously, the phospholipidosis and autophagy assays were found to be useful for testing selected sets of individual compounds, but because of the relatively small assay windows observed with these measures, we did not use those assays for the library screening. The MPH assay was assessed according to assay sensitivity, selectivity, and predictive value. For the general toxicity assay cells were incubated for 72 h with compounds at concentrations of 1, 3, 10, 30, and 100 μM. Compounds were tested in duplicates and different compound concentrations were tested on separate plates. For the mechanism specific assay (JC-10) cells were treated for 60 min at two concentrations, 10 and 100 μM. The output parameters that were used for the assessment were (1) number of Calcein-positive cells; (2) number of MitoTracker-positive cells; (3) total cell number; (4) average Calcein AM intensity; (5) total Calcein AM cell area; (6) MitoTracker intensity; (7) MitoTracker cell area; (8) average nuclear area; (9) nuclear average intensity; (10) JC-10 number of granules/cell; and (11) JC-10 total number of granules.
As described previously, the library included 240 hepatotoxic compounds of which 30 were “safe compounds,” or negative controls, and 2 were random blanks. The remaining 208 compounds were expected to be hepatotoxic, or “positive” based on known effects in clinical and preclinical studies. In addition to the tested compounds, each plate contained 48 wells with same concentrations of DMSO. Assay variability was determined from average and SD of the DMSO controls and each compound concentration.
The results acquired for all compounds tested at 100 μM concentration are presented in Supplementary Figure S3A and S3B. Averages from duplicates are presented for the multiple read-outs. Compounds are grouped into different compound classes based on therapeutic indication (not chemical similarity), based on information provided by library description. Hits were classified as compounds that caused effects greater than ±3 SD from DMSO controls, and showed such effects in more than one parameter. Compounds indicated as hits in Supplementary Figure S3A and S3B are highlighted in red (>3 SD decrease in each parameter as compared to the appropriate DMSO control), or yellow (>3 SD increase in each parameter in comparison to the appropriate DMSO control). We observed dose-dependent, incremental changes for most of the parameters along with an incremental increase in the number of hits across the concentrations. However, many hits were only detected at the highest tested concentration (100 μM). Previous reports have described hepatotoxicity effects observed at 300 μM or even 1 mM concentrations. Our studies were limited by the maximum DMSO concentration allowed. We observed that 1% DMSO did not have an impact on tested parameters while an increase to 3% DMSO resulted in increased variability (>20%) between read-outs from control wells (data not shown). We have tested concentration responses for individual selected compounds up to 1 mM. Among compounds detected as hits were anticancer drugs, neuroleptics, hormonal drugs, antihistamines, statins, cardiac drugs, antifungal drugs, known toxins, and some antibiotics and anti-inflammatory drugs.
Finally, we tested the potential for the multiparametric endpoint data to rank compounds for their potential hepatotoxicity hazard based on concentration-response in the iPSC-derived hepatocyte cell model. Because the vehicle-only effect level was normalized across plates, we used four-parameter maximum-likelihood logistic modeling to fit the data. In accordance with the US EPA guidance for dose–response modeling28 we selected a one SD departure from the control mean as the benchmark response from which a BMC is derived. If the maximum response did not reach this level in the concentration range tested, the “no observable adverse effect level” was recorded as 100 μM, the highest concentration used. Same curve fitting and BMC derivation procedure was applied to all chemicals and endpoints collected. We integrated multiple parameters collected in this study to rank chemicals in the screened library for their overall relative safety rank with respect to liver toxicity hazard (Fig. 8). Each compound's BMC values for 8 different parameters were analyzed and visualized using the ToxPi approach,29 which generates transparent graphical rankings to facilitate decision making. The analysis of the data showed that most nontoxic compounds were ranked high for Relative Activity Index, while most hepatotoxic compounds were ranked low. While in this analysis all parameters were given equal weight, data integration assumptions may be amended in accordance to the user's needs and this analysis should be interpreted as semi-quantitative.
Discussion
There is a need for improved methods to evaluate in vitro toxicity to meet the challenges associated with drug discovery and development to the clinic. This is particularly true for hepatotoxicity, which is a leading cause of compound failure during the preclinical and clinical stages of development.4–6 High-content imaging has been demonstrated to be particular useful for hepatotoxicity testing as it can assess subtle phenotypic changes and provide multiparametric read outs.25,26 In the present work, we have demonstrated the value of iPSC-derived hepatocytes for this use in these types of assays. The morphological, marker expression and functional characteristics exhibited are consistent with those identified in primary hepatocytes (Fig. 1). In addition, iPSC-derived hepatocytes are available in large quantities, stable in culture, and display consistent, reproducible performance.
We have further developed high-content analysis methods using this cell model. A number of phenotypic read-outs and output parameters were characterized and a simple workflow assay protocol have been developed that can be used for defining general and mechanism-specific toxicity of compounds. We have developed an automated multiparametric screening method and optimized protocol for cell treatment, high-content imaging, and image analysis. Using specific examples and a diverse set of hepatotoxic compounds we have demonstrated typical cell responses and how different read-outs can be used in combination to characterize different types of toxicity. In addition to its convenience, this assay system has the potential for use in screening environments.
It is possible that other assays would be beneficial to, and could even further increase, the overall predictivity of the system. Two such assays were evaluated here: phospholipidosis and autophagy. There were clear dose-dependent responses observed with these assays, but improvements are needed to increase the assay precision and robustness for them to be included in a full screen. CYP450 expression or other read-outs relevant to metabolic activity could further increase assay predictivity especially for the compound classes mentioned above. This will be the subject of future studies.
Future experiments will also focus on the use of specific patient-derived cell models for addressing idiosyncratic hepatotoxicity. One of the great benefits of using iPSC-derived hepatocytes is that they can be obtained from different patients representing multiple genotypes. This affords the opportunity to study the sensitivity of different genotypes to hepatotoxic compounds and the prescreening of patients for potential adverse liver reactions to drugs. This could also allow optimization of drugs during development using SAR techniques for minimizing hepatotoxicity.
The iPSC-hepatocyte cell model and high-content imaging-based assays presented here show great promise for providing a highly sensitive, specific, and predictive tool for assessing hepatotoxicity. While a few limitations have been identified, further development of the methods and models used will only increase the utility of this important tool for the development of the next generation of therapeutic compounds.
Supplementary Material
Abbreviations
- BMC
benchmark concentration
- CME
Custom Module Editor
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- iPSC
induced pluripotent stem cells
- MP
mitochondria potential
- MPH
multiparametric hepatotoxicity
- PBS
phosphate-buffered saline
- SAR
structure activity relationship
- SD
standard deviation
- TRITC
tetramethyl rhodamine isothiocyanate
Acknowledgments
The authors would like to thank CDI, in particular Jennifer Luebke-Wheeler, Kristin Block, and Christian Kannemeier for providing iCell hepatocyte characterization data and David Mann for critical review of the article. This work was supported, in part, by the National Institutes of Health (ES015241]; and the United States Environmental Protection Agency [STAR RD83516601].
Disclosure Statement
O.S., J.H., and E.F.C. are employed by Molecular Devices, which sells the ImageXpress Micro XL system and MetaXpress software. I.R. reports no competing financial interests.
References
- 1.Kola I, Landis J: Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 2004;3:711–715 [DOI] [PubMed] [Google Scholar]
- 2.Gunawan BK, Kaplowitz N: Mechanisms of drug-induced liver disease. Clin Liver Dis 2007;11:459–475 [DOI] [PubMed] [Google Scholar]
- 3.Stine JG, Lewis JH: Drug-induced liver injury: a summary of recent advances. Expert Opin Drug Metab Toxicol 2011;7:875–890 [DOI] [PubMed] [Google Scholar]
- 4.Corsini A, Ganey P, Ju C, et al.: Current challenges and controversies in drug-induced liver injury. Drug Saf 2012;35:1099–1117 [DOI] [PubMed] [Google Scholar]
- 5.Lee WM: Acute liver failure in the United States. Semin Liver Dis 2003;23:217–226 [DOI] [PubMed] [Google Scholar]
- 6.Lee WM: Drug-induced hepatotoxicity. N Engl J Med 2003;349:474–485 [DOI] [PubMed] [Google Scholar]
- 7.Hartung T, Daston G: Are in vitro tests suitable for regulatory use? Toxicol Sci 2009;111:233–237 [DOI] [PubMed] [Google Scholar]
- 8.Hartung T: A toxicology for the 21st century—mapping the road ahead. Toxicol Sci 2009;109:18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krewski D, Westphal M, Al Zoughool M, Croteau MC, Andersen ME: New directions in toxicity testing. Annu Rev Public Health 2011;32:161–178 [DOI] [PubMed] [Google Scholar]
- 10.Hartung T: Toxicology for the Twenty-First Century. Nature 2009;460:208–212 [DOI] [PubMed] [Google Scholar]
- 11.Soldatow VY, LeCluyse EL, Griffith LG, Rusyn I: In vitro models for liver toxicity testing. Toxicol Res 2013;2:23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groneberg DA, Grosse-Siestrup C, Fischer A: In vitro models to study hepatotoxicity. Toxicol Path 2002;30:394–399 [DOI] [PubMed] [Google Scholar]
- 13.Wormser U, Ben-Zakine S: The liver slice system: an in vitro acute toxicity test for assessment of hepatotoxins and their antidotes. Toxicol In Vitro 1990;4:449–451 [DOI] [PubMed] [Google Scholar]
- 14.Elferink MG, Olinga P, van Leeuwen EM, et al.: Gene expression analysis of precision-cut human liver slices indicates stable expression of ADME-Tox related genes. Toxicol Appl Pharmacol 2011;253:57–69 [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Lechon MJ, Castell JV, Donato MT: The use of hepatocytes to investigate drug toxicity. Methods Mol Biol 2010;640:389–415 [DOI] [PubMed] [Google Scholar]
- 16.Rodrigues RM, Bouhifd M, Bories G, Sacco MG, Gribaldo L, Fabbri M, Coecke S, Whelan MP: Assessment of an automated in vitro basal cytotoxicity test system based on metabolically-competent cells. Toxicol In Vitro 2013;27:760–767 [DOI] [PubMed] [Google Scholar]
- 17.Anson BD, Kolaja KL, Kamp TJ: Opportunities for use of human iPS cells in predictive toxicology. Clin Pharmacol Ther 2011;89:754–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweikart K, Guo L, Shuler Z, et al.: The effects of jaspamide on human cardiomyocyte function and cardiac ion channel activity. Toxicol In Vitro 2013;27:745–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jehle J, Ficker E, Wan X, et al.: Mechanisms of zolpidem-induced long QT syndrome: acute inhibition of recombinant hERG K(+) channels and action potential prolongation in human cardiomyocytes derived from induced pluripotent stem cells. Br J Pharmacol 2013;168:1215–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris K, Aylott M, Cui Y, Louttit JB, McMahon NC, Sridhar A: Comparison of electrophysiological data from human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs) to functional pre-clinical safety assays. Toxicol Sci 2013;134:412–426 [DOI] [PubMed] [Google Scholar]
- 21.Medine CN, Lucendo-Villarin B, Storck C, et al. : Developing high-fidelity hepatotoxicity models from pluripotent stem cells. Stem Cells Trans Med 2013;2:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rana P, Anson B, Engle S, Will Y: Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol Sci 2012;130:117–131 [DOI] [PubMed] [Google Scholar]
- 23.Reynolds JG, Geretti E, Hendriks BS, et al.: HER2-targeted liposomal doxorubicin displays enhanced anti-tumorigenic effects without associated cardiotoxicity. Toxicol Appl Pharmacol 2012;262:1–10 [DOI] [PubMed] [Google Scholar]
- 24.Thomas N: High-content screening: a decade of evolution. J Biomol Screen 2010;5:1–9 [DOI] [PubMed] [Google Scholar]
- 25.O'Brien PJ, Irwin W, Diaz D, et al. : High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high-content screening. Arch Toxicol 2006;80:580–604 [DOI] [PubMed] [Google Scholar]
- 26.Mennecozzi M, Landesmann B, Harris GA, Liska R, Welan M: Hepatotoxicity screening taking a mode-of-action approach using HepaRG cells and HCA. ALTEX Proc 2012;1:193–204 [Google Scholar]
- 27.Zhang JH, Chung TDY, Oldenburg KR: A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999;4:67–73 [DOI] [PubMed] [Google Scholar]
- 28.United States Environmental Protection Agency: Benchmark Dose Technical Guidance. U.S. EPA, Washington, DC, 2012 [Google Scholar]
- 29.Reif DM, Sypa M, Lock EF, et al.: ToxPi GUI: an interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics 2013;29:402–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu K, Yu J, Suknuntha K, et al.: Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 2011;117:e109–e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Gulbranson DR, Hou Z, et al.: Chemically defined conditions for human iPSC derivation and culture. Nat Methods 2011;8:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann DA, Einhorn S, Block K, et al. : Human iPSC-derived hepatocytes: functional model tissue for in vitro predictive metabolism, toxicity, and disease modeling. Gen Eng Biotech News 2013;33:28–29 [Google Scholar]
- 33.Tone S, Sugimoto K, Tanda K, et al.: Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp Cell Res 2007;313:3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.