Abstract
So far, it has been suggested that phosphoenolpyruvate carboxylases (PEPCs) and PEPC kinases (PPCKs) fulfill several important non-photosynthetic functions. However, the biological functions of soybean PPCKs, especially in alkali stress response, are not yet well known. In previous studies, we constructed a Glycine soja transcriptional profile, and identified three PPCK genes (GsPPCK1, GsPPCK2 and GsPPCK3) as potential alkali stress responsive genes. In this study, we confirmed the induced expression of GsPPCK3 under alkali stress and investigated its tissue expression specificity by using quantitative real-time PCR analysis. Then we ectopically expressed GsPPCK3 in Medicago sativa and found that GsPPCK3 overexpression improved plant alkali tolerance, as evidenced by lower levels of relative ion leakage and MDA content and higher levels of chlorophyll content and root activity. In this respect, we further co-transformed the GsPPCK3 and SCMRP genes into alfalfa, and demonstrated the increased alkali tolerance of GsPPCK3-SCMRP transgenic lines. Further investigation revealed that GsPPCK3-SCMRP co-overexpression promoted the PEPC activity, net photosynthetic rate and citric acid content of transgenic alfalfa under alkali stress. Moreover, we also observed the up-regulated expression of PEPC, CS (citrate synthase), H+-ATPase and NADP-ME genes in GsPPCK3-SCMRP transgenic alfalfa under alkali stress. As expected, we demonstrated that GsPPCK3-SCMRP transgenic lines displayed higher methionine content than wild type alfalfa. Taken together, results presented in this study supported the positive role of GsPPCK3 in plant response to alkali stress, and provided an effective way to simultaneously improve plant alkaline tolerance and methionine content, at least in legume crops.
Introduction
As a versatile crop, alfalfa (Medicago sativa L.) is used for pasture, hay, silage and green-chop, and acts in crop rotation through its positive effects on soil fertility and soil structure [1]. Due to its versatility, high productivity, high feed value and potential roles in soil improvement and soil conservation, alfalfa is grown over a wide range of climatic conditions [1], [2]. However, environmental challenges, especially soil salinity and alkalinity, not only seriously restrict alfalfa yield, but also affect nodules formation and symbiotic nitrogen-fixation capacity [3]. With the global climate change and the global shrinkage of arable lands, a grimmer reality of soil salinity and alkalinity is painted, hence more and more attentions have been paid to exploring alfalfa cultivation on marginal lands.
As reported earlier, of the 831 million hectares salt-alkali soils on the world, saline and alkaline soils underline 397 (47%) and 434 (53%) million hectares, respectively [4], [5]. What’s worse, in northeast China, over 70% of land area is alkaline grassland [6]. Alkaline soil is characterized by high pH, high exchangeable sodium, poor fertility, dispersed physical properties and low water content [7]. Recently, a handful of researches suggested that, compared with salt stress, alkaline stress always caused much stronger inhibition of plant growth and development [8]. Unfortunately, until now, little attention has been paid on the molecular mechanisms of plant adaptation to alkaline stress.
Phosphoenolpyruvate carboxylase (PEPC; EC4.1.1.31) is a kind of tightly controlled cytosolic enzyme which functions in carbon fixation during photosynthesis [9]–[11]. PEPC kinase (PPCK) controls the phosphorylation state and bioactivity of PEPCs. In recent years, PPCK genes have been identified in different higher plants, including two for Arabidopsis [12], three for rice [13], and four for soybean [14]. Recently, several lines of direct evidence supported that PEPCs and PPCKs fulfilled important non-photosynthetic functions, particularly in plant response to environmental challenges. One of the best studied examples was that salt stress remarkably increased PPCK activity [15]–[17]. Further investigation revealed that salt stress not only increased PPCK gene expression levels but also decreased PPCK protein degradation rates [17]. Moreover, PEPC/PPCK activity in Arabidopsis and poplar was also regulated by alkali stress [12], [18].
Besides high stress tolerance, another important desired trait for alfalfa is high nutritional value in terms of essential amino acids. As we know, legume plants are deficient in the sulfur-containing amino acids, namely, methionine and cysteine [19]. Methionine is nutritionally essential for mammals, but at low levels in legume. Unlike plants, mammals could not synthesize methionine; hence they have to obtain it from their diets. In this respect, methionine deficiency obviously limited the nutritional value of legumes; therefore, increasing the methionine content has become another important goal for legumes breeding. To solve this problem, in a previous study, we designed and synthesized the SCMRP gene according to the maize methionine-rich 10 ku zein protein [20], [21]. A similar study reported that overexpression of the maize 10 ku zein gene increased the sulphur-containing amino acids of transgenic potato [22].
For these reasons, in this study, we aimed to generate the transgenic alfalfa not only with higher alkali tolerance but also with higher methionine content. Based on the Glycine soja (G07256) microarray data, we isolated and characterized an alkali stress responsive gene GsPPCK3. Our results demonstrated that GsPPCK3 overexpression in alfalfa improved plant tolerance to alkali stress. In this case, we co-transformed the GsPPCK3 and SCMRP genes into alfalfa, and demonstrated that the transgenic alfalfa displayed not only higher alkali tolerance but also higher methionine content. Taken together, results presented in this study demonstrated the biological function of GsPPCK3 under alkali stress, and provided an effective way to simultaneously improve plant alkaline tolerance and methionine content, at least in legume crops.
Results
Isolation and Sequence Analysis of the GsPPCK3 Gene
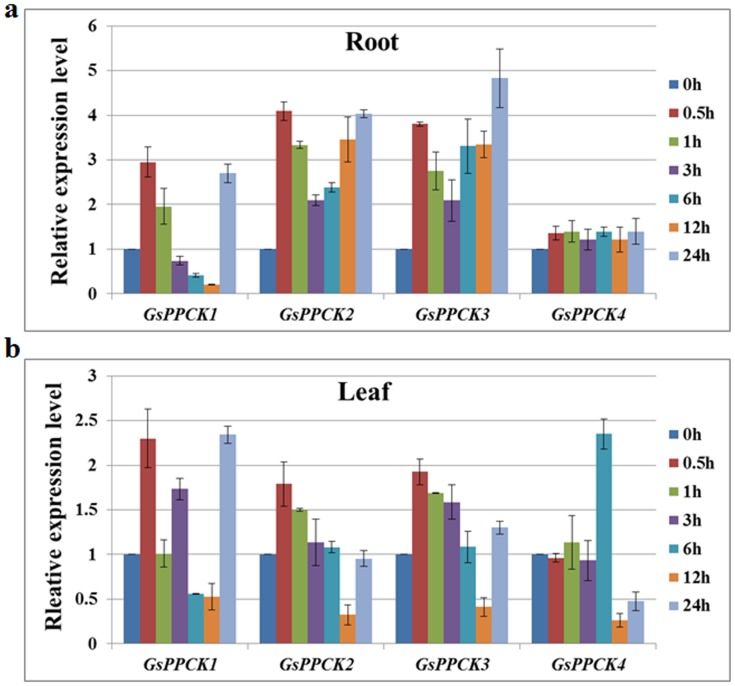
In a previous study, we constructed a transcriptional profile of Glycine soja (G07256) roots and leaves in response to salt-alkali stress (50 mM NaHCO3, pH 8.5), by using the Affymetrix® Soybean Genome Array [23]. Three PEPC kinase genes GsPPCK1, GsPPCK2 and GsPPCK3 were identified as potential stress responsive genes (Fig. 1). Contrarily, the last PPCK gene GsPPCK4 essentially did not respond to salt-alkali stress, only with an increase at 6 h in leaves.
Figure 1. Expression patterns of the Glycine soja PPCK family genes under 50 mM NaHCO3 (pH 8.5) treatment based on the microarray data.
a. Expression patterns of the PPCK family genes under alkali stress in Glycine soja roots. b. Expression patterns of the PPCK family genes under alkali stress in Glycine soja leaves.
In this study, we obtained the full length coding region of GsPPCK3 by using homologous cloning strategy. GsPPCK3 contained a complete open reading frame (ORF) of 915 bp encoding 304 amino acids with an estimated molecular weight (Mr) of 34 000 and a theoretical pI of 5.35. A BLASTP search at NCBI showed that GsPPCK3 shared 90%, 84%, 69% and 61% sequence identity with the Glycine max GmPPCK3 (GeneBank Accession: NP_001238645), GmPPCK2 (NP_001236660), GmPPCK1 (NP_001241581) and GmPPCK4 (NP_001237016), respectively. It is worth noting that GsPPCK3 contained a 30 amino acid length sequence (KLLLASVFLFDIIFGGFCVDGIFGCFVFVG) at its C-terminus, which did not exist in any soybean and Arabidopsis PPCKs. By comparing the DNA and mRNA sequences of GsPPCK3 and GmPPCK3, we found that this 30 aa sequence was encoded by the 90 bp length intron of GmPPCK3.
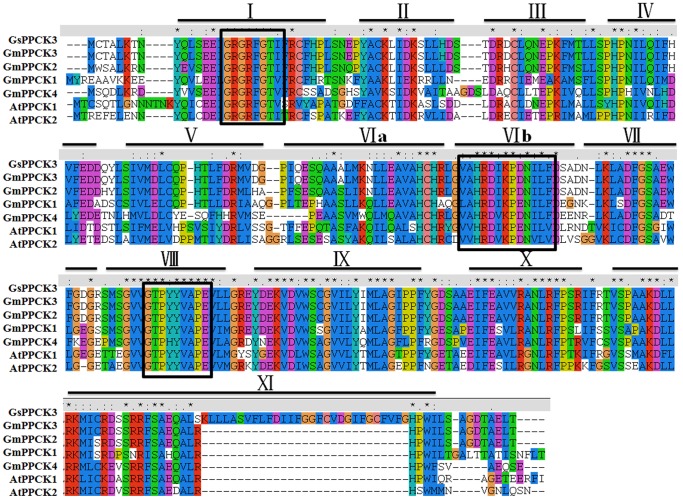
In order to get better understanding of GsPPCK3 structure, we compared the amino acid sequences of GsPPCK3 with PPCK homologs from Glycine max and Arabidopsis thaliana (AtPPCK1 and AtPPCK2) (Fig. 2). Protein sequence analysis revealed that GsPPCK3 comprised a minimal Ser/Thr kinase domain, closely related to the catalytic domain of plant calcium-dependent protein kinases (CDPKs) without the N- and C-terminal extensions. GsPPCK3 contained all of the 11 conserved subdomains required for kinase activity [24], [25] (Fig. 2), along with a protein kinase ATP-binding signature (GxGxxG, residues 16–21) and a Ser/Thr kinase active site signature (VAHRDIKPDNILF, residues 128–140) [26]. Similar to other PPCKs, GsPPCK3 also contained a conserved G-T/S-XX-Y/F-X-APE motif in subdomain VIII, indicating that GsPPCK3 was a potential Ser/Thr kinase rather than a Tyr kinase [27]. Second structure prediction revealed that GsPPCK3 contained two transmembrane domains at the C-terminus (residues 189–207, and residues 263–289), implying the potential membrane localization.
Figure 2. Multiple alignment between GsPPCK3 and homologous PPCKs from Arabidopsis and soybean based on the full-length amino acid sequences.
The 11 subdomains of the catalytic domain were marked as solid lines. The protein kinase ATP-binding signature (GxGxxG), the Ser/Thr kinase active site signature (VAHRDIKPDNILF) and the conserved G-T/S-XX-Y/F-X-APE motif were marked as black solid boxes. Sequences were aligned by using ClustalW, and gaps were introduced to maximize alignment.
Expression Patterns of GsPPCK3 in Response to Alkaline Stress and in Different Tissues
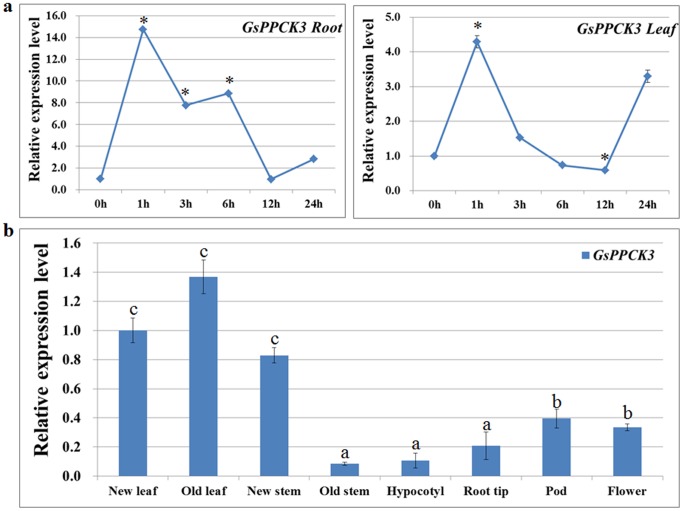
In an attempt to verify the induced expression of GsPPCK3 under alkali stress, we carried out the quantitative real-time PCR analyses. Consistent with the microarray data, GsPPCK3 expression in both leaves and roots of Glycine soja seedlings was greatly and rapidly induced by alkali stress (Fig. 3A). After 50 mM NaHCO3 (pH8.5) treatment, GsPPCK3 displayed an obvious increase and reached a maximum point at 1 h. It is noteworthy that the induction degree of GsPPCK3 in roots was significantly higher than that in leaves. The possible reasons for the stronger response might be that plant roots were the exact sites of perception and injury for stresses, or there were different response mechanisms for GsPPCK3 expression between roots and leaves. Anyway, it is obvious that GsPPCK3 expression was greatly induced by alkali stress, suggesting an important role of GsPPCK3 in plant response to alkali stress.
Figure 3. Expression patterns of GsPPCK3 in Glycine soja.
a. Expression levels of GsPPCK3 were up-regulated by alkali stress in both roots and leaves. Total RNA was extracted from leaves and roots of the 3-week-old Glycine soja seedlings treated with 50 mM NaHCO3 (pH 8.5) for the indicated time points, respectively. Relative transcript levels were determined by quantitative real-time PCR analysis with GAPDH as an internal control. The mean values from three fully independent biological repeats and three technical repeats were shown. *P<0.05; **P<0.01 by Student’s t-test. b. Tissue expression specificity of GsPPCK3 in Glycine soja. Total RNA was extracted from different tissues of Glycine soja seedlings. Significant differences (P<0.05 by Duncan’s Multiple Range Test) were indicated by different lowercase letters.
In order to get better understanding of GsPPCK3 expression patterns, we further investigated the spatial specific expression of GsPPCK3 in Glycine soja seedlings (Fig. 3B). The real-time PCR results showed that, among the eight tissues detected in this study, GsPPCK3 displayed the highest expression level in leaves, which could be explained by the important role of GsPPCK3 in photosynthesis. Contrarily, the GsPPCK3 transcript level in roots was relatively lower, even though it exhibited a greater degree of alkali stress induction in roots. This difference led us to propose the possibility of different mechanisms and different roles of GsPPCK3 between roots and leaves in plant response to alkali stress.
GsPPCK3 Overexpression Confers Enhanced Alkaline Tolerance in Transgenic Alfalfa
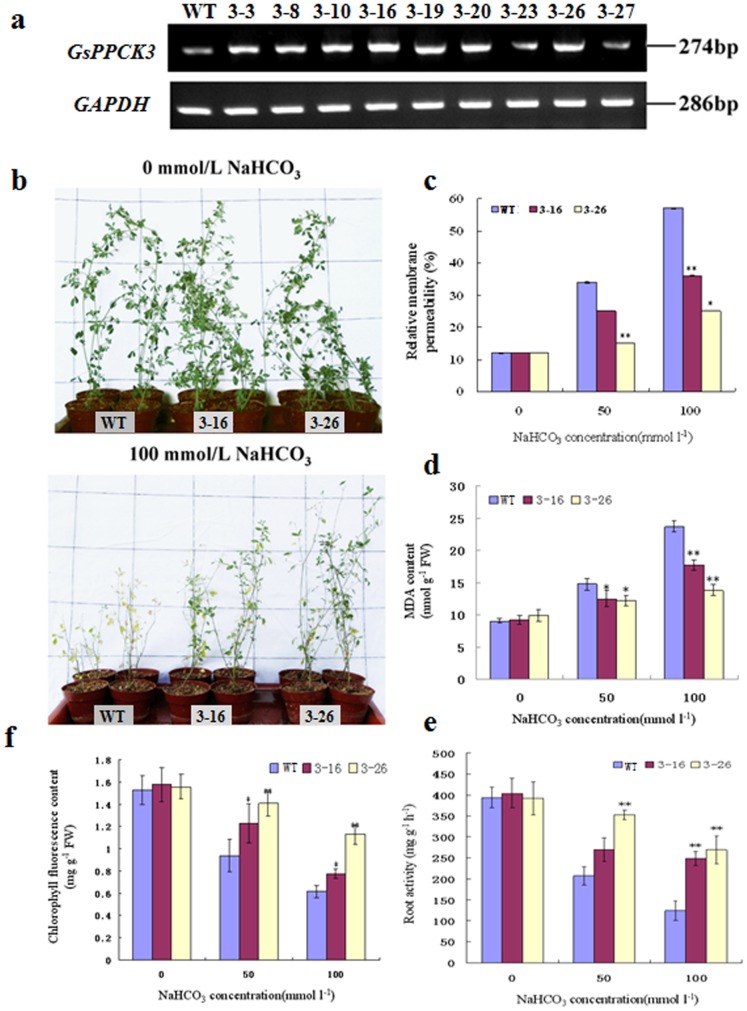
The stress induced expression of GsPPCK3 gave us an insight into its potential role in response to alkali stress. To precisely investigate the biological and physiological function of GsPPCK3 under alkaline stress, we transformed GsPPCK3 into the wild type (WT) M. Sativa through the Agrobacterium tumefaciens-mediated transformation strategy (Figure S1A). By using PCR and semi-quantitative RT-PCR analyses, we identified a total of nine transgenic lines (Fig. 4A), and two of them, with relatively higher expression levels (3–16 and 3–26), were selected to examine the response to alkaline stress.
Figure 4. GsPPCK3 overexpression in alfalfa conferred enhanced alkaline tolerance.
a. Semi-quantitative RT-PCR identification of GsPPCK3 transgenic alfalfa. b. Growth performance of WT and transgenic lines under control conditions or 100 mM NaHCO3 treatment. Photographs were taken 12 days after initial treatment. c. The relative membrane permeability of WT and transgenic plants. d. The MDA content of WT and transgenic plants. e. The root activity of WT and transgenic plants. f. The total chlorophyll content of WT and transgenic plants. For phenotypic analysis of GsPPCK3 transgenic alfalfa, the 3-week-old WT and GsPPCK3 transgenic plants with similar sizes were treated with 1/8 Hoagland nutrient solution containing either 0, or 50, or 100 mM NaHCO3 every 3 days for a total of 12 days.
Under control conditions, transgenic lines showed no obvious differences in growth performance compared with WT. After 50 or 100 mM NaHCO3 treatment for 14 d, both the WT and transgenic lines showed growth retardation in a dose-dependent manner. However, the growth inhibition of transgenic lines was less severe than that of WT (Fig. 4B). In details, WT plants exhibited severe chlorosis and even death, whereas the transgenic lines maintained continuous growth after 100 mM NaHCO3 treatment.
We further compared the relative membrane permeability (Fig. 4C), MDA content (Fig. 4D), chlorophyll content (Fig. 4E) and root activity (Fig. 4F) of WT and GsPPCK3 transgenic lines, respectively. No obvious differences were observed between WT and transgenic lines under control condition. As expected, alkali stress significantly increased the relative membrane permeability and MDA content, but decreased the total chlorohpyll content and root activity of both WT and transgenic plants. However, the transgenic lines showed relatively lower levels of electrolyte leakage and MDA content (Fig. 4C, D), but higher levels of total chlorohpyll content and root activity than WT (Fig. 4E, F) (*P<0.05; **P<0.01 by Student’s t-test). Taken together, these results strongly suggested that GsPPCK3 overexpression improved the alkali tolerance of transgenic alfalfa.
Increased Alkaline Tolerance of GsPPCK3-SCMRP Overexpression Transgenic Alfalfa
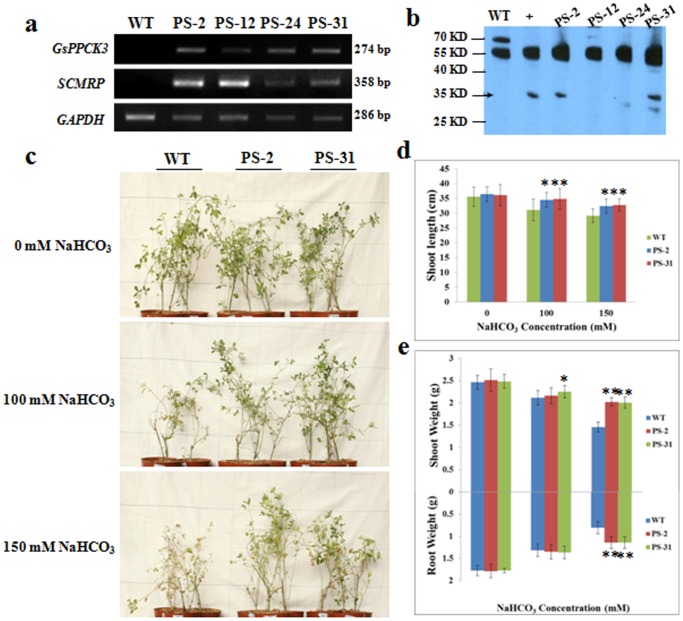
Considering the deficiency in sulfur-containing amino acids, we then co-transformed the GsPPCK3 and SCMRP genes into M. Sativa (Fig. S1B), in an attempt to obtain the transgenic alfalfa with not only higher stress tolerance but also higher methionine content. We identified a total of four transgenic lines by using PCR and semi-quantitative RT-PCR assays (Fig. 5A). Based on western blot analysis, two transgenic lines (PS-2 and PS-31) were selected for further studies (Fig. 5B).
Figure 5. Increased alkaline tolerance of GsPPCK3-SCMRP overexpression transgenic alfalfa.
a. Semi-quantitative RT-PCR analysis of GsPPCK3 and SCMRP expression levels in WT and trangenic lines. b. Western blot identification of GsPPCK3-SCMRP transgenic alfalfa. c. Growth performance of WT and transgenic lines after alkali treatment. Photographs were taken 15 days after initial treatment. d. Shoot length of the WT and transgenic lines. e. Shoot weight and root weight of the WT and transgenic lines. For phenotypic analysis of GsPPCK3-SCMRP transgenic alfalfa, the 4-week-old plants were treated with 1/8 Hoagland nutrient solution containing either 0, or 100, or 150 mM NaHCO3 every 3 days for a total of 15 days.
We firstly compared the alkali tolerance between WT and GsPPCK3-SCMRP transgenic alfalfa. As shown in Fig. 5C, alkali stress obviously inhibited the growth of both WT and transgenic plants; however, transgenic lines displayed significantly taller (Fig. 5D) and more biomass accumulation than WT (Fig. 5E). Specifically, in the presence of 150 mM NaHCO3, the shoot length was 29.14 cm for WT, 32.31 cm for PS-2 and 32.72 cm for PS-31, and the shoot/root weight was 1.46/0.81 g for WT, 1.91/1.08 g for PS-2 and 1.9/1.08 g for PS-31, respectively (*P<0.05; **P<0.01 by Student’s t-test). These results suggested that GsPPCK3 and SCMRP co-transformation promoted seedling growth of the transgenic alfalfa under alkali stress.
Improvement of PEPC Activity, Photosynthetic Rate and Citric Acid Content in Transgenic Alfalfa
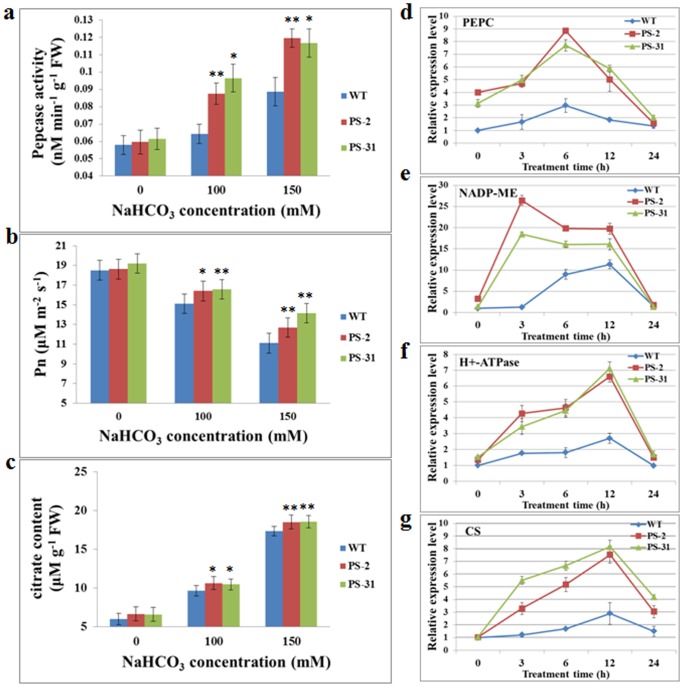
To further elucidate the influence of GsPPCK3 overexpression in alfalfa, PEPC activity, which was regulated by PPCK phosphorylation [28], [29], was determined by measuring the seedling crude extracts. As shown in Fig. 6A, alkaline stress improved the PEPC activity in both WT and transgenic lines, however, an obvious up-regulation of PEPC activity was observed in the transgenic lines. Specifically speaking, PEPC activity of the transgenic lines was 31.64% (PS-2) and 34.78% (PS-31) higher than that of WT, respectively (*P<0.05; **P<0.01 by Student’s t-test).
Figure 6. GsPPCK3 overexpression altered a set of physiological indices and expression levels of stress responsive genes.
a. The PEPC activity of the WT and GsPPCK3-SCMRP transgenic lines under alkali treatment. b. The net photosynthetic rate of the WT and GsPPCK3-SCMRP transgenic lines. c. The citric acid content of the WT and transgenic lines. d. Increased expression levels of PEPC in GsPPCK3-SCMRP transgenic plants under alkali stress (50 mM NaHCO3, pH 8.5). e. Increased expression levels of NADP-ME in GsPPCK3-SCMRP transgenic plants. f. Increased expression levels of H+-ATPase in GsPPCK3-SCMRP transgenic plants. g. Increased expression levels of CS in GsPPCK3-SCMRP transgenic plants. To explore expression patterns of stress-responsive genes, the 4-week-old WT and GsPPCK3-SCMRP transgenic seedlings after shoot cottage were treated with 50 mM NaHCO3 (pH 8.5) for 0, 3, 6, 12 and 24 h, respectively. Relative transcript levels were determined by quantitative real-time PCR with GAPDH as internal reference, and were normalized to WT plants at 0 h. Values represented the means of three fully independent biological replicates, and three technological replicates for each.
It has been suggested that photosynthetic rate decreased proportionally, along with the increase of NaHCO3 concentration [30]. Considering the function of PEPC/PPCK in photosynthesis, we measured the net photosynthesis (Pn) rates of both WT and transgenic lines under alkali stress. The quantitative analysis revealed that Pn of transgenic alfalfa was significantly higher than that of WT, in the presence of 100 or 150 mM NaHCO3 (Fig. 6B) (*P<0.05; **P<0.01 by Student’s t-test). These results suggested that overexpression of GsPPCK3 led to an obvious increase of Pn under alkali stress, maybe by activating PEPCs.
Citric acid content was considered to be an indicator of plant response to pH challenge resulting from alkaline stress [31]. As expected, plants accumulated more citric acids under alkali stress than under control conditions. Compared with WT, the transgenic lines exhibited significantly higher levels of citric acid content, after 100 or 150 mM NaHCO3 treatment (Fig. 6C) (*P<0.05; **P<0.01 by Student’s t-test). Collectively, these results demonstrated that the alleviation of high pH damage caused by alkali stress in transgenic alfalfa might be related to the elevated levels of PEPC activity, photosynthetic rate and citric acid content, due to GsPPKC3 overexpression.
GsPPCK3 Overexpression Up-regulated the Expression Levels of Several Stress Responsive Genes
Previous studies showed that H+-ATPase and NADP-ME maintained the homeostasis of the cytosolic pH value [32], [33]. In view of the increased PEPC activity and citric acid content of transgenic alfalfa, we examined the expression patterns of PEPC, CS (Citrate synthase), H+-ATPase and NADP-ME after 100 mM NaHCO3 treatment. The real-time PCR analysis indicated that expression of PEPC, CS, H+-ATPase and NADP-ME was induced by alkali stress in both WT and transgenic lines. Expectedly, the expression levels in transgenic plants were significantly higher than that in WT (Fig. 6D-G), which explained the up-regulation of the PEPC activity and citrate acid content. These results implied that GsPPCK3 overexpression promoted the accumulation of transcript expression levels of the stress responsive genes, which might be helpful for the intracellular pH regulation under alkali stress.
Increased Methionine Content in Leaves of GsPPCK3-SCMRP Transgenic Alfalfa
In addition to the enhanced alkali tolerance, we also determined the content of 16 amino acids in leaves of both WT and transgenic plants, to verify if GsPPCK3-SCMRP co-transformation increased the methionine content (Table 1). As expected, GsPPCK3-SCMRP transgenic lines displayed significantly higher levels of methionine content than WT (*P<0.05; **P<0.01 by Student’s t-test), without obvious changes for other amino acids. Specifically speaking, the methionine contents were 0.97±0.04% in WT plants, 2.27±0.89% (2.34 folds to WT) in transgenic line PS-2 and 2.23±0.67% (2.29 folds) in line PS-31. Taken together, these results suggested that GsPPCK3-SCMRP co-overexpression not only enhanced the alkaline tolerance, but also increased the methionine content of transgenic alfalfa.
Table 1. Amino acids content of the WT and GsPPCK3-SCMRP transgenic alfalfa.
| Amino acid | WT | Content (%) PS-2 | PS-31 |
| Asp | 6.91±0.39 | 6.84±0.32 | 7.38±0.51 |
| Thr | 6.48±0.00 | 6.27±0.06 | 6.40±0.06 |
| Ser | 3.65±0.05 | 3.58±0.01 | 3.68±0.10 |
| Glu | 8.74±0.09 | 8.69±0.01 | 8.76±0.11 |
| Gly | 19.28±0.20 | 17.53±0.37 | 18.76±0.83 |
| Ala | 9.40±0.07 | 9.28±0.02 | 9.38±0.27 |
| Cys | 3.88±0.05 | 3.99±0.08 | 3.77±0.09 |
| Met | 0.97±0.04 | 2.27±0.89 ** | 2.23±0.67 * |
| Ile | 4.16±0.10 | 4.17±0.14 | 3.96±0.06 |
| Leu | 8.38±0.10 | 8.49±0.18 | 8.13±0.24 |
| Tyr | 1.53±0.27 | 1.38±0.28 | 1.14±0.19 |
| Lys | 6.98±0.10 | 7.18±0.16 | 6.75±0.06 |
| NH3 | 1.62±0.16 | 1.80±0.14 | 1.82±0.29 |
| His | 3.49±0.09 | 3.55±0.16 | 3.58±0.07 |
| Arg | 4.96±0.03 | 5.00±0.07 | 4.86±0.17 |
| Pro | 9.51±0.01 | 9.98±0.08 | 9.41±0.24 |
*P<0.05;
**P<0.01 by Student’s t-test.
Discussion
Saline-alkaline stress, as a kind of widespread environmental stress with significantly negative impact on plant growth, severely reduces crop productivity and affects agricultural production worldwide. It has been suggested that alkaline soil, characterized by high NaHCO3/Na2CO3 content, caused injury to plants not only through salt stress, but also through alkali stress [34]–[36]. Plants could homeostatically maintain the intracellular pH value and ion concentration in a temperate range [32], [37]–[39]. Unfortunately, up to now, most studies emphasized on salt stress [40]–[43], and no slightly definite mechanism was proposed about plant response to alkaline stress.
Glycine soja, the wild ancestor of cultivated soybean (Glycine max), could normally germinate and grow in the alkaline soil with a pH value at 9.02 [44]. In previous studies, we constructed the global transcriptional profile of Glycine soja (G07256) under alkali stress (50 mM NaHCO3, pH 8.5), and three of the four PPCK genes GsPPCK1, GsPPCK2 and GsPPCK3 were identified as putative stress responsive genes (Fig. 1) [23]. Contrarily, GsPPCK4 essentially did not respond to alkali stress, only with an increase at 6 h in leaves. Consistent with previous researches, these results further supported the evolutionary relationship of soybean PPCKs [14], [45]. PPCK1, PPCK2 and PPCK3 shared a high similarity and belonged to the same legume PPCK subfamily, while PPCK4 represented a high divergence, outlier to the legume PPCK.
In a previous study, we isolated and characterized one of the alkali stress responsive PPCK genes GsPPCK1, and found that overexpression of GsPPCK1 in alfalfa significantly improved plant tolerance to alkali stress [46]. In the present study, we focused on the expression pattern and biological function of GsPPCK3. We cloned the full length GsPPCK3, and found that it shared 90%, 84%, 69% and 61% sequence identity with GmPPCK3, GmPPCK2, GmPPCK1 and GmPPCK4, respectively. Protein sequence analysis revealed a unique motif consisting of 30 amino acids in subdomain XI of GsPPCK3, which was not found in any soybean and Arabidopsis PPCKs. This unique sequence was encoded by the 90 bp length intron of GmPPCK3, indicating the existence of different transcripts in Glycine soja. Furthermore, GsPPCK3 protein contained all conserved subdomains required for kinase activity [24], [25] (Fig. 2), including a protein kinase ATP-binding signature, a Ser/Thr kinase active site signature [26] and a conserved G-T/S-XX-Y/F-X-APE motif, as well as two transmembrane domains.
GsPPCK3 expression was greatly and rapidly induced by alkali stress in both leaves and roots (Fig. 3A). Previous researches also suggested the induced expression of PPCKs under environmental stress. For example, salt stress significantly increased the transcript accumulation of SbPPCK1 and SbPPCK2 in sorghum [17]. Moreover, GsPPCK1 was also induced by alkali stress and positively regulated the alkali tolerance of transgenic alfalfa [46]. Therefore, the alkali induced expression indicated an important role of GsPPCK3 in plant response to alkali stress. It is worth noting that the stress induction degree of GsPPCK3 in roots was significantly higher than that in leaves. It is possible that plant roots were the exact sites of perception and injury for stresses, or there were different response mechanisms for GsPPCK3 expression between roots and leaves.
It has been reported that PPCK genes showed spatial expression specificity. For example, OsPPCK1 and OsPPCK3 displayed obviously high expression levels in roots [13], while GmPPCK2 and GmPPCK3 showed high transcription levels in root nodules [14]. Except for root nodules, GmPPCK3 also showed relatively higher expression levels in stems, flowers and young leaves. In this study, we found that GsPPCK3 displayed the highest level in leaves but relatively lower in roots (Fig. 3B). As we know, PEPC undergoes an important function in carbon fixation during photosynthesis, and its activity was largely regulated by PPCK phosphorylation [28], [29]. It is reasonable to speculate that the high levels of GsPPCK3 transcripts in leaves could make sure of the high PEPC activity and thereby the effective photosynthesis of plant leaves. Considering the difference in the transcript levels and alkali induction degrees of GsPPCK3 between roots and leaves, one could speculate that in leaves, most GsPPCK3 products were used for carbon fixation in photosynthesis process, but in roots, GsPPCK3 mainly functioned in alkali response.
To further confirm the biological function of GsPPCK3 in alkali response, we transformed GsPPCK3 into alfalfa and carried out the alkali tolerance assays. We gave four lines of direct evidence showing the increased tolerance and possible mechanisms of GsPPCK3 transgenic alfalfa in response to alkali stress (Fig. 4, 5). Firstly, GsPPCK3 overexpression significantly promoted plant growth under alkali stress. GsPPCK3 transgenic lines displayed much better at plant height, shoot weight and root weight than WT under alkali stress (Fig. 5D, E). Secondly, GsPPCK3 overexpression alleviated the damage caused by alkali stress, as evidenced by an obvious decrease in the relative ion leakage (Fig. 4C) and MDA content (Fig. 4D), but an increase in root activity (Fig. 4E) [37], [47]–[49]. Thirdly, GsPPCK3 overexpression in alfalfa resulted in increased PEPC activity (Fig. 6A), increased chlorophyll content (Fig. 4F) and thereby the increased Pn rate (Fig. 6B), which in turn could promote plant growth under alkali stress. We also observed the higher expression levels of PEPC gene in transgenic alfalfa (Fig. 6D). In this context, we proposed the hypothesis that GsPPCK3 regulated the PEPC activity not only through protein phosphorylation process, but also through gene transcription regulation. Finally, overexpression of GsPPCK3 led to more effective response to high pH stress by increasing the citrate acid content (Fig. 6C) [50]. The up-regulation of CS gene expression (Fig. 6G) in transgenic plants explained the increase of citrate acid content [51], [52]. Meanwhile, we also suggested relatively higher expression level of H+-ATPase in transgenic lines under alkali stress (Fig. 6F), which was also helpful for intracellular pH regulation. Based on these results, we speculated that GsPPCK3 regulated the gene expression and enzyme activity involved in photosynthesis and pH regulation, alleviated damage caused by alkali stress, and finally promoted plant growth under alkali stress.
On the other hand, we also obtained the transgenic alfalfa with not only higher alkali tolerance but also higher methionine content, by co-transforming the GsPPCK3 and SCMRP genes. As a kind of nutritionally essential amino acid, methionine was found to be at low level in legume. In this study, we significantly improved the methionine content of transgenic alfalfa by ectopically expressing the SCMRP gene, which was designed and synthesized according to the maize methionine-rich 10 ku zein protein [20], [21]. Similarly, it has been reported that overexpression of the maize 10 ku zein gene in potato could increase the contents of sulphur-containing amino acids [22]. During the past decades, a series of efforts have been made to increase the methionine content by using genetic engineering methods, for example altering expression levels of the methionine-rich storage proteins [53]–[55], or increasing the soluble content of methionine [56]–[60].
Collectively, here we provided an effective way to simultaneously improve plant alkaline tolerance and methionine content, at least in legume crops. For the first time, we gave exact evidence for a PPCK protein from Glycine soja, and provided insights into a plausible mechanism by which GsPPCK3 positively controlled plant tolerance to alkali stress.
Materials and Methods
Plant Material and Growth Conditions
Glycine soja (G07256) seeds, obtained from Jilin Academy of Agricultural Sciences (Changchun, China), were treated with 98% sulfuric acid for 10 min, washed five times with sterile water, and then kept in complete darkness with humidity for 2–3 days to promote germination. The seedlings were transferred and grown in 1/4 Hoagland solution for 3 weeks at 24–26°C and 16 h light/8 h dark cycles. To explore the expression profiles of GsPPCK3 under alkali stress, the 3-week-old seedlings were treated with 1/4 Hoagland solution containing 50 mM NaHCO3 (pH 8.5) for 0, 1, 3, 6, 12 and 24 h, respectively. Equal amounts of leaves and roots were harvested and stored at −80°C after snap-frozen in liquid nitrogen.
Medicago Sativa L. was kindly obtained from Heilongjiang Academy of Agricultural Sciences (Haerbin, China), and grown in green house under controlled environmental conditions (24–26°C, 16 h light/8 h dark cycles, 600 µmol m−2 s−1, 80±5% relative humidity). To investigate the expression patterns of stress-responsive genes, the 4-week-old seedlings after shoot cottage were treated with 1/4 Hoagland solution containing 50 mM NaHCO3 (pH 8.5) for 0, 3, 6, 12 and 24 h, respectively. Samples were harvested and stored as described above.
Isolation and Sequence Analysis of the GsPPCK3 Gene
Total RNA was extracted from the 3-week-old Glycine soja seedlings, by using RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), and subjected to cDNA synthesis by using SuperScript™ III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). The full-length coding region of GsPPCK3 was PCR amplified with gene specific primers (5′-AAGATAGAATGTGCACAGCCCTAAAG-3′ and 5′-TTCTCAAGTGAGTTCAGCCGTGTC-3′), and cloned into pGEM-T vector (Promega, Madison, WI, USA) for sequencing.
Sequence similarity was examined by using the on-line BLASTP program at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homology searches were executed by BLASTP at Phytozome (http://www.phytozome.net/soybean). TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html) were used to predict the transmembrane domains.
Quantitative Real-time PCR Analyses
Total RNA extraction and cDNA synthesis were operated as described above. Quantitative real-time PCR analyses were performed by using SYBR Premix ExTaq™ II Mix (TaKaRa, Shiga, Japan) on an ABI 7500 sequence detection system (Applied Biosystems, Carlsbad, CA, USA). The glyceraldehyde-3-phosphate dehydrogenase genes in G. soja (Accession: DQ355800) and M. sativa (Accession: Medtr3g085850) were used as internal references, respectively. Expression levels for all candidate genes were calculated by using the 2-ΔΔCT method, and the relative intensities were normalized as described previously [61]. To enable statistical analysis, three fully independent biological replicates and three technical repeats were conducted. Primers used for quantitative real-time PCR were designed using Primer 5 software and listed in Table 2.
Table 2. Gene-specific primers used for quantitative real-time PCR assays.
| Gene name | Gene ID | Primer Sequence (5′ to 3′) | PCR productsize (bp) |
| GsGAPDH | DQ355800 | Forward: GACTGGTATGGCATTCCGTGTReverse: GCCCTCTGATTCCTCCTTGA | 121 |
| GsPPCK3 | Forward: CGCAGAACAAGCCTTGAGTAAGReverse: CCACCACGAGTAGACCACCTT | 264 | |
| MtGAPDH | Medtr3g085850 | Forward: GTGGTGCCAAGAAGGTTGTTATReverse: CTGGGAATGATGTTGAAGGAAG | 286 |
| PEPC | L39371.2 | Forward: CATTGGCTCGGTTGTTCTCCReverse: TCTGTGCGACTTTGATGAGGTC | 159 |
| H+-ATPase | AJ132891 | Forward: GGCAGCCCTCTACCTACAAGTCReverse: AGCAATCATAAAAGCACCCAAT | 121 |
| NADP-MEhttp://www.ncbi.nlm.nih.gov/nucleotide/357520876?report = genbank&log$ = nucltop&blast_rank = 1&RID = 35MT7XMC01R | XM_003630679.1 | Forward: TAGGTGGAGTTCGTCCTTCAGCReverse: AGGTCATAGTATTCCTTCCCAGTTG | 133 |
| Citrate Synthase | HM030734.1 | Forward: TCTATATGGACCTCTTCATGGTGGReverse: TGAGCTTTCGTTTCCTGGCT | 122 |
Generation of Transgenic Alfalfa
In order to investigate the influence of GsPPCK3 and SCMRP on plant stress tolerance and methionine content, we constructed the expression vectors carrying the GsPPCK3 gene alone (Fig. S1A), and the GsPPCK3 and SCMRP genes together (Fig. S1B), respectively. The GsPPCK3 gene was inserted to the bone vector pBEOM (made in our lab) under the control of the cauliflower mosaic virus (CaMV) 35S promoter, with the binding enhancers E12 and omega. The SCMRP gene was under the control of double CaMV35S promoter and omega sequence. The Bar gene was used as the selectable marker. The recombinant vectors were introduced into A. tumefaciens strain EHA105, and then transformed into M. sativa by using the cotyledonary node method. The transformants were selected by using 0.5 mg L−1 glufosinate ammonium, and regenerated shoots were rooted on 1/2 Murashige and Skoog (MS) medium. At last, the glufosinate-positive seedlings were transplanted into soil and grown in green house under controlled conditions.
Presence of the GsPPCK3 and SCMRP genes in the glufosinate-positive plants was confirmed by PCR analysis using CaMV35S promoter specific forward primer and Bar gene specific reverse primer (5′-CCTGTGCCTCCAGGGAC-3′ and 5′-GCGGTCTGCACCATCGTC-3′). GsPPCK3 transcript levels in the PCR-positive plants were analyzed by semi-quantitative RT-PCR analysis using gene specific primers (5′-CCCTCCTTTCACCTCACC-3′ and 5′-GAACCGAAGTCCGCCAGT-3′). Expression levels of the SCMRP gene were examined by using a pair of specific primers (5′-CAGCAGGGTCTCGCTTCACT-3′ and 5′-GCAGATTCCAATGCCACAAT-3′). The alfalfa GAPDH gene was used as an internal control. And the PCR- and RT-PCR positive seedlings were subjected to western blot analysis with specific polyclonal antibody to the C-terminus of GsPPCK3 protein (CHPWILSAGDTAELT).
Phenotypic Analysis of Transgenic Alfalfa Under Alkali Stress
The lignified WT and transgenic alfalfa plants were propagated by stem cuttings, and about 2 weeks later, adventitious roots appeared. The seedlings were transplanted into plastic culture pots filled with a mixture of peat moss: soil (1∶1; v/v), irrigated with 1/8 Hoagland nutrient solution and grown in green house under controlled conditions. For phenotypic analysis of GsPPCK3 transgenic alfalfa, the 3-week-old WT and transgenic plants with similar sizes were exposed to alkali stress by irrigating with 1/8 Hoagland solution containing either 0, or 50, or 100 mM NaHCO3 every 3 days for a total of 12 days. For phenotypic analysis of GsPPCK3-SCMRP transgenic alfalfa, the 4-week-old plants were exposed to alkali stress by irrigating with 1/8 Hoagland solution containing either 0, or 100, or 150 mM NaHCO3 every 3 days for 15 days.
The total chlorophyll content was determined in 80% (v/v) acetone extract according to the method of Arnon [62]. The relative electrolyte leakage was measured using a conductivity meter (DDSJ-308A, Shanghai, China) as described previously [63]. The malon dialdehyde (MDA) content was determined according to the protocol described by Peever et al. [64]. The biological activity of roots from alkali-treated WT and transgenic plants was assessed by measuring root dehydrogenase activity using triphenyltetrazolium chloride (TTC) reduction technique as described [65]. The citric acid content was determined by using a spectrophotometer (UV-2550, Shimadzu, Japan) at the absorbance of 490 nm, according to the method of Zhu [66].
The net photosynthesis rate was determined by using the LI-6400 chamber (LI-COR Biosciences, Lincoln, NE, USA). During the experiments, the light intensity was 500 µmol photons m−2 s−1, and the CO2 concentration was in the range of 350 to 400 µmol mol−1. The flow rate was adjusted to 500 µmol s−1, and the leaf temperature was maintained at 25°C. For statistical analysis, three independent experiments were conducted with at least 5 independent plants per genotype and experiment.
PEPC activity was measured by the coupled spectrophotometric method at 340 nm and 30°C as previously described by Gonzalez [67]. Total proteins of the WT and OX alfalfa leaves were extracted by using the 0.1 M Tris-HCl (pH 7.5) solution, containing 20% (v/v) glycerol, 1 mM EDTA, 10 mM MgCl2, 10 µg mL−1 chymostatin, 10 µg mL−1 bestatin, 10 µg mL−1 leupeptin, 1 mM PMSF, 1 µg mL−1 microcystin-L/R (L and R are two variable amino acids in the structure of microcystin), and 14 mM β-mercaptoethanol. Protein concentration was determined according to the method of Bradford (1976) [68]. All of the above numerical data were subjected to statistical analyses using EXCEL 2007 and/or IBM SPSS statistics 19, and analyzed by Student’s T-test and/or Duncan’s Multiple Range Test.
Determination of Amino Acid Content
Leaves of WT and transgenic alfalfa were fixated at 105°C for 15 min, and then dried to constant weight at 80°C. About 100 mg dried leaves were crushed, suspended in 6 M HCl, and then hydrolyzed at 110°C for 22 h with nitrogen replacement. The hydrolysate was dried under reduced pressure, dissolved in 0.02 M HCl, and subjected to the amino acid analyzer (Hitachi, Japan).
Supporting Information
Schematic representation of the constructs for Agrobacterium tumefaciens-mediated transformation into Medicago sativa. a, Schematic representation of the GsPPCK3 overexpression construct. b, Schematic representation of the GsPPCK3-SCMRP overexpression construct.
(TIF)
Acknowledgments
We thank members of the lab for discussions and comments on this manuscript. We thank Prof. Wei Ding of Northeast Agricultural University (Heilongjiang, China) for technical assistance on analyzing the net photosynthesis rate.
Funding Statement
This work was supported by Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005); National Natural Science Foundation of China (31171578); National Major Project for Cultivation of Transgenic Crops (2011ZX08004-002); Scientific Research Foundation of Graduate School of Heilongjiang Province (YJSCX2012-047HLJ) and National Science Foundation for Fostering Talents in Basic Research of China (J1210069). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chang S, Liu N, Wang X, Zhang Y, Xie Y (2012) Alfalfa carbon and nitrogen sequestration patterns and effects of temperature and precipitation in three agro-pastoral ecotones of northern China. PLoS ONE 7: e50544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagavathiannan MV, Begg GS, Gulden RH, Van Acker RC (2012) Modelling the dynamics of feral alfalfa populations and its management implications. PLoS ONE 7: e39440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Postnikova OA, Shao J, Nemchinov LG (2013) Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54: 1041–1055. [DOI] [PubMed] [Google Scholar]
- 4. Jin H, Plaha P, Park JY, Hong CP, Lee IS, et al. (2006) Comparative EST profiles of leaf and root of Leymus chinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci 170: 1081–1086. [Google Scholar]
- 5. Wang Y, Ma H, Liu G, Xu C, Zhang D, et al. (2008) Analysis of gene expression profile of Limonium bicolor under NaHCO3 stress using cDNA microarray. Plant Mol Biol Rep 26: 241–254. [Google Scholar]
- 6. Kawanabe S, Zhu TC (1991) Degeneration and conservation of Aneurolepisium chinense grassland in northern China. Journal of Japan Grassland Science 37: 91–99. [Google Scholar]
- 7. Vestin JLK, Nambu K, van Hees PAW, Bylund D, Lundström US (2006) The influence of alkaline and non-alkaline parent material on soil chemistry. Geoderma 135: 97–106. [Google Scholar]
- 8. Xu W, Jia L, Baluška F, Ding G, Shi W, et al. (2012) PIN2 is required for the adaptation of Arabidopsis roots to alkaline stress by modulating proton secretion. J Exp Bot 63: 6105–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guillet C, Aboul-Soud MAM, Le Menn A, Viron N, Pribat A, et al. (2012) Regulation of the fruit-specific PEP carboxylase SlPPC2 promoter at early stages of tomato fruit development. PLoS ONE 7: e36795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu J, Fan X, Zhang X, Xu D, Mou S, et al. (2012) Evidence of coexistence of C3 and C4 photosynthetic pathways in a green-tide-forming alga, Ulva prolifera . PLoS ONE 7: e37438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chollet R, Vidal J, O’Leary M (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Ann Rev Plant Physiol Mol Bio 47: 273–298. [DOI] [PubMed] [Google Scholar]
- 12. Chen ZH, Jenkins GI, Nimmo HG (2008) pH and carbon supply control the expression of phosphoenolpyruvate carboxylase kinase genes in Arabidopsis thaliana . Plant Cell Environ 31: 1844–1850. [DOI] [PubMed] [Google Scholar]
- 13. Fukayama H, Tamai T, Taniguchi Y, Sullivan S, Miyao M, et al. (2006) Characterization and functional analysis of phosphoenol pyruvate carboxylase kinase genes in rice. Plant J 47: 258–268. [DOI] [PubMed] [Google Scholar]
- 14. Xu W, Sato SJ, Clemente TE, Chollet R (2007) The PEP-carboxylase kinase gene family in Glycine max (GmPpcK1–4): an in-depth molecular analysis with nodulated, non-transgenic and transgenic plants. Plant J 49: 910–923. [DOI] [PubMed] [Google Scholar]
- 15. Peng Y, Cai J, Wang W, Su B (2012) Multiple inter-kingdom horizontal gene transfers in the evolution of the phosphoenolpyruvate carboxylase gene family. PLoS ONE 7: e51159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Mauriño S, Monreal JA, Alvarez R, Vidal J, Echevarría C (2003) Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 216: 648–655. [DOI] [PubMed] [Google Scholar]
- 17. Monreal JA, Arias-Baldrich C, Pérez-Montaño F, Gandullo J, Echevarría C, et al. (2013) Factors involved in the rise of phosphoenolpyruvate carboxylase-kinase activity caused by salinity in sorghum leaves. Planta 237: 1401–1413. [DOI] [PubMed] [Google Scholar]
- 18. Wang HM, Wang WJ, Wang HZ, Wang Y, Xu HN, et al. (2013) Effect of inland salt-alkaline stress on C4 enzymes, pigments, antioxidant enzymes, and photosynthesis in leaf, bark, and branch chlorenchyma of poplars. Photosynthetica 51: 115–126. [Google Scholar]
- 19. Avraham T, Badani H, Galili S, Amir R (2005) Enhanced levels of methionine and cysteine in transgenic alfalfa (Medicago stativa L.) plants overexpressing the Arabidopsis cystathionine γ-synthase gene. Plant Biotechnol J 3: 71–79. [DOI] [PubMed] [Google Scholar]
- 20. Zhai H, Bai X, Zhu YM, Chen XH (2009) Protokaryotic expression of SCMRP gene and preparation of polyclonal antibody. Journal of Northeast Agricultural University 40: 60–65. [Google Scholar]
- 21. Kirihara JA, Petri JB, Messing J (1988) Isolation and sequence of a gene encoding a methionine-rich 10 ku zein protein from maize. Gene 2: 359–370. [DOI] [PubMed] [Google Scholar]
- 22. Li L, Liu S, Hu Y, Zhao W, Lin Z (2001) Increase of sulphur-containing amino acids in transgenic potato with 10 ku zein gene from maize. Chinese Sci Bull 46: 482–484. [Google Scholar]
- 23. Ge Y, Li Y, Zhu YM, Bai X, Lv DK, et al. (2010) Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol 10: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanks SK, Hunter T (1995) Protein kinases 6: the eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596. [PubMed] [Google Scholar]
- 25. Yang T, Chaudhuri S, Yang L, Chen Y, Poovaiah BW (2004) Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J Biol Chem 279: 42552–42559. [DOI] [PubMed] [Google Scholar]
- 26. Hardie DG (1999) Plant protein serine/threonine kinases: classification and function. Annu Rev Plant Physiol Mol Biol 50: 97–131. [DOI] [PubMed] [Google Scholar]
- 27. Stone JM, Walker JC (1995) Plant protein kinase families and signal transduction. Plant Physiol 108: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vidala J, Cholletb R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230–237. [Google Scholar]
- 29. Bakrim N, Prioul JL, Deleens E, Rocher JP, Arrio-Dupont M, et al. (1993) Regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase (A cardinal event influencing the photosynthesis rate in Sorghum and Maize). Plant Physiol 101: 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Melack JM, Kilham P (1974) Photosynthetic rates of phytoplankton in East African alkaline, saline lakes. Limnol Oceanogr 19: 743–755. [Google Scholar]
- 31. Hughes JA, West NX, Parker DM, van den Braak MH, Addy M (2000) Effects of pH and concentration of citric, malic and lactic acids on enamel, in vitro. J Dent 28: 147–152. [DOI] [PubMed] [Google Scholar]
- 32. I Kurtz (1987) Apical Na+/H+ antiporter and glycolysis-dependent H+-ATPase regulate intracellular pH in the rabbit S3 proximal tubule. J Clin Invest 80: 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martinoia E, Rentsch D (1994) Malate compartmentation-responses to a complex metabolism. Ann Rev Plant Physiol Mol Bio 45: 447–467. [Google Scholar]
- 34. Li RL, Shi FC, Fukuda K (2010) Interactive effects of various salt and alkali stresses on growth, organic solutes, and cation accumulation in a halophyte Spartina alterniflora (Poaceae). Environ Exp Bot 68: 66–74. [Google Scholar]
- 35. Li RL, Shi FC, Fukuda K (2010) Interactive effects of salt and alkali stresses on seed germination, germination recovery, and seedling growth of a halophyte Spartina alterniflora (Poaceae). South Afr J Bot 76: 380–387. [Google Scholar]
- 36. Shi DC, Wang DL (2005) Effects of various salt-alkali mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant Soil 271: 15–26. [Google Scholar]
- 37. Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445. [DOI] [PubMed] [Google Scholar]
- 38. Oh DH, Lee SY, Bressan RA, Yun DJ, Bohnert HJ (2010) Intracellular consequences of SOS1 deficiency during salt stress. J Exp Bot 61: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H, Wu Z, Han J, Zheng W, Yang C (2012) Comparison of ion balance and nitrogen metabolism in old and young leaves of alkali-stressed rice plants. PLoS ONE 7: e37817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681. [DOI] [PubMed] [Google Scholar]
- 41. Charkazi F, Ramezanpour SS, Soltanloo H (2010) Expression pattern of two sugar transporter genes (SuT4 and SuT5) under salt stress in wheat. Plant Omics J 3: 194–198. [Google Scholar]
- 42. Jemâa E, Saïda A, Sadok B (2011) Impact of indole-3-butyric acid and indole-3-acetic acid on the lateral roots growth of Arabidopsis under salt stress conditions. AJAE 2: 18–24. [Google Scholar]
- 43. Ibraheem O, Dealtry G, Roux S, Bradley G (2011) The effect of drought and salinity on the expressional levels of sucrose transporters in rice (Oryza sativa Nipponbare) cultivar plants. Plant Omics J 4: 68–74. [Google Scholar]
- 44. Ge Y, Zhu YM, Lv DK, Dong TT, Wang WS, et al. (2009) Research on responses of wild soybean to alkaline stress. Pratacultural Science 26: 47–52. [Google Scholar]
- 45. Sullivan S, Jenkins GI, Nimmo HG (2004) Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiol 135: 2078–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei ZW, Zhu YM, Hua Ye, Cai H, Ji W, et al. (2013) Transgenic alfalfa with GsPPCK1 and its alkaline tolerance analysis. Acta Agro Sin 39: 68–75. [Google Scholar]
- 47. Long Y, Kong D, Chen Z, Zeng H (2013) Variation of the linkage of root function with root branch order. PLoS ONE 8: e57153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399. [DOI] [PubMed] [Google Scholar]
- 49. Simon EW (1974) Phospholipids and plant membrane permeability. New Phytol 73: 377–420. [Google Scholar]
- 50. Kooten O, Snel JFH (1990) The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth Res 25: 147–150. [DOI] [PubMed] [Google Scholar]
- 51. Shi DC, Yin SJ, Yang GH, Zhao KF (2002) Citric acid accumulation in an alkali-tolerant plant Puccinellia tenuiflora under alkaline stress. Acta Botanica Sinica 44: 537–540. [Google Scholar]
- 52. Zhang X, Wei L, Wang Z, Wang T (2013) Physiological and molecular features of Puccinellia tenuiflora tolerating salt and alkaline-salt stress. J Integr Plant Biol 55: 262–276. [DOI] [PubMed] [Google Scholar]
- 53. Lee TT, Hou RC, Chen LJ, Su RC, Wang CS, et al. (2003) Enhanced methionine and cysteine levels in transgenic rice seeds by the accumulation of sesame 2S albumin. Biosci Biotechnol Biochem 67: 1699–1705. [DOI] [PubMed] [Google Scholar]
- 54. Lee M, Toro-Ramos T, Fraga M, Last RL, Jander G (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54: 310–320. [DOI] [PubMed] [Google Scholar]
- 55. Liu HY, Quampah A, Chen JH, Li JR, Huang ZR, et al. (2013) QTL mapping based on different genetic systems for essential amino acid contents in cottonseeds in different environments. PLoS ONE 8: e57531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tabe L, Molvig L, Droux M, Hell R (2010) Overexpression of serine acetlytransferase produced large increases in O-acetylserine and free cysteine in developing seeds of a grain legume. J Exp Bot 61: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hacham Y, Mattiyaho I, Schuster G, Amir R (2008) Overexpression of mutated forms of aspartate kinase and cystathionine γ-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. Plant J 54: 260–271. [DOI] [PubMed] [Google Scholar]
- 58. Peñagaricano F, Souza AH, Carvalho PD, Driver AM, Gambra R, et al. (2013) Effect of maternal methionine supplementation on the transcriptome of bovine preimplantation embryos. PLoS ONE 8: e72302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanafy MS, Rahman SM, Nakamoto Y, Fujiwara T, Naito S, et al. (2013) Differential response of methionine metabolism in two grain legumes, soybean and azuki bean, expressing a mutated form of Arabidopsis cystathionine γ–synthase. J Plant Physiol 170: 338–345. [DOI] [PubMed] [Google Scholar]
- 60. Nguyen HC, Hoefgen R, Hesse H (2012) Improving the nutritive value of rice seeds: elevation of cysteine and methionine contents in rice plants by ectopic expression of a bacterial serine acetyltransferase. J Exp Bot 63: 5991–6001. [DOI] [PubMed] [Google Scholar]
- 61. Willems E, Leyns L, Vandesompele J (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem 379: 127–129. [DOI] [PubMed] [Google Scholar]
- 62. Daniel I Arnon (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris . Plant Physiol 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK (1998) HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis . Plant Cell 10: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peever TL, Higgins VJ (1989) Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum . Plant Physiol 90: 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. DP Knievel (1973) Procedure for estimating ratio of live to dead root dry matter in root core samples. Crop Science 13: 124–126. [Google Scholar]
- 66. Zhu JL (2012) Quantitative determination of citrate by spectrophotometry method. Chinese Journal of Analysis Laboratory 31: 115–117. [Google Scholar]
- 67. Gonzalez DH, Iglesias AA, Andreo CS (1984) On the regulation of phosphoenolpyruvate carboxylase activity from maize leaves by L-malate. Effect of pH. J Plant Physiol 116: 425–434. [DOI] [PubMed] [Google Scholar]
- 68.Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 2 48–254. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the constructs for Agrobacterium tumefaciens-mediated transformation into Medicago sativa. a, Schematic representation of the GsPPCK3 overexpression construct. b, Schematic representation of the GsPPCK3-SCMRP overexpression construct.
(TIF)