Abstract
Protein acetylation is mediated by histone acetyltransferases (HATs) and deacetylases (HDACs), which influence chromatin dynamics, protein turnover and the DNA damage response. ATM and ATR mediate DNA damage checkpoints by sensing double-strand breaks and single-strand-DNA–RFA nucleofilaments, respectively. However, it is unclear how acetylation modulates the DNA damage response. Here we show that HDAC inhibition/ablation specifically counteracts yeast Mec1 (orthologue of human ATR) activation, double-strand-break processing and single-strand-DNA–RFA nucleofilament formation. Moreover, the recombination protein Sae2 (human CtIP) is acetylated and degraded after HDAC inhibition. Two HDACs, Hda1 and Rpd3, and one HAT, Gcn5, have key roles in these processes. We also find that HDAC inhibition triggers Sae2 degradation by promoting autophagy that affects the DNA damage sensitivity of hda1 and rpd3 mutants. Rapamycin, which stimulates autophagy by inhibiting Tor, also causes Sae2 degradation. We propose that Rpd3, Hda1 and Gcn5 control chromosome stability by coordinating the ATR checkpoint and double-strand-break processing with autophagy.
HATs and HDACs target histones and non-histone proteins1–4 and regulate chromosome dynamics. They also influence the DNA damage response through acetylation of key DNA repair and checkpoint proteins2. HDACs can be classified into three classes on the basis of sequence similarity5. HDAC inhibition is a promising therapeutic strategy against cancer6, and certain inhibitors—such as valproic acid (VPA)7—affect class I and II HDACs.
The DNA damage checkpoint response is mediated by two PI3 kinases, ATR and ATM (Mec1 and Tel1 in yeast, respectively)8. ATR is assisted by ATRIP (Ddc2 (also known as Lcd1) in yeast) and, in response to DNA damage, activates a signal transduction pathway that coordinates cell cycle events with DNA repair and controls apoptosis in mammals. In yeast, the Rad53 (CHK2 (also known as CHEK2) in humans) protein kinase has a pivotal role in transducing ATR signalling8. Double-strand breaks (DSBs) are dangerous DNA lesions that can be repaired by different recombination processes, depending on the cell cycle phase9. In G2, DSBs are processed into single-strand DNA (ssDNA) and engaged intohomologous recombination-mediated repair pathways9,10. Although several DNA repair proteins are acetylated, the functional significance of these modifications is mostly unknown.
Protein acetylation has been also implicated in promoting degradation of certain proteins through autophagy2,11. Autophagy is a highly conserved process involved in protein and organelle turnover and results in their vacuolar (lysosomal in mammals) degradation. Crosstalk between ubiquitination and autophagy has been reported12,13. Autophagy is triggered by a variety of stimuli, including nutrient starvation and TOR1 inhibitors, some of which are currently into clinical trials for cancer therapy14–16.
Here we report a connection between the ATR pathway, DSB repair, protein acetylation and autophagy.
VPA counteracts the DNA damage response
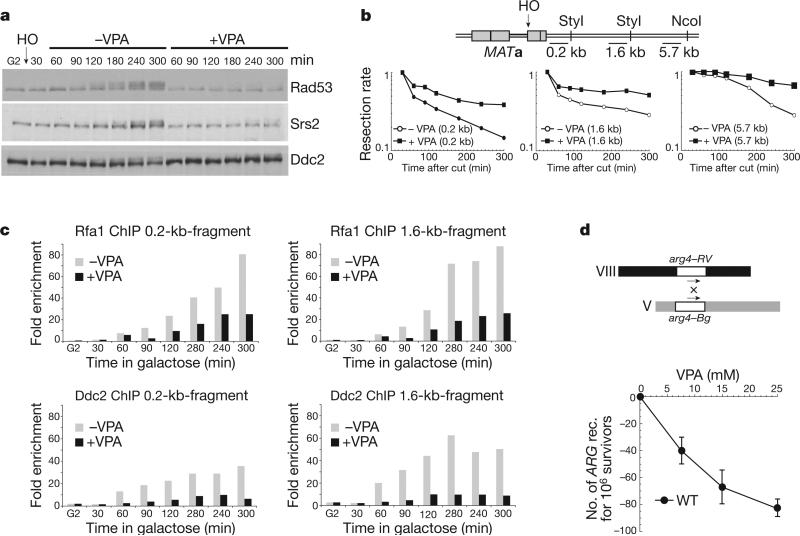
We investigated how HDAC inhibition by VPA affects the DNA damage response in budding yeast. VPA treatment per se did not activate Rad53 (data not shown), but counteracted Rad53 phosphorylation after exposure to 4NQO (an ultraviolet-mimetic drug) in G1 and G2 cells or exposure to MMS (a DNA-alkylating agent) in S phase cells (Supplementary Fig. 1a). It is unlikely that VPA limits the accumulation of checkpoint activators, because inhibition of protein synthesis did not influence Rad53 phosphorylation (Supplementary Fig. 1b). As cycloheximide treatment did not restore checkpoint activation, it is also unlikely that VPA enhanced negative checkpoint regulators (Supplementary Fig. 1b). We then analysed the effect of VPA in cells experiencing a single and irreparable DSB at a specific chromosomal locus. We overexpressed HO, a yeast nuclease that recognizes a specific DNA sequence17 (Fig. 1a–c and Supplementary Fig. 2). Checkpoint activation after DSB formation requires Cdc28 (CDK1 in mammals) activity and DSB resection, which generates RPA–ssDNA nucleofilaments and recruitment of the Ddc2–Mec1 complex8,10. Ten kilobases of ssDNA must accumulate to trigger Rad53 activation, which occurs 90 min after DSB formation17. After HO induction in G2, VPA counteracted Rad53 phosphorylation (Fig. 1a). VPA also affected Mec1-dependent Ddc2 and Srs2 phosphorylation8,18. We next measured DSB resection at three loci (0.2, 1.6 and 5.7 kb from the break site) (Fig. 1b and Supplementary Fig. 2a). Resection rates were reduced compared to untreated conditions. Without VPA, resection of the 5.7-kb fragment was obvious at 120 min, whereas with VPA resection was still impaired after 300 min. Hence, VPA-treated cells failed to accumulate the 10 kb of ssDNA needed for Rad53 activation. We then measured the recruitment of Rfa1 and Ddc2 to the DSB region with or without VPA by chromatin immunoprecipitation (ChIP) analysis (Fig. 1c and Supplementary Fig. 2b). VPA counteracted Rfa1 and Ddc2 recruitment at the 0.2-kb and 1.6-kb fragments, indicating that DSB processing and signalling are crippled in VPA. As DSB resection is a key step in homologous recombination, VPA should also impair recombination frequencies. Spontaneous ectopic recombination frequencies19 were indeed reduced by VPA treatment (Fig. 1d). Hence, VPA counteracts DSB resection and signalling, thus affecting homologous recombination and the signal transduction response mediated by Mec1.
Figure 1. VPA treatment counteracts DNA double-strand-break processing.
a–c, RFA1::FLAG DDC2::MYC cells were arrested in G2 and released in YP galactose to induce HO endonuclease. After 30 min, the culture was split in two: +VPA and −VPA. a, Samples were processed for western blot using anti-Rad53, Srs2 and Ddc2 antibodies. b, A schematic diagram showing probe locations with respect to the HO cut site. The resection rate was calculated as the rate of HO cut band disappearance. c, Fold enrichment of the 0.2-kb fragment was calculated after ChIP of Rfa1–Flag, Ddc2–Myc. d, VPA effect on ectopic recombination (rec.) in wild-type (WT) cells. Error bars represent standard deviation (s.d.) calculated from four independent experiments.
Sae2 and Exo1 are degraded in VPA-treated cells
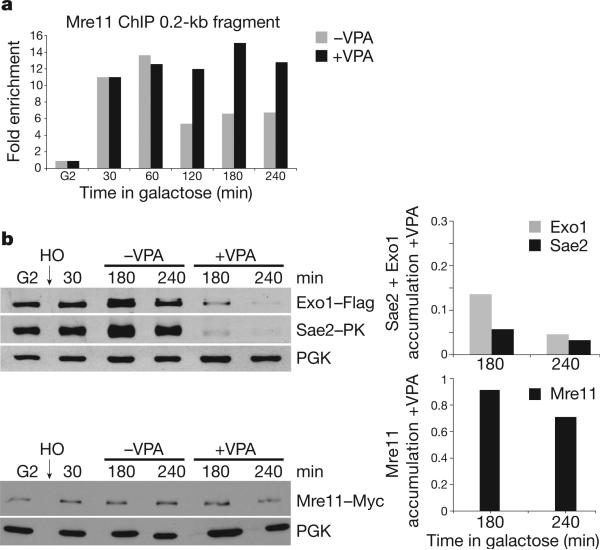
Next we analysed the early events mediating DSB processing (Fig. 2). Mre11 is the first factor recruited to a DSB to activate Tel120. Mre11 indirectly influences DSB resection and its removal from the DSB region depends on Sae2, a CDK1 target involved in DSB processing20,21. Exo1, Dna2 and Sgs1 (BLM in human cells), are also implicated in resection22–24. We reasoned that VPA could limit the recruitment of Mre11 at DSBs25. The timing of Mre11 loading at the 0.2-kb fragment was comparable with or without VPA, but Mre11 association persisted in VPA-treated cells (Fig. 2a and Supplementary Fig. 2c). Hence, VPA does not counteract DSB processing by preventing Mre11 recruitment.
Figure 2. VPA affects Sae2 and Exo1 but not Mre11 protein levels.
a, MRE11::MYC cells were treated as in Fig. 1a. Cell samples were processed for ChIP analysis and the fold enrichment of the 0.2-kb fragment after ChIP of Mre11–Myc without (−VPA) or with (+VPA) VPA was calculated. b, EXO1::FLAG SAE2::PK MRE11::MYC cells were grown as in a. Cell samples were taken and processed for western blot analysis using anti-Flag, PK and Myc antibodies.
VPA treatment affected Sae2 and Exo1 protein levels. After 180 min of HO induction in VPA, Sae2 and Exo1 were barely detectable whereas Mre11 was not affected (Fig. 2b). Hence, VPA affects Sae2 and Exo1 turnover, although with different kinetics as the decrease in Exo1 level was delayed compared to Sae2 (Fig. 2b and data not shown). These results account for the VPA-dependent accumulation of Mre11 because Sae2 is needed for Mre11 displacement at the DSB region15,20, but may also explain the VPA-induced DSB resection defect as both Sae2 and Exo1 influence DSB resection.
VPA stimulates autophagy
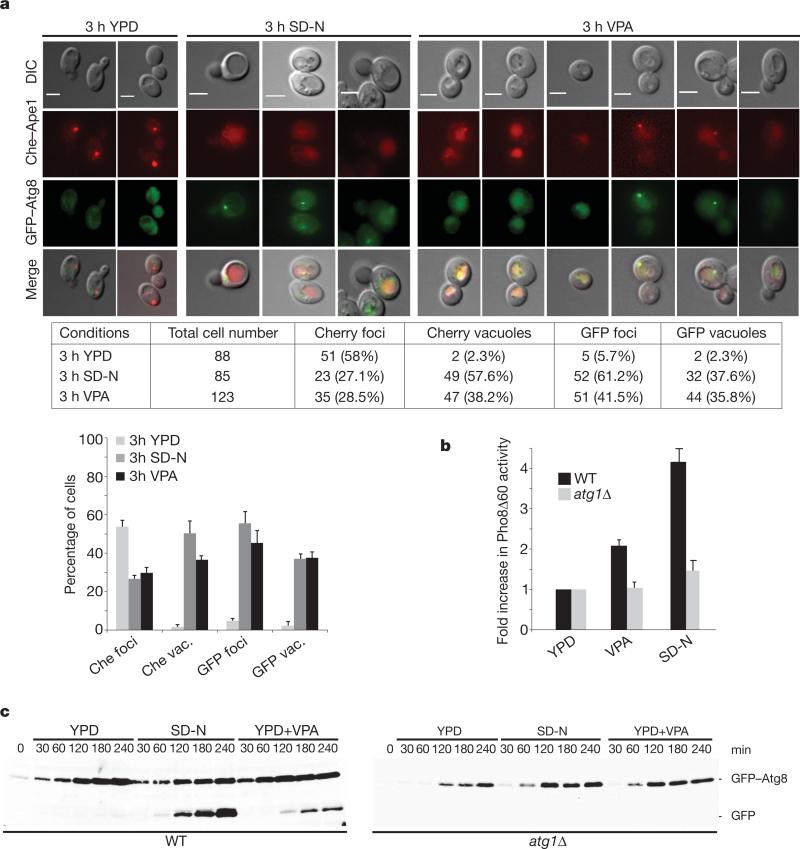
We tested whether VPA induces autophagy in yeast as in mammals26. Autophagy induction correlates with: (1) vacuolar staining of Cherry–Ape1, an aminopeptidase specific for the CVT (cytoplasm to vacuole targeting) subpathway, and perivacuolar foci and vacuolar staining of GFP–Atg8, an autophagosome component27; (2) increased enzymatic activity of Pho8Δ60, an autophagy marker28; and (3) processing of GFP–Atg829.
Yeast cells grown in YPD medium (Fig. 3a) showed mostly Cherry–Ape1 foci but very little Cherry vacuolar staining. The foci may reflect Ape1 oligomer formation after synthesis29. Under conditions of nitrogen starvation (SD-N medium), cells exhibiting fluorescent vacuolar staining increased, whereas the foci diminished. These results reflect starvation-induced autophagy27. VPA treatment partially mimicked starvation conditions. YPD cells showed few GFP–Atg8 foci and little vacuolar staining. Conversely, starved cells expressed GFP–Atg8 foci and showed vacuolar staining. This trend was recapitulated in VPA. We then measured the Pho8Δ60 activity (Fig. 3b) in wild-type cells and in mutants in ATG1, encoding an essential autophagy kinase30. YPD wild-type and atg1 mutants exhibited basal Pho8Δ60 activity. Under starvation or in VPA, Pho8Δ60 activity increased in wild-type cells but not in atg1 mutants. We then analysed GFP–Atg8 processing (Fig. 3c). Whereas YPD wild-type cells did not undergo GFP–Atg8 cleavage, starved and VPA cells exhibited an Atg1-dependent GFP–Atg8 processing. These results indicate that VPA induces autophagy.
Figure 3. GFP–Atg8 Cherry–Ape1 cellular distributions in VPA-treated cells.
a, Cherry::APE1 GFP::ATG8 cells were grown and shifted to YPD, nitrogen starvation (SD-N) or YPD+VPA medium for 3 h. Samples were processed for microscopy. The table shows numbers corresponding to the experiment. Percentage of fluorescence signals is presented and error bars represent the s.d. obtained from three independent experiments. DIC, differential interference contrast. Scale bars, 3 μm. b, Pho8Δ60 and Pho8Δ60 atg1Δ cells were grown as in a. Pho8Δ60 activity was calculated by measuring alkaline phosphatase levels. Error bars represent s.d. calculated from five independent experiments. c, GFP::ATG8 and GFP::ATG8 atg1Δ cells were grown as in a. Cell samples were processed for western blot using anti-GFP antibody. Quantification is presented in Supplementary Fig. 3.
VPA-induced Sae2 acetylation and degradation
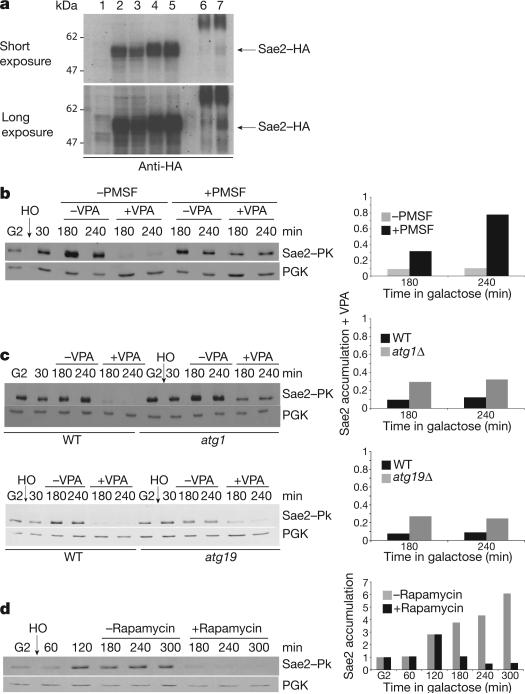
Next we tested whether Sae2 and/or Exo1 were acetylated. We immunoprecipitated overexpressed HA–Sae2 in cells ± VPA with anti-HA and subsequently ± anti-acetyl-Lysine antibodies. Recovered Sae2 increased in VPA (Fig. 4a), indicating that Sae2 is acetylated. We failed to detect acetylated Exo1, although acetylated Exo1 has been previously described2. Autophagy is the preferred pathway for the degradation of oligomeric complexes that cannot be recognized by the ubiquitin–proteasome system and form toxic aggregates13,31. Certain proteins are specifically shunted into the autophagic pathway when they are hyperacetylated11. Hence, Sae2 might be degraded through autophagy, as its level declines in VPA, it is acetylated and forms complexes32. We tested whether Sae2 disappearance in VPA was dependent on autophagy (Fig. 4b–d). Phenylmethylsulphonyl fluoride (PMSF) inhibits serine proteases but also blocks autophagy by inhibiting vacuolar proteases33. PMSF treatment counteracted Sae2 disappearance in VPA (Fig. 4b). We also tested whether genetic inactivation of autophagy would affect Sae2 levels. Deletion of ATG1 or ATG19 (specific for the CVT pathway)30 partially compensated Sae2 destabilization in VPA (Fig. 4c). Finally, rapamycin, which induces autophagy by inhibiting Tor134, also destabilized Sae2 in an ATG1-dependent manner (Fig. 4d and data not shown). Thus, in VPA, autophagy contributes to Sae2 degradation perhaps through acetylation as has been reported for the Huntingtin protein11.
Figure 4. Sae2 in VPA-treated cells.
a, 4HA–Sae2 was immunoprecipitated ± VPA with anti-HA and subsequently ± anti-acetyl-Lysine. Eluate was analysed using anti-HA. Lane 1: input Sae2; 2: input Sae2–HA –VPA; 3: as in 2 but + VPA; 4: 3 μl elution Sae2–HA – VPA after anti-HA immunoprecipitation (IP; input AcK-IP); 5: double amount of 4; 6: IP anti-AcK elution from anti-HA IP of Sae2–HA – VPA; 7: as in 6 but + VPA. b, SAE2::PK erg6Δ cells were treated as in Fig. 2a. After 30 min induction, VPA and PMSF were added or not. Samples were processed for western blot using anti-PK. c, Wild-type SAE2::PK, SAE2::PK atg1Δ and SAE2::PK atg19Δ cells were grown as in b. After 30 min induction, VPA was added or not and samples treated as in b. d, Wild-type SAE2::PK cells were grown as in b. After 120 min induction, rapamycin (200 ng ml−1) was added or not and samples were treated as in b.
Gcn5, Rpd3 and Hda1 control Sae2 levels
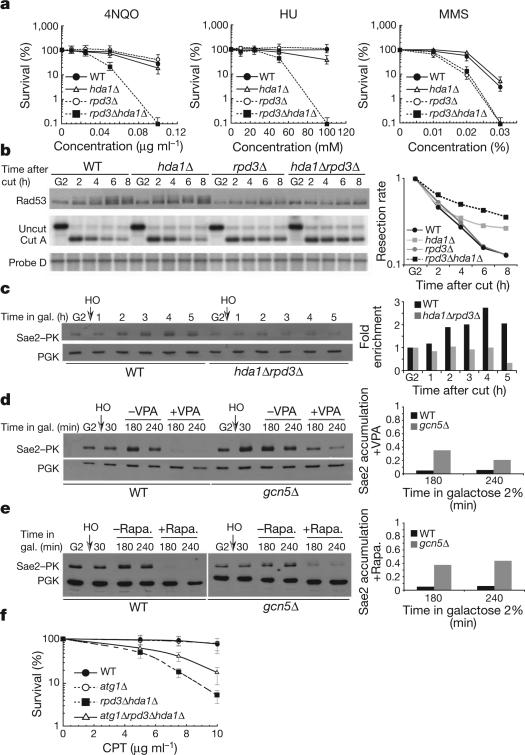
We tested whether VPA-induced phenotypes can be recapitulated in rpd3 and hda1 mutants, altered in two class I and II HDACs, respectively35. rpd3 hda1 double mutants showed hypersensitivity to 4NQO and hydroxyurea (HU; a DNA synthesis inhibitor) compared to single mutants and wild-type cells, whereas only rpd3 cells were hypersensitive to MMS (Fig. 5a). Damage-induced recombination was reduced in hda1 rpd3 mutants (data not shown). Hence, Rpd3 and Hda1 may partially substitute for each other to respond to DNA damage and to assist homologous recombination, and may be targeted by VPA. We analysed Rad53 phosphorylation in G1 or G2 arrested wild-type, hda1, rpd3 and hda1 rpd3 cells treated with 4NQO, MMS and HU and found that the double mutants failed to promote robust checkpoint activation (data not shown). Thus, both Hda1 and Rpd3 influence Rad53 activation. In response to DSB formation, rpd3 and hda1 rpd3 cells showed a severe and equivalent defect in Rad53 phosphorylation (Fig. 5b), and rpd3 cells were resection defective although less than double mutants. Hence, Hda1 and rpd3 influence DSB processing and signalling, although to a different extent. Sae2 failed to accumulate at wild-type levels in hda1 rpd3 cells after HO induction in G2 (Fig. 5c). Thus, the impairment in checkpoint activation and DSB processing and the Sae2 instability observed in VPA can be recapitulated in rpd3 hda1 mutants. The action of Rpd3 and Hda1 is counteracted by the Gcn5 HAT (SAGA in mammals)36. We found that, in the absence of Gcn5, VPA and rapamycin-mediated destabilization of Sae2 were attenuated (Fig. 5d, e).
Figure 5. Gcn5, rpd3 and Hda1 influence Sae2 levels and cell survival in atg1 mutants in response to DNA damage.
a, Survival of wild-type, rpd3Δ, hda1Δ and rpd3Δ hda1Δ strains after 4NQO, MMS and HU treatment. Error bars represent s.d. calculated from seven independent experiments. b, HO was induced in G2 wild-type, hdalΔ, rpdΔ and hdalΔ rpdΔ cells and western and Southern blot analyses were performed. c, Wild-type SAE2::PK and hdalΔ rpdΔ SAE2::PK cells were grown as in b and western blot was performed as in Fig. 4b. d, e, Wild-type SAE2::PK and gcn5Δ SAE2::PK strains were grown as in b and after 30 min of HO induction either VPA (d) or rapamycin (e) was added or not. Western blot was performed. f, Percentage of viability of wild-type, atg1Δ, rpd3Δ hdalΔ and atg1Δ rpd3Δ hdalΔ strains. Error bars represent s.d. calculated from four independent experiments.
The observations described earlier lead to the expectation that autophagy might contribute to the hdal rpd3 sensitivity to DNA damaging agents. atg1 and wild-type cells exhibited comparable survival rates in response to camptothecin (CPT) treatment whereas hdal rpd3 cells exhibited hypersensitivity to CPT (Fig. 5f). ATG1 ablation partially counteracted CPT-induced lethality in hdal rpd3 cells. Hence, in hdal rpd3 mutants, the Atg1-mediated autophagic response contributes to cell lethality, perhaps by destabilizing key DNA damage response factors.
Discussion
We showed that class I and II HDACs influence the DNA damage response at three levels (Supplementary Fig. 4a): checkpoint activation throughout the cell cycle, DSB processing in G2/M and degradation of key recombination protein(s). The following considerations indicate that HDACs mediate a global DNA damage response. Firstly, besides Sae2, Cdk1, Ku, MRN, Blm2 and Rfa1 (our unpublished observations) are also acetylated. Secondly, Sae2 acetylation might influence its own accumulation, thus resembling certain phenotypes of sae2Δ cells such as the inability to remove Mre11 from the DSB site37. However, in contrast to sae2 mutants, checkpoint activation and DSB resection are markedly impaired by HDAC inhibition, indicating that Sae2 is not the only relevant target. Accordingly, also Exo1 is degraded in VPA, perhaps as a consequence of Sae2 destabilization. Thirdly, the DSB resection defects caused by HDAC inhibition might explain the lack of checkpoint signals (RPA filaments) in G2/M but not the checkpoint impairment in G1 and G2 cells treated with the ultravioletmimetic drug 4NQO38, as 4NQO-induced checkpoint signalling does not require DSB resection. Because all the damaging agents used lead to the accumulation of RPA filaments through different mechanisms, perhaps Rfa1 acetylation also influences checkpoint signalling.
The Tip60 HAT positively influences ATM39. This observation seems at odds with the findings that class I and II HDACs stimulate ATR. It is possible that HATs and HDACs have both positive and negative roles depending on the checkpoint subpathway. However, as ATR activation depends on the processing of DSBs (that represent ATM signals), it is possible that HAT-mediated ATM activation is a non-direct consequence of ATR inhibition. We note that after HDAC inhibition ATM/Tel1-mediated histone H2A phosphorylation is not affected (data not shown), probably because Mre11 maintains Tel1 active by remaining loaded at the DSB. Moreover, the fact that Gcn5/ SAGA promotes Sae2 degradation implies that this HAT indirectly negatively influences ATR signalling, perhaps by generating acetylated substrates for the autophagic pathway and/or by directly promoting autophagy.
Histone 3 lysine 9, 14, 18, 23 (converted to glycines) and histone 4 lysine 5, 8, 12, 16 (converted to arginines) mutants, altered in H3 and H4 acetylation, still activate the checkpoint (Supplementary Fig. 1c), thus indicating that H3 and H4 histone acetylation does not have a relevant role for checkpoint activation.
HDAC inhibition induces autophagy throughunknown processes15,26 and we show that HDAC impairment destabilizes Sae2 through an autophagic pathway. However, the magnitude of autophagy induction after HDAC inhibition is not as strong as that in nitrogen-starved cells. Tor1 counteracts autophagy34 and we showed that rapamycin affects Sae2 turnover. The finding that Gcn5/SAGA ablation counteracts Sae2 degradation in VPA- and rapamycin-treated cells pinpoints the HAT activity involved in this regulatory process. Intriguingly, gcn5 mutants are sensitive to rapamycin. Moreover, dna2 mutants are altered in DSB resection and require TOR1 overexpression40 for suppression. A tantalizing hypothesis is that an excess amount of Tor1 rescues dna2 mutants by counteracting authophagy-mediated Sae2 destabilization.
We propose that (Supplementary Fig. 4b) after DSB formation, the broken chromosome arm is relocated close to the nuclear envelope41. Rpd3 and Hda1 will keep Sae2 in the deacetylated form that influences Mre11 dynamics at the DSB site37. Sae2 is then released from the DSB site, perhaps as a multimeric form32 and Gcn5-mediated acetylation shunts it into autophagy-mediated degradation. This last step might be needed to counteract extensive DSB resection and/or simply to eliminate Sae2 once the first step of DSB processing has been accomplished; we note that exposure to reactive oxygen species, ultraviolet and ionizing radiation, besides damaging DNA, can cause protein damage and protein–DNA crosslinking42, and certain damaged repair proteins or crosslinked proteins might have to be destroyed to prevent cellular problems. Cells may use specific autophagy subpathways rather than recycle all the cellular components, including those that are not damaged. This notion is supported by the observation that Sae2 degradation depends on Atg19, a factor specific for certain types of selective autophagy.Inany case, triggering unprogrammed autophagy-mediated turnover of key repair proteins, either by inhibiting HDACs and/or Tor1 would contribute to DNA damaging sensitivity. CtIP is ubiquitinated43 and Sae2 levels under normal conditions increase after proteosome inhibition with MG132 (data not shown). These observations, besides indicating that Sae2 also undergoes ubiquitination, may account for the partial rescue of Sae2 levels in atg mutants. Crosstalk between ubiquitination and autophagy has previously been described12. Future work will address the contribution of both pathways to DSB metabolism.
METHODS SUMMARY
Strains are listed in Supplementary Table 1. Growth conditions, synchronization and HO induction have previously been described10. VPA was used at 10 mM unless otherwise indicated. Nitrogen starvation (SD-N) medium, viability and recombination analysis have previously been described19,28. Error bars represent s.d. calculated from at least three independent experiments. FACS analysis, TCA extraction and SDS–PAGE have previously been described44. For immunodetection of Rad53 we used EL7 and F9 antibodies45. For Myc, HA, PK, PGK1, Flag and GFP we used the 9E10, 12CA5, V5-TAG, 22C5 and M2 antibodies, respectively. Protein quantification was normalized with respect to PGK1 and accumulation was calculated as the ratio of VPA- or rapamycin-treated to untreated cells. Resection experiments were previously described10. Purified genomic DNA was digested with StyI (0.2-, 1.6-kb fragments) or NcoI (5.7 kb) and treated for Southern blot analysis. The density of the HO-cut band at t = 30 min (Supplementary Fig. 2a) was set to 100%. The total amount of DNA loaded in each sample was normalized by re-probing the blots with probe Control D located 170 kb from the HO site. ChIP analysis was previously described46. The fold enrichment of fragments located 0.2 kb and 1.6 kb from the DSB was calculated as the ratio between the value of the fragment of interest and the value of the fragment used as control (ARS305). The number obtained was divided for the same ratio calculated for the whole cell extract samples of each time point. Primers for resection and ChIP experiments are the same as used previously47. Samples for microscopic analysis were fixed in 4% formaldehyde for 5 min at room temperature (21 °C) and washed in cold 1 × PBS. Images were taken with an Olympus BX51 fluorescent microscope. Oil immersion ×100 objective UPlan APO, NA 1.4 was used. We used an 800 ms exposure time for Cherry and 400 ms for GFP. Alkaline phosphatase activity was measured using the Pho8Δ60 assay as described28.
Supplementary Material
Acknowledgments
Acknowledgements We thank S. Piatti, M. P. Longhese, M. Grunstein, J. K. Tyler, A. Pellicioli, M. Costanzo, R. Brost, M. Vogelauer, D. Klionsky, T. Roberts and C. Bertoli for reagents and technical suggestions, J. Barlow for DNA damage foci analysis, A. Sartori and S. Ferrari for communicating unpublished results, C. Lucca, D. Branzei, R. Bermejo and the members of our laboratories for comments. Work in M.F. laboratory was supported by grants from the Italian Association for Cancer Research and partially from Telethon, European Community (GENICA) and the Italian Ministry of Health. T.R. was supported by fellowships from FRM and EMBO and I.C. was supported by a short fellowship from HFSP and from FIRC. This work was also supported by GM50237 (to R.R.), GM67055 (to R.R.) and GM088413 (to K.B.).
Footnotes
Author Contributions T.R. and F.V. performed the experiments in Figs 1, 2, 4 and 5, T.R. performed those in Fig. 3 and Supplementary Fig. 3, F.V. and I.C. those in Supplementary Figs 1 and 2. I.C. contributed to Fig. 1, G.S. to Fig. 5, O.A.B. to Fig. 4. K.A.B., A.O. and D.P. provided advice and technical support for imaging. T.R., F.V., I.C. and M.F conceived the experiments. T.R., F.V., G.S., R.R., S.M. and M.F. analysed the results. T.R., F.V., G.S. and M.F. wrote the paper, S.M. and M.F. conceived the project.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Bird AW, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 3.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 4.Scott KL, Plon SE. Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:4522–4531. doi: 10.1128/MCB.23.13.4522-4531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 7.Gottlicher M, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 9.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamark T, Johansen T. Autophagy: links with the proteasome. Curr. Opin. Cell Biol. 2009;22:192–198. doi: 10.1016/j.ceb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc. Natl Acad. Sci. USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 18.Liberi G, et al. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu J, Shao CJ, Chen FR, Ng HK, Chen ZP. Autophagy induced by valproic acid is associated with oxidative stress in glioma cell lines. Neuro Oncol. 2010;12:328–340. doi: 10.1093/neuonc/nop005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shintani T, Reggiori F. Fluorescence microscopy-based assays for monitoring yeast Atg protein trafficking. Methods Enzymol. 2008;451:43–56. doi: 10.1016/S0076-6879(08)03204-7. [DOI] [PubMed] [Google Scholar]
- 28.Noda T, Klionsky DJ. The quantitative Pho8D60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 29.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 30.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 31.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 32.Kim HS, et al. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 34.Kamada Y, Sekito T, Ohsumi Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 2004;279:73–84. doi: 10.1007/978-3-642-18930-2_5. [DOI] [PubMed] [Google Scholar]
- 35.Robyr D, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 36.Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 37.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl Acad. Sci. USA. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiorentino DF, Crabtree GR. Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase. Mol. Biol. Cell. 1997;8:2519–2537. doi: 10.1091/mbc.8.12.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellicioli A, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiorani S, Mimun G, Caleca L, Piccini D, Pellicioli A. Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle. 2008;7:493–499. doi: 10.4161/cc.7.4.5323. [DOI] [PubMed] [Google Scholar]
- 46.Lucca C, et al. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene. 2004;23:1206–1213. doi: 10.1038/sj.onc.1207199. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Haber JE. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:104–112. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.