Abstract
Two groups of lipidated steroid saponins including seven new compounds (2, 3, 5, and 7–10) were isolated from the widely used botanical, wild yam (Dioscorea villosa), employing a fractionation protocol of metabolomic mining. This methodology has very recently led to the isolation of 14 diarylheptanoids from the same plant. Together with these lipidated steroid saponins, they establish additional new markers for Dioscorea villosa. The lipidation of steroids with analogue long-chain fatty acids containing different degrees of unsaturation generates entire series of compounds which are difficult to purify and analyze. The structures of the two series of lipidated steroid saponins (series A and B) were demonstrated by a combination of 1D and 2D NMR as well as GC-MS after chemical modification. Series A was determined to be a mixture of lipidated spirostanol glycosides (1–5), while series B (6–10) proved to be a mixture of five lipidated clionasterol glucosides. The latter group represents the first derivatives of clionasterol to be found in D. villosa. The discovery of this specific structural type of aliphatic esters of steroid saponins expands the characterization of the secondary metabolome of D. villosa. It also may inspire biological studies which take into account the lipophilic character and significantly altered physiochemical characteristics of these otherwise relatively polar phytoconstituents.
Keywords: Dioscorea villosa, wild yam, metabolomic mining, lipidated steroid saponins, residual complexity
1. Introduction
Species of the genus Dioscorea (family Dioscoreaceae) are widely used as botanical dietary supplements. These plants are well known for containing steroidal saponins, mainly belonging to the spirostanol and furostanol classes, and these have been used as chemical marker compounds for quality control of the botanical products [1]. Wild yam, the rhizomes and roots of Dioscorea villosa L., is an important source of diosgenin [2], a phytoestrogen that has been investigated thoroughly from both chemical and biological perspectives [3–5]. The methods aimed at targeted isolation and purification of the major steroid saponins in D. villosa were developed by exploring various chromatographic techniques, such as high-speed countercurrent and centrifugal partition chromatographs as well as HPLC [1,6,7]. To date, based on a comprehensive literature survey, twelve steroidal saponins and two flavan-3-ol glycosides have previously been isolated as major secondary metabolites of wild yam [1,2,6–8]. Our group recently reported 14 diarylheptanoids [9]. These diarylheptanoids were first isolated and characterized from wild yam based on a new fractionation methodology using 1D qHNMR and 2D NMR profiles along the preparative fractionation pathway [9]. Primary fractionation of the methanolic extract using a MeOH/H2O solvent gradient on a preparative C18 solid phase extraction (SPE) cartridge effectively enriched the diarylheptanoids into three fractions (4–6), which were further purified by VLC, MPLC, and HPLC. Mining these 1H NMR spectral profiles of the last two fractions (10 and 11) led to the new observation of similar patterns (i.e., signals of steroid glycosides) dominant in almost all of these relatively lipophilic fractions. Accordingly normal phase TLC fraction monitoring was used throughout the separation process, and a polar solvent system of chloroform/methanol (8:1 v/v) was found to be most suitable for the last two fractions (10 and 11) to give TLC spots with Rf values around 0.5. However, the polarity of this solvent system is much higher than what would commonly be used for lipophilic fractions that elute from the C18 SPE cartridge with 90% and/or pure MeOH. Subsequent purification of these two fractions finally afforded two series of compounds, each of which showed a single spot on TLC in a variety of solvent systems. HPLC analysis using a C18 column resulted in no distinctive absorption at 210 nm with a UV detector, and remained undetected even in an ESI-LC-MS system. In addition to the characteristic resonances of steroid saponins, the 1H NMR spectra of these two series of compounds exhibited signals that were reasonably assigned to a mixture of homologous aliphatic residues, suggesting that both series were residual complex mixture of lipidated steroid saponins containing, e.g., fatty acid residues. This feature assignment was also in line with the much more lipophilic chromatographic behavior of the compounds, in particular on the C18 absorbent. To elucidate their structures, 2D NMR and GC-MS were employed. The proportion of each constituent in each of the two samples was determined using a GC-MS method. This resulted in the characterization of two series of lipidated steroid saponins (1–10), including seven new compounds (2, 3, 5, and 7–10), which overall represents the first report of this metabolite class from D. villosa [10–12]. The occurrence of lipidated steroid saponins in the genus Dioscorea was first reported in the International Congress on Natural Products Research (ICNPR) 2012 and covered two of such compounds from D. cayenensis (1 and 4) [10]. Four additional analogues had been discovered in 2005 and 2010 from two other families, the Valerianaceae (Valeriana officinalis) and Brassicaceae (Sisymbrium irio), respectively [11,12]. Lipidated steroid saponins possess one polar sugar residue(s) in the middle and a non-polar long-chain aliphatic acid and a steroid residue in two ends, which altogether explains their unusual chromatographic behavior considering to general compound class. The present metabolomic mining approach yielded these lipidated steroid saponins and also the previously reported diarylheptanoids. Both findings widen the chemical profile of wild yam and other Dioscorea plants and facilitate the characterization of residual complexity of D. villosa extracts. Discovery of new types of secondary metabolites also potentially offers new chemical leads for the development of standardization and quality control of wild yam and other Dioscorea botanical products. Herein, we present the details of the isolation methodology, the structure elucidation of the previously unreported lipophilic steroid saponin metabolites (2, 3, 5, and 7–10) of D. villosa, and their potential broader impact on the understanding of the biological mechanisms of action of yam botanicals in general.
2. Experimental
2.1. General experimental procedures
NMR spectra were obtained on Bruker AVANCE-400 (5 mm broadband ATM probe), AVANCE-600 (5 mm TXI CryoProbe), or AVANCE-900 (5 mm ATM CryoProbe) NMR spectrometers (Bruker, Zürich, Switzerland) using pyridine-d5 (D 99.5%, Cambridge Isotope Laboratories, Inc., Andover, MA) as the solvent. The chemical shifts of the compounds were referenced to the residual solvent signals (δH 8.740 and δC 150.35 for α-H and α-C, respectively). Offline NMR data processing was performed with MestReNova software version 8.0.0-10524 (Mestrelab Research, Santiago de Compostela, Spain). All NMR data were acquired with default Bruker pulse sequences. The 1H NMR data were processed with double zero-filling and Lorentz-Gauss resolution enhancement (LB −1.8 Hz and GB 1.0 Hz) prior to Fourier transformation. Calculations for the 1H NMR iterative Full Spin Analysis (HiFSA) were performed with the PERCH software package v.2010.1 (PERCH Solutions Ltd., Kuopio, Finland). GC-MS experiments were carried out on an Agilent 7000A Triple Quad MS and a 7890A GC system with an Agilent 7683B autosampler (Agilent Technologies, Santa Clara, CA, USA). Two tandem Agilent J&W HP-5MS GC columns (each 30 m × 0.25 mm × 0.25 µm) were used for optimal separation. The samples dissolved in hexane or chloroform were introduced into the GC-MS system in split mode (1:1), and the inlet temperature was set as 230 °C. The needle was washed 5 times each before aspiration and after injection with solvents chloroform/hexane for methylated fatty acids and hexane/chloroform for acetylated sugars (5 µL), respectively. In order to avoid carry over, one solvent blank sample was inserted after each sample being analyzed, using the same conditions. Helium (99.999%) was used as a carrier gas at 1 mL/min. The oven temperature was initially set to 120 °C, and linearly raised to 310 °C in 19 min, then held for 5 min isothermally. The transfer temperature was set at 280 °C, 70 eV was applied in the Electron Impact source voltage, and the temperature was set at 220 °C. The mass range for MS full scan was 100 ~ 600 amu.
Silica gel (230–400 mesh, Macherey-Nagel), C18 reversed-phase silica gel (Macherey-Nagel), Sephadex LH-20 (Sigma), and HW-40F gel (Tosoh) were used for VLC and MPLC. General fraction monitoring of preparative chromatographic separations was done by TLC analysis with precoated TLC plates (250 µm thickness, K6F Si gel 60, EM Science, Germany). The chromatograms were visualized by spraying 5% H2SO4 in EtOH on the dried plates, followed by heating until the spots appeared. Solvents used for LC were HPLC grade and purchased from Thermo Fisher Scientific Inc. (Waltham, USA). The solvents used in GC-MS were chromatographic grade and purchased from Sigma-Aldrich Co. (St. Louis, USA). All monosaccharide and fatty acid standards were also purchased from this vendor.
2.2. Plant material
Authentic rhizomes/roots of in-house cultivated D. villosa L. were collected in the UIC Dorothy Bradley Atkins Medicinal Plant Garden in October 2010. A voucher specimen of the commercial sample used for the isolation work (accession number: BC630) has been deposited at the Field Museum of Natural History Herbarium, Chicago, IL. Harvested rhizomes/roots of wild Dioscorea villosa L. (4.5 kg) were purchased from Mountain Rose Herbs in August 2011 and were subject to pharmacognostic ID testing companying it with the in-house cultivated plant material.
2.3. Extraction and isolation
Authentic, in-house cultivated D. villosa (BC601, 5 g) was extracted with MeOH to give 980.8 mg crude extract, of which 128.5 mg was subjected to an SPE VLC using 6 g of C18 reversed-phase silica gel as packing material. An eleven-step gradient of methanol/water (0:10 to 10:0, v/v) was used as the mobile phase, and the 1H NMR spectra and TLC analysis of all 11 fractions were examined in order to systematically detect their content. Fractions 10 and 11 exhibited the lowest polarity corresponding to their C18 chromatographic behavior and the highest abundance of aliphatic resonances in their 1H NMR spectra. However, their normal phase TLC analysis required a much more polar solvent system consisting of chloroform/methanol (8:1 v/v) regarding to their non-polar chromatographic behavior, thus indicating the presence of an unprecedented structural class. Fractions 10 and 11 were combined to give one enriched lipidated fraction for future preparative analysis.
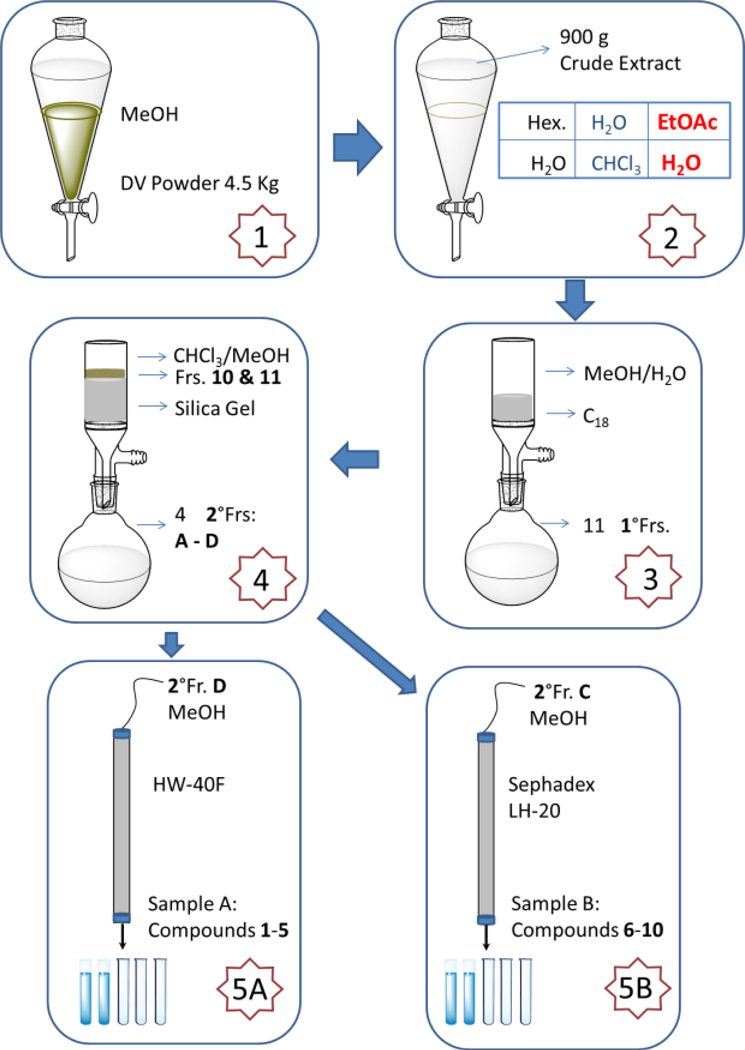
For isolation, the dried and milled rhizomes/roots of D. villosa (BC630, 4.5 kg) were extracted three times with methanol to yield 900 g of crude extract. This material was suspended in water/methanol (9:1) and successively extracted at room temperature with hexane, CHCl3, EtOAc, and n-BuOH. Using the aforementioned enrichment strategy, the EtOAc extract (43.0 g) was subjected to a C18 SPE VLC, affording the enriched lipidated fraction (7.0 g), which was chromatographed on a silica gel VLC eluting with a CHCl3/MeOH gradient (100:1 to 10:1, v/v) to give 4 secondary subfractions (A to D). Subfraction C was further fractionated by MPLC on Sephadex LH-20 gel, eluting with MeOH to afford three tertiary subfractions and sample B (8.1 mg; series B: 6–10). Subfraction D was chromatographed over MPLC of HW-40F gel, eluting with MeOH to afford sample A (2.0 mg; series A: 1–5) as well as four tertiary subfractions. TLC fraction monitoring as described above was used throughout the separation process.
Sample A (1–5): white, amorphous powder; 1H and 13C NMR data, see Tables 1 and 2; GC-MS data, see Figures 3 and 4.
Table 1.
1H and 13C NMR Spectroscopic Data of the Mixtures of Lipidated Steroid Saponins 1–5 (Series A) and 6–10 (Series B) in Comparison with Diosgenina
| Pos | A, 1–5 | B, 6–10c | Diosgenind | |||
|---|---|---|---|---|---|---|
| δH, mult. (J in Hz)b | δC, mult. | δH, mult. (J in Hz) | δC, mult | δH, mult.(J in Hz) | δC, mult | |
| 1α | 1.1257, ddd (13.96, 13.37, 3.82) | 38.12, CH2 | 1.198, m | 38.11, CH2 | 1.1386, ddd (13.96, 13.37, 3.82) | 38.26, CH2 |
| 1β | 1.8323, ddd (13.37, 3.65, 3.54) | 1.852, m | 1.8356, ddd (13.37, 3.65, 3.54) | |||
| 2α | 2.2160, ddddd (12.72, 4.22, 3.82, 3.54, 2.33) | 30.77, CH2 | 2.234, m | 30.80, CH2 | 2.1075, ddddd (12.72, 4.22, 3.82, 3.54, 2.33) | 3.09, CH2 |
| 2β | 1.9400, dddd (13.96, 12.72, 7.34, 3.65) | 1.598, m | 1.8106, dddd (13.96, 12.72, 11.07, 3.65) | |||
| 3 | 3.9678, dddd (11.36, 7.34, 4.84, 4.22) | 79.14, CH | 3.9653, dddd (12.01, 10.22, 4.07, 3.93) | 78.90, CH | 3.8630, dddd (11.36, 11.07, 4.79, 4.22) | 71.70, CH |
| 4α | 2.7704, ddddd (12.99, 11.36, 2.80, 2.33, 2.17) | 39.60, CH2 | 2.7277, ddd (13.54, 4.07, 2.15) | 39.85, CH2 | 2.6186, ddddd (12.99, 11.36, 2.80, 2.33, 2.17) | 43.97, CH2 |
| 4β | 2.8476, ddd (12.99, 4.84, 2.44) | 2.5116, ddd (13.54, 12.01, 2.54) | 2.6471, ddd (12.99, 4.79, 2.44) | |||
| 5 | 141.40, C | 141.47, C | 142.43, C | |||
| 6 | 5.3504, dddd (5.14, 2.44, 2.17, 0.77) | 122.24, CH | 5.4053, ddd (br d) (4.95, 3.29, 2.54) | 122.37, CH | 5.4135, dddd (5.14, 2.44, 2.17, 0.77) | 121.49, CH |
| 7α | 1.5505, ddd (17.46, 10.52, 0.77) | 32.82, CH2 | 1.948, m | 32.68, CH2 | 1.5558, ddd (17.46, 10.52, 0.77) | 32.28, CH2 |
| 7β | 1.9032, dddd (17.46, 5.18, 5.14, 2.80) | 1.593, m | 1.9465, dddd (17.46, 5.18, 5.14, 2.80) | |||
| 8 | 1.6550, dddd (11.06, 11.00, 10.52, 5.18) | 32.20, CH | 1.269, m | 32.57, CH | 1.6386, dddd (11.06, 11.00, 10.52, 5.18) | 32.21, CH |
| 9 | 0.9658, ddd (12.58, 11.00, 4.58) | 50.84, CH | 1.002, m | 50.92, CH | 0.9751, ddd (12.58, 11.00, 4.58) | 50.86, CH |
| 10 | 37.65, C | 37.48, C | 37.49, C | |||
| 11α | 1.5213, ddd (4.89, 4.58, 2.85) | 21.63, CH2 | 1.542, m | 21.81, CH2 | 1.5302, ddd (4.89, 4.58, 2.85) | 21.67, CH2 |
| 11β | 1.4960, ddd (13.25, 12.58, 4.28) | 0.927, m | 1.4646, ddd (13.25, 12.58, 4.28) | |||
| 12α | 1.1572, ddd (13.25, 12.32, 4.89) | 40.40, CH2 | 2.041, m | 40.47, CH2 | 1.1624, ddd (13.25, 12.32, 4.89) | 40.43, CH2 |
| 12β | 1.7395, ddd (12.32, 4.28, 2.85) | 1.229, m | 1.7501, ddd (12.32, 4.28, 2.85) | |||
| 13 | 40.97, C | 43.00, C | 40.95, C | |||
| 14 | 1.1051, ddd (13.85, 11.06, 5.72) | 57.18, CH | 1.171, m | 56.81,CH | 1.1075, ddd (13.85, 11.06, 5.72) | 57.22, CH |
| 15α | 2.0712, ddd (11.78, 7.88, 5.72) | 32.72, CH2 | 1.863, m | 25.01, CH2 | 2.0545, ddd (11.78, 7.51, 5.72) | 32.68, CH2 |
| 15β | 1.4843, ddd (13.85, 11.78, 6.11) | 1.688, m | 1.4608, ddd (13.85, 11.78, 6.29) | |||
| 16 | 4.5706, ddd (8.42, 7.88, 6.11) | 81.59, CH | 1.431, m | 29.01, CH2 | 4.5649, ddd (8.54, 7.51, 6.29) | 81.57, CH |
| 1.308, m | ||||||
| 17 | 1.8445, dd (8.42, 6.36) | 63.42, CH | 1.431, m | 57.38, CH | 1.8376, dd (8.54, 6.36) | 63.36, CH |
| 18 | 0.8610, s | 16.84, CH3 | 0.706, s | 12.60, CH3 | 0.8785, s | 16.89, CH3 |
| 19 | 1.0803, s | 19.90, CH3 | 0.996, s | 19.69, CH3 | 1.0630, s | 20.08, CH3 |
| 20 | 1.9820, (6.97, 6.36) | 42.46, CH | 1.132, m | 36.87, CH | 1.9794, dq (7.00, 6.36) | 42.44, CH |
| 21 | 1.1681, d (6.97) | 15.53, CH3 | 1.034, m | 19.52, CH3 | 1.1642, d (7.00) | 15.54, CH3 |
| 22 | 109.74, C | 1.44, m | 34.96, CH2 | 109.71, C | ||
| 23α | 1.6683, ddd (13.28, 3.33, 3.06) | 32.62, CH2 | 1.293, m | 26.87, CH2 | 1.6686, ddd (13.28, 3.33, 3.06) | 32.79, CH2 |
| 23β | 1.7224, ddd (13.28, 11.30, 6.74) | 1.7204, ddd (13.28, 11.30, 6.74) | ||||
| 24α | 1.6009, dddd (19.45, 11.30, 9.15, 3.06) | 29.75, CH2 | 1.02, m | 46.54, CH | 1.5904, dddd (19.45, 11.30, 9.15, 3.06) | 29.73, CH2 |
| 24β | 1.5730, dddd (19.45, 8.93, 6.74, 3.33) | 1.5713, dddd (19.45, 8.93, 6.74, 3.33) | ||||
| 25 | 1.6117, ddddq (9.77, 9.15, 8.93, 5.59, 1.45) | 31.08, CH | 1.307, m | 29.96, CH | 1.5999, ddddq (11.24, 9.15, 8.93, 6.51, 4.66) | 31.07, CH |
| 26a | 3.6031, dd (10.67, 1.45) | 67.35, CH2 | 0.891, d (6.93) | 19.91, CH3 | 3.6009, dd (10.77, 4.66) | 67.31, CH2 |
| 26b | 3.5227, dd (10.67, 9.77) | 3.5163, dd (11.24, 10.77) | ||||
| 27 | 0.7121, d (5.59) | 17.81, CH3 | 0.911, d (7.80) | 20.41, CH3 | 0.7108, d (6.51) | 17.81, CH3 |
| 28 | 1.691, m | 25.01, CH2 | ||||
| 29 | 0.9330, t (7.48) | 12.43, CH3 | ||||
Reason of chemical shift and J determinations: four decimal places for δ values from HiFSA analyses; three decimal places for 1H NMR δ data; two decimal places for 13C NMR δ data and J values from HiFSA analyses
Measured in pyridine-d5 at 600 MHz
Measured in pyridine-d5 at 400 and 100 MHz, respectively
Measured in pyridine-d5 at 900 and 225 MHz, respectively
Table 2.
1H and 13C NMR Spectroscopic Data of the Sugar and Fatty Acid Residues of Lipidated Steroid Saponins 1–5 (Series A) and 6–10 (Series B)a
| Pos | A, 1–5 | B, 6–10c | ||
|---|---|---|---|---|
| δH, mult. (J in Hz)b | δC, mult.c | δH, mult. (J in Hz) | δC, mult. | |
| 1′ | 5.0273, d (6.82) | 101.18, CH | 5.0285, d (8.04) | 103.33, CH |
| 2′ | 4.3008, dd (7.97, 6.82) | 78.16, CH | 4.1138, dd (8.36, 8.04) | 72.20, CH |
| 3′ | 4.0087, dd (7.97, 6.76) | 72.26, CH | 4.0855, dd (9.71, 8.36) | 75.64, CH |
| 4′ | 4.2887, dd (6.76, 4.79) | 79.88, CH | 4.2868, dd (9.71, 6.23) | 79.23, CH |
| 5′ | 4.0121, ddd (4.79, 2.56, 1.21) | 75.43, CH | 4.1056, ddd (6.23, 4.46, 0.25) | 75.64, CH |
| 6′ | 4.9703, dd (11.65, 1.21) | 64.96, CH2 | 5.0347, dd (12.12, 0.25) | 65.18, CH2 |
| 4.8206, dd (11.65, 2.56) | 4.8722, dd (12.12, 4.46) | |||
| 1″ | 6.4079, d (br s) (1.49) | 102.65, CH | ||
| 2″ | 4.8231, dd (3.29, 1.49) | 73.05, CH | ||
| 3″ | 4.6514, dd (9.37, 3.29) | 73.33, CH | ||
| 4″ | 4.3794, dd (t-like) (9.44, 9.37) | 74.63, CH | ||
| 5″ | 5.0012, dd (9.44, 6.14) | 70.05, CH | ||
| 6″ | 1.7932, d (6.14) | 19.16, CH3 | ||
| FA-1 | 174.09, C | 174.05, C | ||
| FA-2 | 34.94, CH2 | 2.406, m | 34.68, CH2 | |
| FA-3 | 25.83, CH2 | 25.86, CH2 | ||
| FA-olefinic carbons | 5.48–5.56, m | 128.5–131.5 × CH | 5.48–5.56, m | 128.5–131.5 CH |
| FA-14 | 1.20–1.34, m | 32.62, CH2 | 1.20–1.34, m | 32.61, CH2 |
| FA-15 | 1.20–1.34, m | 23.43, CH2 | 1.20–1.34, m | 23.41, CH2 |
| FA-16 | 0.8873, t (7.19) | 14.77, CH3 | 0.9008, t (6.82) | 14.74, CH3 |
Four decimal places were determined for δH and two decimal places were assigned for both δC and J
Measured in pyridine-d5 at 600 MHz
Measured in pyridine-d5 at 400 and 100 MHz, respectively
Figure 3.
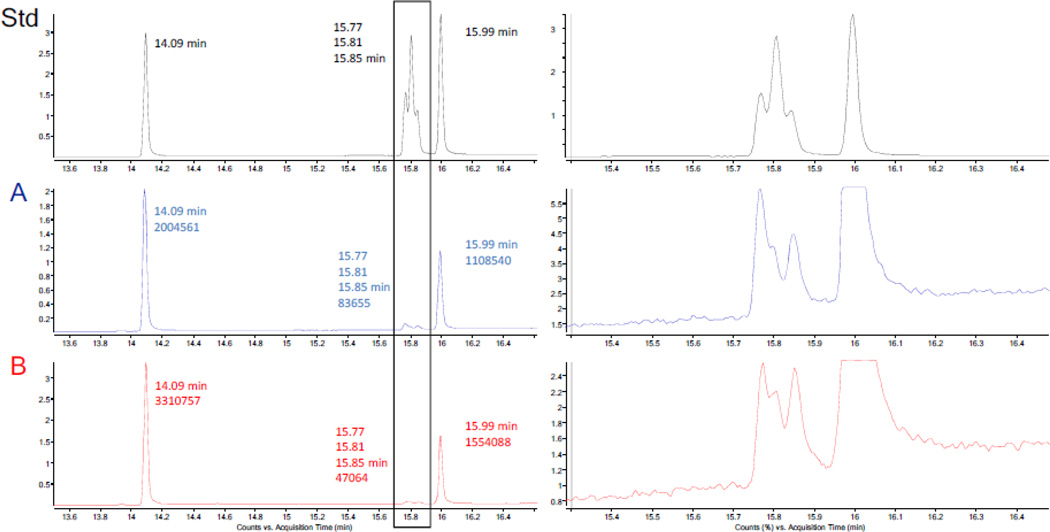
The GC-MS analysis of the methyl esters of the five standard fatty acids (Std) and the hydrolysates of samples A and B: the left panel represents the full view of the GC-MS TIC chromatogram, the retention times, and the integrals in case of samples A and B; the right panel shows expansion of the GC-MS TIC chromatogram of the three unsaturated fatty acid methyl esters.
Figure 4.
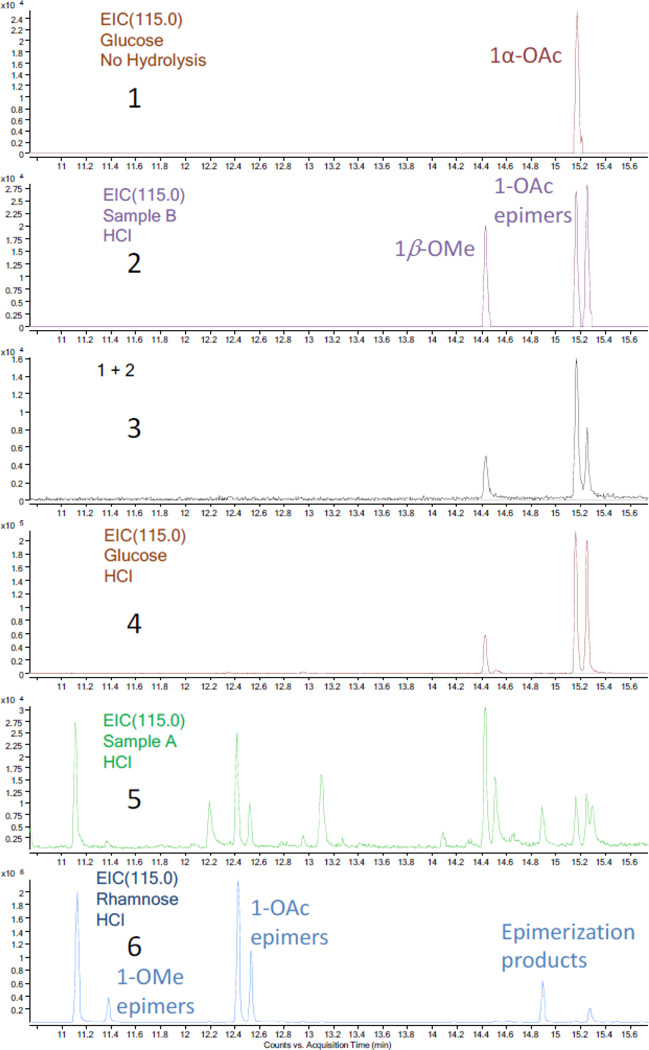
Comparison of the GC-MS EIC chromatograms (m/z 115.0) of the acetylated products (10.8 to 15.6 min). 1, acetylated glucose standard without the HCl hydrolysis procedure; 2, acetylated product of hydrolysate of sample B; 3, mixture of 1 and 2, giving a higher ratio for second peak; 4, acetylated glucose standard with the HCl hydrolysis procedure; 5, acetylated product of hydrolysate of sample A (low intensity); 6, acetylated rhamnose standard with the HCl hydrolysis procedure.
Sample B (6–10): white, amorphous powder; 1H and 13C NMR data, see Tables 1 and 2; GC-MS data, see Figures 3 and 4.
2.4. Preparation of the methyl esters of aliphatic acids and acetylated sugars
Independent aliquots of each of the sample A and B (0.5 mg) were dissolved in 1 mL MeOH, followed by the addition of 1 mL 1.2 M NaOH solution. The reaction vessels were sealed and stored at 100 °C for 30 min. Then, 4 mL HCl solution (50% 6 N HCl and 50% MeOH) was added into the mixture for methylation at 80 °C for 10 min. Finally, 6 mL of hexanes were used to extract the methyl esters of the aliphatic acids from the hydrolysates of both sample A and B.
A second aliquot of each of sample A and B (0.5 mg) was dissolved in 1 mL aliquots of MeOH. To each of these was added 4 mL of 1,4-dioxane and 5 mL of 12 N HCl solution. The reaction mixtures were heated under reflux (110 °C) for 2 h and then cooled to room temperature. H2O was then added, prior to extraction with CHCl3. The H2O fractions were transferred to 25 mL round bottom flasks and dried under vacuum. After drying, pyridine (4 mL), 4-(dimethylamino)pyridine (4 mg) and an excess amount of acetic anhydride were successively added under Ar gas protection to each flask, and the reactions were kept at room temperature overnight. Finally, chloroform (20 mL) was used to extract the acetylated sugars after adding 20 mL H2O to quench the reactions. The same procedures, except for the acid hydrolysis, were used for the acetylation of the monosaccharide standards.
3. Results and Discussion
3.1. New Purification Strategy and Structural Analysis
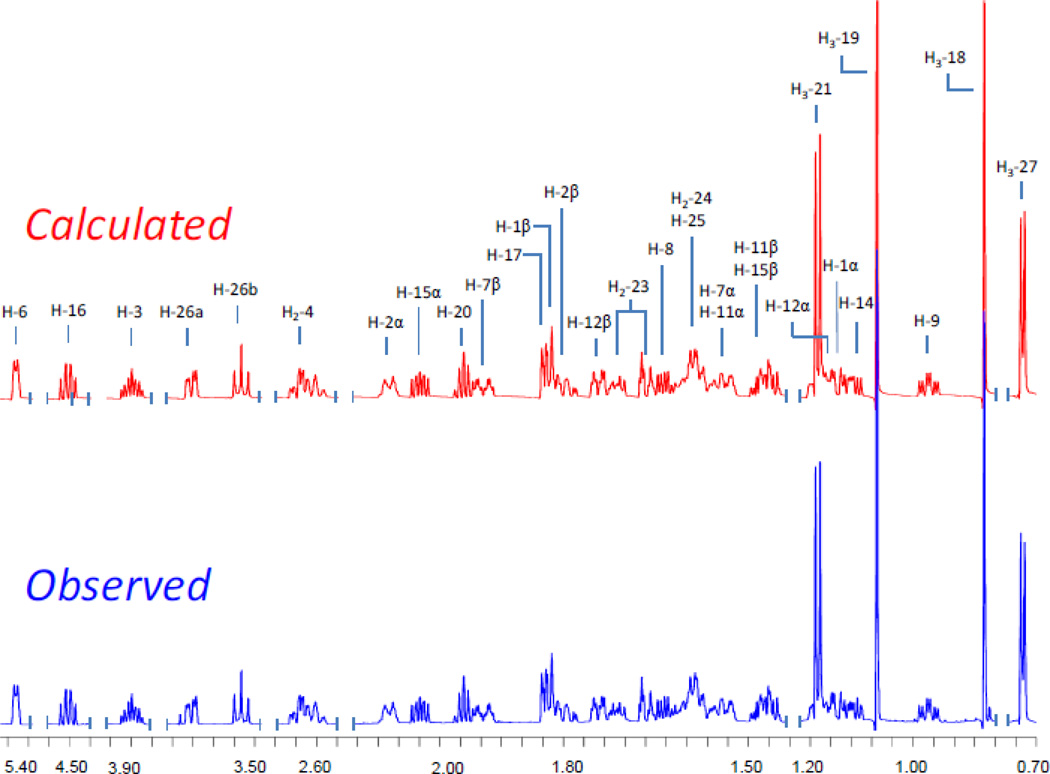
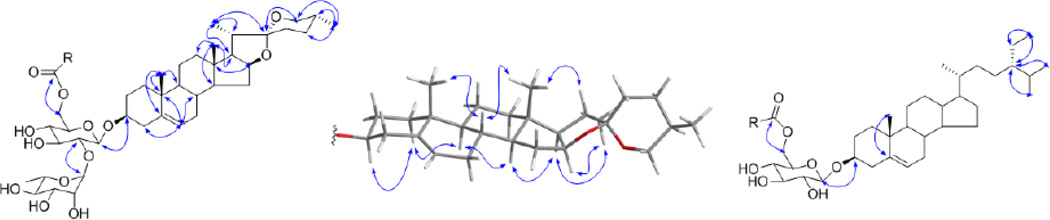
The purification scheme that yielded the lipidated steroid saponins started with the EtOAc partition of the MeOH extract of D. villosa roots/rhizomes (Scheme 1). A suspension of this material in MeOH-H2O (1:9, v/v) was loaded on a vacuum C18 SPE cartridge. Eleven primary fractions were collected by elution with a gradient of a MeOH-H2O from 0:10 to 10:0 v/v [12]. The most non-polar fractions, 10 and 11, were combined for further purification by VLC, MPLC, and HPLC. Purification and spectroscopic as well as chemical analysis eventually afforded 10 lipidated steroid saponins (1–10) which were yielded as two series of mixtures. Seven of the compounds were new (2, 3, 5, and 7–10) and contain either the spirostanol or clionasterol aglycone as the core of the molecule. The structures of the steroid saponin moieties were determined by extensive 1D and 2D NMR spectroscopic analysis, while the identities of aliphatic acid residues were ascertained by GC-MS analysis after hydrolysis and methylation. In order to support the structures with unambiguous assignments of their relative stereochemistry, the highly complex 1H NMR spectra were analyzed by Iterative Full Spin System Analysis (HiFSA) to generate full HiFSA profiles (see subsection General in the Experimental Section) for diosgenin and the sugar residues of both series A and B (Figure 1 and S3 in Supplementary data). Moreover, determination of all δ and J values (Table 1) by HiFSA to 0.0001 ppm and 0.01 Hz precision, respectively, provides valuable data for future unambiguous structure dereplication of the entire compound class of Dioscorea steroids.
Scheme 1.
Summary of the five-step purification strategy. Step 1, the dried and milled rhizomes/roots of D. villosa (4.5 kg) were extracted three times with MeOH to yield 900 g of crude extract; Step 2, the crude extract was suspended in water/methanol (9:1) and successively extracted at room temperature with hexane, CHCl3, and EtOAc; Step 3, the EtOAc partition was subjected to a C18 VLC, affording 11 subfractions; Step 4, the enriched lipidated steroid in contained in subfractions 10 and 11 were chromatographed using NP silica gel VLC, eluting with a CHCl3/MeOH gradient, to give four secondary subfractions (A to D); Step 5A, subfraction C was further fractionated by MPLC on Sephadex LH-20 gel, eluting with MeOH to afford sample B; Step 5B, subfraction D was chromatographed by MPLC using HW-40F gel, eluting with MeOH to afford sample A.
Figure 1.
Result of the 1H NMR iterative full spin analysis and assignment of diosgenin at 900 MHz.
3.2. Structure Elucidation
Series A was obtained as a white, amorphous powder. The 1H NMR spectrum of A (Table 1) exhibited the typical resonances of a steroid saponin and a long-chain aliphatic moiety. The five methyl groups belonging to the steroid aglycone and one sugar residue were clearly observed at δH 0.712 (3H, d, 5.6 Hz), 0.861 (3H, s), 1.080 (3H, s), 1.168 (3H, d, 7.0 Hz), and 1.793 (3H, d, 6.1 Hz), together with the down-fielded resonances between δH 3.4 and 6.5, of which the majority belongs to the sugar residues [7]. The presence of a long-chain aliphatic moiety was indicated by the highly overlapped triplet methyl signal at δH 0.887 and methylene resonances between δH 1.20 and 1.34 [12]. Accordingly, the 13C DEPT-Q NMR spectrum of series A (Table 1) exhibited six methyl group resonances at δC 14.77, 15.53, 16.84, 17.81, 19.16 and 19.90, as well as two anomeric carbon resonances at δC 101.18 and 102.65, and a series of methylene resonances at δC 29.0~30.5 [7,12]. The aforementioned data, together with the ketal carbon resonance at δC 109.74 and the trisubstituted olefin carbons at δC 122.24 and 141.40, indicated the presence of a Δ5 spirostanol moiety as the aglycone core of the molecule. Based on the detailed analysis of the coupling constants (Table 2), the two sugar residues showing the anomeric carbons at δC 101.18 and 102.65 were designated as β-glucose and α-rhamnose, respectively. Both were connected between C-2′ of glucose and C-1″ of rhamnose based on the evidence from the HMBC spectrum (Figure 2) [7]. The sugar residues were showing to be attached to C-3 of the spirostanol via C-1′ of glucose on the basis of the HMBC correlation of H-3/C-1′ (Figure 2). The carbonyl carbon of the long-chain acyl group at δC 174.09 was connected with the C-6′ of the glucose residue through an ester bond, which also was confirmed by the HMBC correlations from H2-6′ to the single carbonyl carbon (Figure 2). In order to establish the structure of the acyl moiety, a small aliquot of sample A was subjected to alkaline hydrolysis and acid catalyzed methylation, followed by GC-MS analysis [13]. Upon comparison with data in the NIST database, the acyl moiety of series A was found to consist of five different constituents: hexadecanoyl, octadecanoyl, 9Z-octadecenoyl, 9Z,12Z-octadecadienoyl and 9Z,12Z,15Z-octadecatrienoyl. A mixture of the corresponding five standards was comparatively analyzed by GC-MS using the same method to give identical retention times and MS fragment patterns as observed with series A (Figure 3). The ratio of the acylated analogues in series A was determined by GC-MS integration as follows: 62.7% hexadecanoic acid, 1.4% 9Z,12Z-octadecadienoic acid, 0.4% 9Z-octadecenoic acid, 0.8% 9Z,12Z,15Z-octadecatrienoic acid, and 34.7% octadecanoic acid derivatives of steroid glycosides. The respective integrals of the three unsaturated fatty acid methyl esters were determined under the assumption that the three corresponding peaks are symmetrical. The relative configuration of the aglycone was determined by analysis of its ROESY spectrum (Figure 2), and was further confirmed by its HiFSA profile which provided a full set of and precise coupling constants (Figure 1). Hence, series A was identified as a mixture of five lipidated steroid saponins: 5-en-spirostanol-2′-O-rha-3-O-glucoside-6′-O-hexadecanoate, 5-en-spirostanol-2′-O-rha-3-O-glucoside-6′-O-octadecanoate, 5-en-spirostanol-2′-O-rha-3-O-glucoside-6′-O-9Z-octadecenoate, 5-en-spirostanol-2′-O-rha-3-O-glucoside-6′-O-9Z,12Z-octadecadienoate, and 5-en-spirostanol-2′-O-rha-3-O-glucoside-6′-O-9Z,12Z,15Z-octadecatrienoate.
Figure 2.
Key 2D NMR correlations of series A (HMBC, left; ROESY, center) and B (HMBC, right).
Similarly, series B was also determined to be the mixture of lipidated steroid saponins on the basis of its 1D NMR spectroscopic analysis. The 13C broad-band decoupled and DEPT NMR spectra showed the diagnostic resonances for seven methyl groups (δC 12.43, 12.60, 14.74, 19.52, 19.69, 19.91 and 20.41), one anomeric carbon (δC 103.33), one double bond (δC 122.37 and 141.47), and one carbonyl carbon (δC 174.05), representing the characteristic skeleton of lipidated clionasterol glucoside [11,12]. This was confirmed by the key HMBC correlations showing in Figure 2. Detailed coupling constant analysis provided evidence for the assignment of the β-glucose residue (Table 2), which was linked with C-3 of clionasterol according to the HMBC correlation of H-1′/C-3. On comparison with series A, the proton and carbon resonances belonging to position C-2′ of series B were shifted upfield by 0.187 and 5.96 ppm, respectively. This substituent chemical shift reflects the absence of the glycosidation effect of the rhamnose moiety linked to position C-2′ in series A. In contrast, the CH-3′ resonances were shifted downfield by 0.077 (1H) and 3.38 (13C) ppm, respectively, again as a result of the missing γ-gauche effect from the glycosidation with rhamnose moiety linked to C-2′in series A. The only carbonyl carbon of the molecule was also linked with the C-6′ of glucose through an ester bond, as evidenced by the HMBC correlations of H2-6′/C-1″. The acyl groups corresponding to the same five congeneric constituents were determined by the same GC-MS methodology used for sample A [13]. The same five fatty acid residues were detected and their ratios were determined to be 67.4:0.4:0.3:0.3:31.6, based on the same symmetrical peak assumption that was employed in analysis of series A. The relative configuration of the aglycone, and especially the stereochemistry of C-24, were determined by comparison of its 1D NMR data with that of structurally related analogues [12]. Thus, series B was characterized as a mixture of the following five compounds: 5-en-clionasterol-3-O-glucoside-6′-O-hexadecanoate, 5-en-clionasterol-3-O-glucoside-6′-O-octadecanoate, 5-en-clionasterol-3-O-glucoside-6′-O-9Z-octadecenoate, 5-en-clionasterol-3-O-glucoside-6′-O-9Z,12Z-octadecadienoate, and 5-en-clionasterol-3-O-glucoside-6′-O-9Z,12Z,15Z-octadecatrienoate.
3.3 1H NMR Full Spin Analysis
To provide unambiguous stereochemical assignment and facilitate the future structural dereplication of congeneric steroid saponins from Dioscorea species, but also in order to support development of qHNMR standardization protocols, the precise 1H NMR profiles of the newly characterized botanical markers were generated by means of HiFSA, using the PERCH software tool [9,14,15]. The long-chain acyl groups contained in all five constituents consist of long aliphatic chains spin-spin coupling and essentially identical chemical shifts for the majority of methylene protons. Altogether, this leads to overcrowded resonances in the range of δH 0.8~2.5 ppm, especially an intensive signal around 1.2 ppm. It is important to note that due to mathematical and computational constraints, the resonances of these aliphatic chains currently cannot be simulated [16]. This situation and the essential lack of structural uniformation actually provide justification for blocking out the (CH2)n signals around δH 1.2 ppm during the HiFSA workflow. Accordingly, the HiFSA calculation focused on the aglycone (series A) and the sugar moieties (series A and B). In the case of series A, resonances of the downfield (> 2.5 ppm) and upfield regions (< 2.5 ppm) were simulated and iterated separately on the basis of their substructures, which included well-dispersed signals in the downfield region and heavily overlapped signals in the upfield region. In order to prevent interference from the acyl groups, the analysis was started with commercial diosgenin, serving as a template for the HiFSA simulation of the spin-spin coupling system of the aglycone of series A (Figure 1). The chemical shifts and coupling constants of the resonances in the downfield region of series A (> 2.5 ppm) were obtained from a separate HiFSA profile, while those in the upfield region (< 2.5 ppm) were obtained by the analysis of its 1D and 2D NMR data together with the HiFSA profile of diosgenin. The full spin analysis of the downfield region (> 2.45 ppm) of series B was also achieved (S2, Supplementary data) and supported the assignment of the sugar moiety. The molecular structures of these selected compounds were used as starting points to analyze each discrete spin system and predict the basic 1H NMR parameters (δ and J). Then, these predicted NMR parameters were optimized through iterative spin system calculations using the PERCHit iterator, until the quantum-mechanical simulations replicated the experimental 1H NMR spectra (S2 and S3, Supplementary data). The final simulated HiFSA spectra exhibited excellent agreement with the observed spectra, for all spectral lines and line intensities, with the exception of the aliphatic signals of the acyl groups mentioned above for the reasons. The final achieved total root-mean-square deviation (RMSD, “residual”) was less than 0.1%. These results further validated the elucidated structures and their relative configuration. The simulated HiFSA spectra represent highly precise fingerprints, which can be used to unambiguously identify the marker compounds and distinguish their resonances from those of impurities by comparison with the experimental spectra. This enables a qHNMR-based determination of content [14] and sample purity [17,18] as well as purity-activity investigations [19]. The digital HiFSA spectra of these secondary metabolomic markers can also serve as references for future metabolomic standardization of wild yam botanical products [14].
3.4 GC-MS Analysis of Sugar Residues
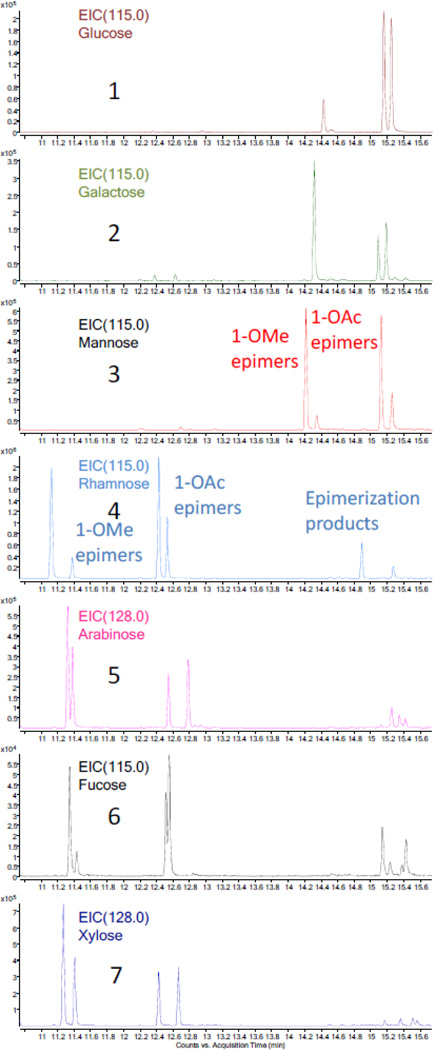
For the purpose of confirming the identity of the sugar residues determined by the analysis of the 1H NMR coupling constants, a GC-MS method (see Experimental Section) was employed to detect the acetylation products of the sugars hydrolyzed from series A and B in comparison with sugar standards. The acetylated hydrolysate of series B showed three peaks in the GC-MS EIC (m/z 115.0, the most abundant fraction ion) chromatogram. Applying the same method to the acetylated product of a D-glucose standard gave only the second peak, which was confirmed by a spiking experiment (Figure 4). The three peaks acquired from sample B were assigned to the derivatives of acetylated glucose with 1β-OMe, 1α-OAc, and 1β-OAc substitution after comparing their MS data with those in the NIST database. The difference between the GC-MS results of two glucose residues can be reasonably ascribed to the acidic hydrolysis in methanol before acetylation, representing the only step which can plausibly introduce the OMe group. In order to confirm this conclusion, the same hydrolysis and acylation procedures used for series B were applied to a D-glucose standard, and indeed the three peaks identical to those obtained from sample B were detected. Series A and the other six sugar standards were also hydrolyzed and subsequently acetylated for GC-MS analysis (Figures 4 and 5). The acetylation products of each of the seven sugar standards displayed three or four disperse peaks in the GC-MS TIC chromatogram, and these clusters could be divided into two groups, the 1-OMe and the 1-OAc derivatives. The 1-OMe derivatives eluted much earlier than 1-OAc derivatives. This characteristic feature and the wide range of retention times together with the high sensitivity of the GC-MS instrument led to an accurate identification of the sugar residues and allowed working with very limited amount of sample.
Figure 5.
Comparison of the GC-MS EIC chromatograms (m/z 115.0 or 128.0) of the seven acetylated sugar standards after applying the HCl hydrolysis procedure (10.8 to 15.6 min). 1–7 represent glucose, galactose, mannose, rhamnose, arabinose, fucose, and xylose, respectively.
3.5 Final Conclusions
The observations make a case that non-polar fractions can still contain steroid saponins and are worthwhile further investigation. Attention to detail is required, especially, their 1H NMR spectra show strongly overlapped and unwelcome signals around 1.2 ppm. Such signals may reflect the presence of ubiquitous aliphatic compounds or lipidated steroid saponins or both. This is a form of hidden residual complexity which can make the secondary metabolomes of botanicals even more complex. Apparently, the plant biosynthesizes a whole series of homologous saturated/unsaturated fatty acid esters. This explains why this portion of the metabolome is so difficult to separate and analyze, and also emphasizes why metabolomic mining is a useful approach to overcome these challenges. Together with its comprehensive application in other research areas [20–23], the success of metabolomic mining methodologies on wild yam so far led to the discovery of two unprecedented compound classes as new biomarkers: the diarylheptanoids [9] and the lipidated steroid saponins.
Steroid saponins are commonly viewed as the only secondary metabolites in wild yam, and that they are important constituents that potentially contribute to the efficacy of the widely-used yam dietary supplements. However, these saponins are equally well known for their poor bioavailability, which results from low intestinal permeability and leads to a significant gap in explaining their alleged therapeutic potential [24]. For example, dioscin [1], an important steroid saponin from wild yam, has been shown to exhibit only very low oral bioavailability (0.2%) after high oral dosing (90 mg/Kg) in rats [25,26]. More generally, poor bioavailability has been demonstrated for analogous steroid saponins [24,27–29]. While these compounds cam can be considered soaps, due to their lipophilic steroid cores and hydrophilic carbohydrate tails, their overall polarity is dominated by the extensive hydroxylation. Accordingly, these molecules show rather polar partition behavior and findings of poor bioavailability are expected.
The lipidation of the hydrophilic carbohydrate adds a significant lipophilic moiety to the molecule, and turns them into dual soaps. In fact, the resulting major change in polarity explains the unexpected chromatographic behavior observed in the present study. It shall be noted that the compound clusters A and B were usually highly retained on C18 reversed phase absorbent.
Transferred to PK parameters, this has two important implications. First, the lipidated steroids are much more lipophilic than their underivatized counterparts. Second, their structurally diverse lipophilic chains imitate the chemistry of membrane bilayers and, in analogy to the altered behavior on C18 silica, can be expected to undergo lipophilic interact with these biological structures.
Taken together, the physicochemical characterization of the lipidated Dioscorea steroids are significantly altered when compared to the parent spirostanols and clionasterols. These observations fuel the hypothesis that these compounds might exhibit a more favorable PK behavior and may potentially represent pro-drug of steroid agents in terms of bioavailability. Investigations regarding the PK behaviors of the new compounds are under way in our laboratory.
Supplementary Material
Acknowledgements
The authors thank Dr. B. Ramirez from UIC-CSB for providing access to the 600 and 900 MHz NMR spectrometers. The authors are also thankful to Dr. C. Simmler, team colleague at UIC, for affording the figure of graphical abstract. M. Totura is acknowledged for maintaining the Atkins Medicinal Plant Garden in UIC. This research was supported by grant P50AT000155 (UIC/NIH Botanical Center), co-funded by the National Center for Complementary and Alternative Medicine (NCCAM) and the Office of Dietary Supplements (ODS), both of the National Institutes of Health. Construction of the UIC-CSB NMR facility and purchase of the 600 and 900 MHz NMR spectrometers were funded by NIHGMS grant P41 GM068944 awarded to Dr. P. G. W. Gettins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Residual Complexity and Bioactivity, Part 21 (see S1, Supplementary Data)
References
- 1.Yoon KD, Chin YW, Yang MH, Choi J, Kim J. Application of high-speed countercurrent chromatography-evaporative light scattering detection for the separation of seven steroidal saponins from Dioscorea villosa. Phytochem Anal. 2012;23:462–468. doi: 10.1002/pca.2342. [DOI] [PubMed] [Google Scholar]
- 2.Marker RE, Turner DL, Ulshafer PR. Sterols. CIV. Diosgenin from certain American plants. J Am Chem Soc. 1940;62:2542–2543. [Google Scholar]
- 3.Dragomirescu A, Muresan A, Alexa E, Andoni M. A HPLC evaluation of genistein – an estrogen-mimetic phytoestrogen – in Glycine max (soy) beans. Acta Endocrinol-Buch. 2009;5:41–47. [Google Scholar]
- 4.Marker RE, Wagner RB, Ulshafer PR, Wittbecker EL, Goldsmith DPJ, Ruof CH. Steroidal sapogenins. J Am Chem Soc. 1947;69:2167–2230. doi: 10.1021/ja01201a032. [DOI] [PubMed] [Google Scholar]
- 5.Aradhana, Rao AR, Kale RK. Diosgenin – a growth stimulator of mammary gland of ovariectomized mouse. Indian J Exp Biol. 1992;30:367–370. [PubMed] [Google Scholar]
- 6.Yoon KD, Kim J. Preparative separation of dioscin derivatives from Dioscorea villosa by centrifugal partition chromatography coupled with evaporative light scattering detection. J Sep Sci. 2008;31:2486–2491. doi: 10.1002/jssc.200800136. [DOI] [PubMed] [Google Scholar]
- 7.Hayes PY, Lambert LK, Lehmann R, Penman K, Kitching W, De Voss JJ. Complete 1H and 13C assignments of the four major saponins from Dioscorea villosa (wild yam) Magn Reson Chem. 2007;45:1001–1005. doi: 10.1002/mrc.2071. [DOI] [PubMed] [Google Scholar]
- 8.Sautour M, Miyamoto T, Lacaille-Dubois MA. Steroidal saponins and flavan-3-ol glycosides from Dioscorea villosa. Biochem Syst Ecol. 2006;34:60–63. [Google Scholar]
- 9.Dong SH, Nikolić D, Simmler C, Qiu F, van Breemen RB, Soejarto DD, Pauli GF, Chen SN. Diarylheptanoids from Dioscorea villosa (wild yam) J Nat Prod. 2012;75:2168–2177. doi: 10.1021/np300603z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali Z, Khan IA. Two new glycosides of spirostane-fatty acid conjugate from Dioscorea cayenensis. Planta Med. 2012;78:PI430. [Google Scholar]
- 11.Al-Qudah MA, Abu Zarga MH. Chemical constituents of Sisymbrium irio L. from Jordan. Nat Prod Res. 2010;24:448–456. doi: 10.1080/14786410903388025. [DOI] [PubMed] [Google Scholar]
- 12.Pullela SV, Choi YW, Khan SI, Khan IA. New acylated clionasterol glycosides from Valeriana officinalis. Planta Med. 2005;71:960–961. doi: 10.1055/s-2005-864179. [DOI] [PubMed] [Google Scholar]
- 13.Cai G, Pauli GF, Wang Y, Jaki BU, Franzblau SG. Rapid determination of growth inhibition of Mycobacterium tuberculosis by GC–MS/MS quantitation of tuberculostearic acid. Tuberculosis. 2013 doi: 10.1016/j.tube.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Napolitano JG, Gödecke T, Rodriguez-Brasco MF, Jaki BU, Chen SN, Lankin DC, Pauli GF. The tandem of full spin analysis and qHNMR for the quality control of botanicals exemplified with Ginkgo biloba. J Nat Prod. 2012;75:238–248. doi: 10.1021/np200949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napolitano JG, Lankin DC, Graf TN, Friesen JB, Chen SN, McAlpine JB, Oberlies NH, Pauli GF. HiFSA fingerprinting applied to isomers with near-identical NMR spectra: the silybin/isosilybin case. J Org Chem. 2013 doi: 10.1021/jo302720h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laatikainen R, Tiainen M, Korhonen SP, Niemitz M. Computerized analysis of high-resolution solution-state spectra. eMagRes. 2011 [Google Scholar]
- 17.Qiu F, Friesen JB, McAlpine JB, Pauli GF. Design of countercurrent separation of Ginkgo biloba terpene lactones by nuclear magnetic resonance. J Chromatogr A. 2012;1242:26–34. doi: 10.1016/j.chroma.2012.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauli GF, Gödecke T, Jaki BU, Lankin DC. Quantitative 1H NMR. Development and potential of an analytical method: an update. J Nat Prod. 2012;75:834–851. doi: 10.1021/np200993k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu F, Cai G, Jaki BU, Lankin DC, Franzblau SG, Pauli GF. Quantitative purity-activity relationships of natural products: the case of anti-tuberculosis active triterpenes from Oplopanax horridus. J Nat Prod. 2013;76:413–419. doi: 10.1021/np3007809. [DOI] [PubMed] [Google Scholar]
- 20.Horgan RP, Broadhurst DI, Walsh SK, Dunn WB, Brown M, Roberts CT, North RA, McCowan LM, Kell DB, Baker PN, Kenny LC. Metabolic profiling uncovers a phenotypic signature of small for gestational age in early pregnancy. J Proteome Res. 2011;10:3660–3673. doi: 10.1021/pr2002897. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Yang B, Sun H, Zhang A. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal Chem. 2012;84:428–439. doi: 10.1021/ac202828r. [DOI] [PubMed] [Google Scholar]
- 22.Kim N, Kim K, Choi BY, Lee D, Shin YS, Bang KH, Cha SW, Lee JW, Choi HK, Jang DS, Lee D. Metabolomic approach for age discrimination of Panax ginseng using UPLC-Q-Tof MS. J Agric Food Chem. 2011;59:10435–10441. doi: 10.1021/jf201718r. [DOI] [PubMed] [Google Scholar]
- 23.Lee SH, Woo HM, Jung BH, Lee J, Kwon OSP, H S, Choi MH, Chung BC. Metabolomic approach to evaluate the toxicological effects of nonylphenol with rat urine. Anal Chem. 2007;79:6102–6110. doi: 10.1021/ac070237e. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Wang J, Gao W, Man S, Guo H, Zhang J, Liu C. Formulation and in vitro absorption analysis of Rhizoma paridis steroidal saponins. Int J Pharm. 2013;441:680–686. doi: 10.1016/j.ijpharm.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Li K, Tang Y, Fawcett JP, Gu J, Zhong D. Characterization of the pharmacokinetics of dioscin in rat. Steroids. 2005;70:525–530. doi: 10.1016/j.steroids.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Li K, Wang Y, Gu J, Chen X, Zhong D. Determination of dioscin in rat plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2005;817:271–275. doi: 10.1016/j.jchromb.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 27.Chen LL, Yuan WW, Hu ZF, Qi J, Zhu DN, Yu BY. Determination and pharmacokinetics of DT-13 in rat plasma by LC-MS. J Pharm Biomed Anal. 2011;56:650–654. doi: 10.1016/j.jpba.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Okawara M, Tokudome Y, Todo H, Sugibayashi K, Hashimoto F. Enhancement of diosgenin distribution in the skin by cyclodextrin complexation following oral administration. Biol Pharm Bull. 2013;36:36–40. doi: 10.1248/bpb.b12-00467. [DOI] [PubMed] [Google Scholar]
- 29.Cai F, Sun L, Gao S, Yang Y, Yang Q, Chen W. A rapid and sensitive liquid chromatography-tandem mass spectrometric method for the determination of timosaponin B-II in blood plasma and a study of the pharmacokinetics of saponin in the rat. J Pharm Biomed Anal. 2008;48:1411–1416. doi: 10.1016/j.jpba.2008.09.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.