Abstract
Phosphatase and Tensin Homologue on chromosome Ten (PTEN) is a tumor suppressor and an antagonist of the phosphoinositide-3 kinase (PI3K) pathway. We identified a 576-amino acid translational variant of PTEN, termed PTEN-Long, that arises from an alternative translation start site 519 bp upstream of the ATG initiation sequence, adding 173 N-terminal amino acids to the normal PTEN open reading frame. PTEN-Long is a membrane permeable lipid phosphatase that is secreted from cells and can enter other cells. As an exogenous agent, PTEN-Long antagonized PI3K signaling and induced tumor cell death in vitro and in vivo. By providing a means to restore a functional tumor suppressor protein to tumor cells, PTEN-Long may have therapeutic uses.
PTEN (Phosphatase and tensin homologue on chromosome ten) is a tumor suppressor that is mutated in multiple types of cancer (1,2). PTEN encodes a dual-specificity phosphatase, whose primary substrate is phosphatidylinositol 3,4,5 trisphosphate (PIP3) (3–7). Through this activity, PTEN antagonizes phosphoinositide 3-kinase (PI3K) signaling and thereby affects a myriad of cellular processes including growth, proliferation, and survival (8–11). In mice, loss of Pten in tumors or the tumor microenvironment results in neoplastic growth (12–14) indicating that PTEN’s tumor suppressive functions are not confined to tumor cells alone.
Inspection of the PTEN mRNA transcript (Fig. 1A) revealed an alternative translation initiation codon (CUG) at bp 513 that was 5’ of and in-frame with the canonical translation initiation codon (AUG) at bp 1032. Alternative translation beginning at bp 513 was predicted to encode a 576-amino acid translational variant, which we termed PTEN-Long. PTEN-Long contains a 173-amino acid domain at its N-terminus followed by the classical 403 amino acids of PTEN (Fig. S1)(15). Search of the Catalogue of Somatic Mutations in Cancer (http://www.sanger.ac.uk/genetics/CGP/cosmic/) database found five somatic missense mutations specific for PTEN-Long in tumor samples (Fig. S1). Alignment of the predicted 173 amino acid sequence of the PTEN-Long unique region with predicted protein sequences from other species indicated that it is evolutionarily conserved (Fig. S2).
Fig. 1.
PTEN-Long is a translational variant of PTEN with lipid phosphatase activity. (A) Shared PTEN/PTEN-Long mRNA (NM_000314.4; orange) with predicted coding regions of PTEN (red) and PTEN-Long (blue). (B) Phosphatase activity assay. 40 nM PTEN and PTEN-Long, assayed with di-C8-PIP3 for lipid phosphatase activity, error bars indicate SEM of three replicates (p<0.001, ANOVA), Regression lines to Michaelis-Menten kinetics with Vmax and 95% Confidence interval (CI) shown in inset moles PO4/min/moles enzyme. This is a representative experiment of 3. (C) Immunoblot of whole cell lysates from wildtype and Pten null embryonic stem cells and cancer cell lines of known PTEN status. PTEN-Long (Gly-2) recognizes an epitope specific to the PTEN-Long unique region. PTEN (138G6) recognizes an epitope that is common to both PTEN and PTEN-Long. (D) Reciprocal immunoprecipitations of proteins from whole cell lysates of HEK293 cells with PTEN-Long specific antibody, probed with an antibody that recognizes an epitope that is common to PTEN/PTEN-Long and vice versa. Input lanes are from longer exposure of the same membrane. Arrows indicate PTEN-Long. (E) Immunoblots of four, randomly selected PTEN wild-type sets of matched breast tumor (T)/normal breast (N) pairs.
To determine if PTEN-Long is translated in cells we transfected human breast (BT549) and glioblastoma (U87MG) cells that lack PTEN with a cDNA expression vector containing the PTEN-Long and PTEN open reading frames. Immunoblots of these lysates revealed two forms of PTEN: a ~75-kD form, PTEN-Long, and a ~55-kD species that corresponded to the conventional translated protein. Expression of PTEN-Long was increased by mutating the alternate start site (CTG) to a classical start site (ATG) (16), was not diminished when the canonical ATG was mutated to ATA, and was lost if the alternatively translated region was deleted or a frame shift was introduced between the alternate start site at 513 and the classic start site at 1032 (Fig. S3). PTEN-Long showed a different enzymatic profile across the tested range of lipid substrate concentrations (soluble di-C8-phosphatidylinositol-3,4,5-trisphosphate (di-C8-PIP3)) (Fig. 1B, Fig. S4) (p<0.001 by ANOVA ) (17). A missense mutation in the phosphatase domain of PTEN-Long (G302R), analogous to PTEN (G129R), a tumor mutation, decreased the phosphatase activity of the protein (Fig. S5)(1, 18, 19). Immunoblots of U87MG cells overexpressing equivalent amounts of transfected PTEN, PTEN-Long, or their mutated analogues confirmed that, similar to PTEN, PTEN-Long decreased signaling through the PI3K pathway in a phosphatase-dependent manner (Fig. S6). Immunoblots of mouse embryonic stem cells and human cancer cell lines with antibodies either specific for PTEN-Long or an epitope shared by PTEN and PTEN-Long showed that a ~75 kD PTEN-Long band was present in wild-type but not cells lacking PTEN (Fig. 1C). Immunoprecipitations with PTEN or PTEN-Long antibodies confirmed that PTEN-Long contains canonical PTEN peptides and is therefore a translational variant of PTEN (Fig. 1D, Fig. S7). Amounts of PTEN-Long were reduced in primary human breast tumors compared to those of matched normal breast samples (Fig. 1E, Fig. S8, Table S1). Immunohistochemistry of brain tissue from a mouse glioblastoma caused by the deletion of Pten, p53, and overexpression of PDGF revealed that PTEN-Long was more abundant in the tumor microenvironment than in the tumor or normal tissue (Fig. S9)(20). PTEN-Long was similarily abundant in the tumor microenvironment in 4 of 50 samples from primary breast cancers (Fig. S10).
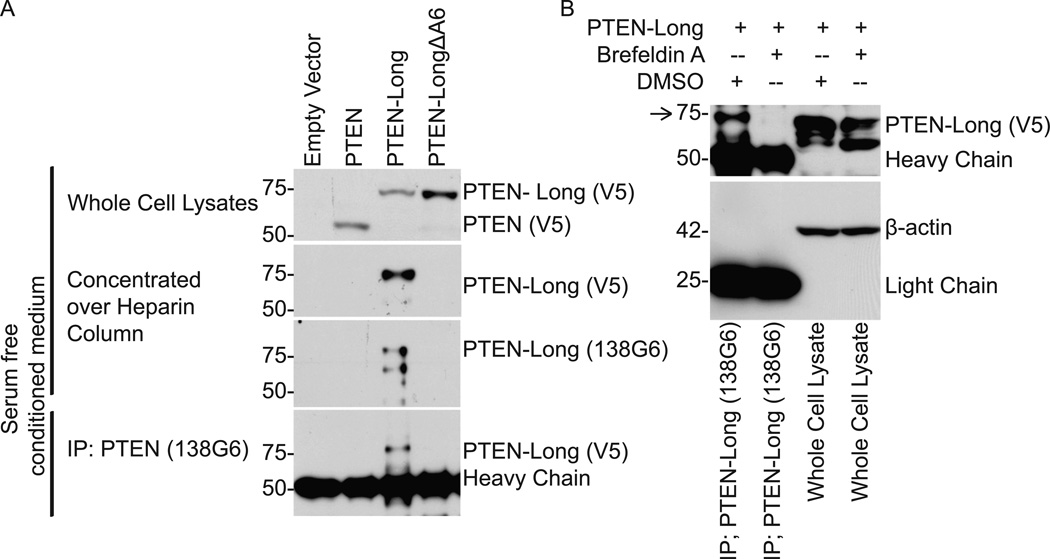
Computer modeling indicated that PTEN-Long contained a secretion signal sequence with a predicted cleavage site at amino acid 22 (Fig. S11)(21). Thus, we tested whether PTEN-Long might exist outside of cells. We overexpressed V5 epitope tagged PTEN, PTEN-Long, and a mutant with an altered signal sequence predicted to abolish secretion (PTEN-LongΔA6) (Fig. S12) in human embryonic kidney (HEK293) cells. We concentrated the proteins from conditioned medium by immunoprecipitation with PTEN (138G6) antibody or with heparin columns (Fig. S13). With antibodies that recognized the V5 tag or the endogenous C terminal region of PTEN and PTEN-Long, we detected PTEN-Long but not PTEN nor PTEN-LongΔA6 in conditioned medium (Fig. 2A, Fig. S14). Brefeldin-A, an inhibitor that interferes with retrograde transport of vesicles in the endoplasmic reticulum, inhibited the secretion of PTEN-Long (Fig.2B)(22). Endogenous PTEN-Long from HEK293 cells bound to concanavalin A-sepharose, a hallmark of a secreted glycoprotein (Fig. S15). Furthermore, PTEN-Long but not PTEN was present in both human plasma and serum (Fig. S16). A screen for PTEN interacting proteins identified multiple heparan-sulfate-modified cell surface proteins in the glypican and syndecan families (Fig. S17)(23). Thus PTEN-Long appears to be secreted from cells and to interact with cell surface proteins.
Fig. 2.
Secretion of PTEN-Long. (A) PTEN, PTEN-Long, and PTEN-LongΔA6 from serum free conditioned medium of transfected HEK293 cells eluted from a heparin column or immunoprecipitated with C-Terminal PTEN Antibody 138G6. (B) Cellular (lanes 3 and 4) or secreted PTEN-Long from culture medium (lanes 1 and 2) of transfected HEK293 cells treated with or without brefeldin A at 1 μg/ml. Arrow indicates PTEN-Long.
We noted the presence of a poly-arginine stretch in the unique region of PTEN-Long that was evolutionarily conserved (Fig. S2) and bore a resemblance to the poly-basic residues of the cell-penetrating element of the HIV transactivator of transcription (TAT) protein (24, 25). To determine whether this sequence conferred similar properties to PTEN-Long, we constructed a PTEN-LongΔR6 construct in which these six arginines were deleted (Fig. S18). After treating cells with 100 nM purified Red Fluorescent Protein (RFP)-V5/His, PTEN-Long-RFP-V5/His, or PTEN-LongΔR6-RFP-V5/His, we detected PTEN-Long-RFP but not RFP nor PTEN-LongΔR6-RFP in the cells by fluorescence microscopy (Fig. 3A, Fig. S19). We confirmed this observation through subcellular fractionation (Fig. 3B, Fig. S20). Fusion of the PTEN-Long unique region (PL) to red fluorescent protein (RFP) enabled cellular uptake of the fusion protein (Fig. S21). Therefore the PL domain facilitates cell penetration of tethered peptide sequences. We next tested whether PTEN-Long affected cellular signaling as an exogenous agent. Treatment of cells in culture for fifteen minutes with purified PTEN-Long reduced basal phosphorylation of the protein kinase AKT on Thr 308 in the majority of cell lines (Fig. 3C, Fig. S22–23). When starved U87MG cells were incubated with PTEN-Long for 10 minutes, subsequent stimulation of phosphorylation of AKT Thr 308 by insulin or epidermal growth factor (EGF) was inhibited. Inhibition of PI3K signaling appeared to be dependent upon the poly-arginine sequence of PTEN-Long because the PTEN-LongΔR6 mutant did not block Insulin or EGF-induced phosphorylation of AKT (Fig. S24). Dose response experiments in cells deprived of serum and treated for 24 hours showed that PTEN-Long decreased PI3K signaling as indicated by the decreased phosphorylation of AKT, FOXO, and PRAS40, and induced cell death as indicated by cleavage of caspase 3 (Fig. 3,D–E). We confirmed this effect on cell survival by treating U87MG and MDA-MB-468 cells grown in 0.1% serum with various doses of PTEN-Long or PTEN-LongΔR6 (Fig. 3F).
Fig. 3.
Entry of PTEN-Long into cells and inhibition of PI3K signaling. (A) Direct fluorescence of U87MG cells 30 min after treatment with 100 nM of either red fluorescent protein (RFP), PTEN-Long-RFP, or PTEN-LongΔR6-RFP. (B) Subcellular fractionation of MDA-MB-468 cells 1 hour after 25 nM treatment with indicated purified proteins. C and N indicate cytoplasmic and nuclear fractions, respectively. Offset, immunoblot of input protein samples used to treat the cells. (C) Effect of 25 nM PTEN-Long at 15 minutes upon pAKT(308). (D) Immunoblots of U87MG and (E) MDA-MB-468 cell lysates 24 hours after treatment with indicated doses of PTEN-Long. (F) Cell viability assay. U87MG and MDA-MB-468 cells were treated with the indicated doses of PTEN-Long or PTEN-LongΔR6 for 24 hours before scoring of the fraction of cells with trypan blue staining. Assayed in quadruplicate, error bars indicate SEM.
Upon treatment of mice with PTEN-Long we observed in blood a transient increase in glucose concentration (Fig. S25) and PTEN-Long (Fig. S26). Similar to what we observed in culture, PTEN-Long was detected in mouse tissues, including a xenografted breast tumor (MDA-MB-468), and could alter cell signaling (Fig. S27–29). PTEN-Long derivatives lacking phosphatase or cell penetrating activity were unable to cause these signaling changes. TAT-PTEN only recapitulated some of the effects of PTEN-Long indicating that the PTEN-Long unique region may have other functions beyond the cell permeability conferred by TAT (Fig. S30).
We further explored the effect of PTEN-Long in murine tumor models. We engrafted U87MG cells subcutaneously into athymic nude mice and allowed them to grow until their tumor volume reached approximately 0.2 cm3. Animals were then injected intraperitoneally with PTEN-Long (4 mg/kg) or a control preparation. PTEN-Long caused tumor regression after four days of treatment (Fig. 4A, Fig. S31). Similar regressions were also observed in multiple other xenograft models with the exception of the human colon cancer tumor cell line HCT116 (Fig. S32–33). Although both PTEN-Long and PTEN-Long G302R were detected within tumor tissues after five days of treatment, only the wild-type protein affected PI3K signaling or induced apoptosis (Fig. 4B). Immunohistochemistry of serial sections of tumor xenografts confirmed exogenous PTEN-Long within tumor cells and a concomitant reduction of pAKT staining. We also treated syngeneic allografts from a mouse model of glioblastoma (GBM). After treatment with PTEN-Long (4 mg/kg), tumors derived from Pten−/−, p53−/−, PDGF overexpressing GBM cells underwent complete tumor regression within five days, whereas treatment with PTEN-LongΔR6 did not prevent tumor growth (Fig. 4C) (20). Mutant PTEN-Long proteins derived from three cancers (Fig. S1) in which PTEN-Long underwent somatic mutation all showed reduced ability to affect signaling in vitro and in vivo, suggesting there may be selective pressure to mutate PTEN-Long during tumor development (Fig. S34). Thus PTEN-Long protein appears to alter PI3K signaling in vivo to inhibit tumor growth and this effect is dependent on its entrance into cells.
Fig. 4.
Effects of PTEN-Long on signaling and tumor development in mice. (A) Graph of U87MG tumor volumes as measured by calipers and treated with either PTEN-Long (4 mg/kg) or an equal volume of mock purified material (N= 5 mice/treatment, error bars indicate +/−SD). (B) Immunoblots of markers of PI3K signaling in U87MG xenografts treated for 5 days with PTEN-Long or phosphatase mutant PTEN-Long(G302R). (C) Pten−/− p53−/− PDGF overexpressing cells derived from the genetically engineered mouse model of GBM were allografted into the flank of syngeneic hosts and treated with 4 mg/kg of PTEN-Long or PTEN-LongΔR6 for 5 days. Tumor volume was assessed daily by caliper measurements and on days 0 and 5 with xenogen imaging system as shown. After 5 days of treatment the mice in the PTEN-LongΔR6 (red arrows) cohort were then treated with PTEN-Long (green arrows) followed with caliper measurements. Blue arrows indicate initial PTEN-Long treatment (days 0–4).
PTEN-Long is a translational variant of PTEN that, like classical PTEN, acts as an antagonist of the PI3K pathway. As a secreted product of a tumor suppressor gene capable of entering cells, endogenous PTEN-Long may contribute to an organism’s maintenance of signaling, tissue homeostasis and suppression of cancer. Recombinant PTEN-Long and derivative fusion proteins may be useful for delivering proteins in a variety of experimental and clinical settings.
Supplementary Material
Acknowledgments
We thank L. Greene, T. Ludwig, D. Yamashiro, K. Olive and members of the Parsons laboratory for their assistance. Supported by NIH CA082783 NCI CA097403, NIH R01NS066955, the Avon Foundation, Octoberwomen Foundation. BH is supported by 2T32 CA09503. Columbia University has applied for patents on PTEN-Long. NM_000314.4 is curated by GenBank. Additional data are present in the supporting online material.
Footnotes
Supplementary Materials
Materials and Methods
Figures S1–S34
Tables S1
References (26–29)
References and Notes
- 1.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;4:356. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 4.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 5.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Leslie NR, Downes CP. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J. 2004;382:1. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 8.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 9.Sun H, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6199. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furnari F, Lin H, Huang H, Cavenee W. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12479. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, et al. The PTEN/MMAC1 Tumor Suppressor Induces Cell Death That Is Rescued by the AKT/Protein Kinase B Oncogene. Cancer Res. 1998;58:5667. [PubMed] [Google Scholar]
- 12.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 1998;19:348. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 13.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1563. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online.
- 16.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnachie G, Pass I, Walker S, Downes C. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 2003;371:947. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers M, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13513. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr, V, Val; W, Trp, and Y, Tyr
- 20.Lei L, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLos ONE. 2011;6:20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:122. [PubMed] [Google Scholar]
- 22.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992;116:1071. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elangovan B, Subramanian T, Chinnadural G. Heterologous basic domain substitutions in the HIV-1 Tat protein reveal an arginine-rich motif required for transactivation. EMBO. 1991;10:2311. doi: 10.1002/j.1460-2075.1991.tb07768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarze S, Ho A, Vocero-Akbani A, Dowdy S. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 26.Xia W, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puc J, et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Saal L, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7564. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lois, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:5556. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.