Abstract
Defective DNA replication can result in genomic instability, cancer, and developmental defects. To understand the roles of DNA damage response (DDR) genes on carcinogenesis in mutants defective for core DNA replication components, we utilized the Mcm4Chaos3/Chaos3 (“Chaos3”) mouse model which, by virtue of an amino acid alteration in MCM4 that destabilizes the MCM2-7 DNA replicative helicase, has fewer dormant replication origins and an increased number of stalled replication forks. This leads to genomic instability and cancer in most Chaos3 mice. We found that animals doubly mutant for Chaos3 and components of the ATM double strand break response pathway (Atm, p21/Cdkn1a, Chk2/Chek2) had decreased tumor latency and/or increased tumor susceptibility. Tumor latency and susceptibility differed between genetic backgrounds and genders, with females demonstrating an overall greater cancer susceptibility to Atm and p21 deficiency than males. ATM deficiency was semilethal in the Chaos3 background and impaired embryonic fibroblast proliferation, suggesting that ATM drug inhibitors might be useful against tumors with DNA replication defects. Hypomorphism for the 9-1-1 component Hus1 did not affect tumor latency or susceptibility in Chaos3 animals, and tumors in these mice did not exhibit impaired ATR pathway signaling. These and other data indicate that under conditions of systemic replication stress, the ATM pathway is particularly important both for cancer suppression and viability during development.
Keywords: DNA replication, DNA damage checkpoints, minichromosome maintenance (MCM) proteins, 9-1-1, MCM2-7 helicase, cancer, replication stress
INTRODUCTION
Genomic studies have shown that many individual genes are spontaneously mutated or misregulated at low frequencies in cancers, but together comprise disruptions in a few key pathways 1-3. Alterations in DNA checkpoint and repair pathways are particularly significant. The BRCA1 and BRCA2 genes are altered in over 1/3 of serous ovarian and basal type breast cancer cases, highlighting the importance of the homologous recombination (HR) pathway of DSB repair 2, 4. During HR repair, DSBs are bound by the MRN (MRE11/RAD50/NBS1) damage sensor complex, the Ataxia Telangiectasia Mutated (ATM) serine/threonine kinase becomes activated via autophosphorylation and, in conjunction with mediator proteins such as BRCA1, signals to downstream transducer and effector kinases to elicit checkpoint and repair responses (reviewed by 5, 6). DDR pathways are responsible for helping maintain genomic stability and suppressing tumorigenesis 7. To control cell cycle progression under conditions of DNA damage or replication stress, DDR genes also target components of the DNA replication machinery, including the Minichromosome maintenance 2-7 (MCM2-7) replicative helicase complex. MCM2 is a direct target of ATR (ATM and RAD9-related), and MCM3 is a target of ATM 8, 9.
Whereas the relationship between defects in various DNA repair systems to cancer is well studied, this is not the case for DNA replication - the process during which the greatest opportunity for mutations exists. Accumulating evidence points to associations between deficiencies of the core DNA replication machinery and cancer. For example, mice bearing mutations in the proofreading functions of the major replicative polymerases δ and ε exhibit mutator phenotypes and cancer predisposition 10-13. Furthermore, Pol ε is frequently mutated in human colorectal cancers 14. In addition to DNA polymerases, mutations in components of the pre-replication complex (pre-RC) have been linked to cancer susceptibility. These complexes assemble at replication origins during G1 phase (but not during S phase), and a subset of these components constitute the CDC45/MCM2-7/GINS (CMG) replicative helicase complex that unwinds DNA in front of the replisome during S phase 15-17. The highly conserved MCM2-7 heterohexameric complex is an essential component of the pre-RC and constitutes the core of the replicative helicase (reviewed in 18). Whereas Mcm2-7 are essential genes, hypomorphic alleles in mice cause GIN, cancer susceptibility, and cell proliferation defects 19-21, as does overexpression and haploinsufficiency 22-24.
To better understand the in vivo impact of the DDR on cancer incidence and tumor latency under conditions of increased replication stress, we utilized the Mcm4Chaos3/Chaos3 (“Chaos3”) mouse model that bears a single amino acid mutation in MCM4 (Phe345Ile). Chaos3 mice have dramatically elevated GIN, and depending on the strain background, Chaos3 mice are predisposed to various cancers including mammary tumors, histiocytic sarcoma, lymphoma, and bone tumors 19, 24, 25. The Chaos3 mutation destabilizes the MCM2-7 helicase by disrupting MCM4:MCM6 interaction, somehow triggering a post-transcription decrease in the levels of all MCM2-7 mRNA and proteins 24-26. This reduces the number of dormant replication origins available as backups to replicate DNA near stalled replication forks. These defects contribute to elevated chromosome breakage and segregation defects in Chaos3 mouse embryonic fibroblasts (MEFs) 25. Studies of diploid S. cerevisiae engineered to carry the identical Chaos3 amino acid change in MCM4 indicated that the defective helicase causes replication fork collapse, leading to DSBs that require repair by HR 27. Consistent with replication fork damage leading to DSBs that trigger HR, Chaos3 MEFs have increased levels of RAD51 and BLM foci 25. Additionally, they exhibit upregulation of p53/TRP53 and p21, indicative that cell cycle checkpoint responses are activated in these cells 28.
DDR pathways aid proper DNA replication by stabilizing transiently stalled forks to prevent the dissociation of replisome components, promoting replication restart, and facilitating fork movement on difficult-to-replicate templates. The ATM pathway is activated in response to DSBs, while the ATR pathway is activated by RPA-coated ssDNA at stalled replication forks. However, there is clearly overlap and cross-signaling between the pathways 29. Failure to safeguard genome integrity during DNA replication is associated with increased cancer predisposition 30, 31.
Despite intact DDR pathways, the elevated GIN in Chaos3 mice eventually result in recurrent segmental copy number alterations that apparently drive carcinogenesis, with a mean latency of 12 months in the case of mammary tumors 19, 32. Here, we exploit this model, in conjunction with mutations in DDR genes, to better understand cellular responses to endogenous replication stress on an organismal level and the impact on carcinogenesis in vivo.
RESULTS
We generated Chaos3 mice that were also deficient for the ATM pathway (Atm or Chk2), ATR pathway (Hus1), or the cyclin-dependent kinase inhibitor p21 that is downstream of both signaling pathways (Figure 1a). At the time of crossing, Mcm4Chaos3 (abbreviated hereafter as Mcm4C3, or just “C3” in the figures) was congenic in strain C3H/HeBFeJ (C3H), but the other mutations were on different strain backgrounds (see Materials and Methods). C3H-Mcm4C3/C3 females develop exclusively mammary adenocarcinomas, but males of that genotype and strain background were not reported to be tumor prone 19. In a mixed genetic background however, other tumor types in females arise (including lymphoma and histiocytic sarcoma) 19, 25. Additionally, males of mixed strain background were also found to be tumor prone, though the sample size was small and most mice were not aged past 14 months 19. Here, mutant and control mice of both sexes were aged for eighteen months or until they showed signs of disease, after which a complete necropsy was performed. The results for each set of compound mutants are described below.
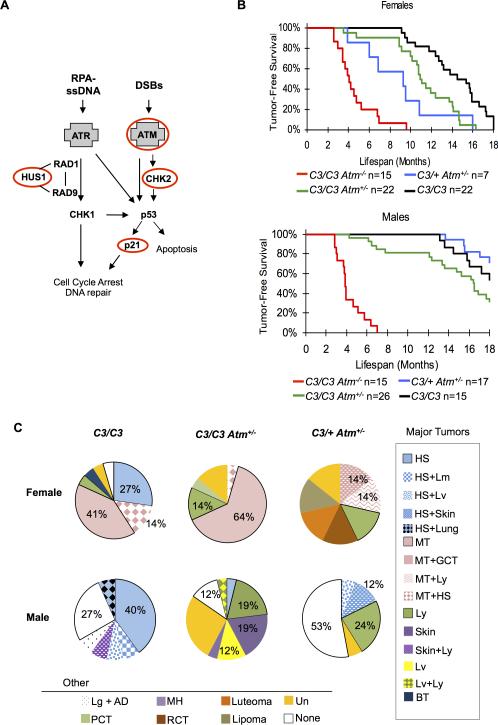
Figure 1. Atm deficiency impacts Chaos3 tumor latency and tumor susceptibility.
(A) DNA damage response pathways. Key genes in DDR pathways are shown with the ATR and ATM DNA damage sensors emphasized in gray boxes. Genes perturbed in this study are indicated by red ovals. (B) Kaplan-Meier graphs of the indicated genotypes and sexes. Mcm4C3/C3 Atm+/− and Mcm4C3/+ Atm+/− females have significantly decreased tumor latency compared to Mcm4C3/C3 alone (see statistics in Supplementary Table 2). Mcm4C3/C3 Atm+/− males neared statistical significance for decreased tumor latency, and Mcm4C3/+ Atm+/− male tumor latency was similar to Mcm4C3/C3 alone (Supplementary Table 2). C3 = Mcm4C3. (C) Tumor spectra of selected genotypes. HS=histiocytic sarcoma, MT=mammary tumor, BT=bone tumor, Ly=lymphoma, None=healthy (no detectable cancer), PCT=plasma cell tumor, RCT=round cell tumor, GCT=granulosa cell tumor, Lv=liver, MH=myeloid hyperplasia, AD=adrenal ganglioneuroma, Un=unknown tumor type. Note that tumor spectrum is affected by genotype and gender, and that Mcm4C3/+ Atm+/− females are more susceptible to cancer than males. C3 = Mcm4C3.
ATM deficiency impacts viability, cell proliferation, tumor latency, and tumor susceptibility of Chaos3 mice
Activation of ATM in response to DSBs triggers several key downstream events. It directly phosphorylates H2AX residing at (and near) the site of DNA breaks. It also phosphorylates downstream targets such as CHK2 to activate the DNA damage checkpoint, leading to cell cycle delay or apoptosis (Figure 1a) 33. ATM deficiency is associated with the development of lymphomas and leukemias in humans and mice. Atm−/− mice develop thymic lymphomas at 2-4 months of age 34, 35,36, 37. We analyzed 648 weaned offspring from mouse crosses bearing Atm and Mcm4 genotype combinations, but only 25 of the expected 65 double homozygotes were obtained (P=6.03*10-6) (Supplementary Figure 1a). To investigate the nature of the semi-lethal phenotype, we examined mid-late gestation embryos from timed matings that would yield double mutants and controls. Mcm4C3/C3 Atm−/− embryos were present at expected ratios at and prior to E15.5 (χ2 P=0.97 and P=0.65, respectively; Supplementary Table 1), but at E18.5 they were smaller than control littermates and/or apparently dead or dying (Supplementary Figure 1b). To better understand the basis for the embryonic lethality at the cellular level, cell proliferation assays were conducted on MEFs of various genotypes. Complete absence of ATM dramatically decreased growth rate regardless of Chaos3 genotype, but Atm heterozygosity also reduced proliferation in Mcm4C3/C3 but not Mcm4C3/+ MEFs (Supplementary Figure 1c). The results suggest that reduced cell proliferation is not entirely responsible for the synthetic lethality of Atm−/− Mcm4C3/C3 embryos.
The early-onset lymphoma susceptibility caused by complete ATM deficiency obscured the detection of potential effects on mammary tumorigenesis. Nearly all Mcm4C3/C3 Atm−/− and Mcm4C3/+Atm−/− mice succumbed to lymphoma at ~2-4 months of age (Supplementary Table 2, Supplementary Table 3), compared to much longer tumor latency in Mcm4C3/C3 animals (Figure 1b, Supplementary Table 2). While several studies have reported that heterozygosity for Atm null mutations (alone or in conjunction with ApcMin or p53 mutations) had no effect on mouse spontaneous tumor frequencies 38-41, a role for ATM in mammary tumor prevention was evident in Mcm4C3/C3 Atm+/− and Mcm4C3/+ Atm+/− animals. Females of these genotypes had median mammary tumor latencies of 10.95 and 9.3 months, respectively, both significantly shorter than Mcm4C3/C3 alone (14.95 months; respectively: LRMCT P=0.001, P=0.0027; GBWT P=0.0031, P=0.0005). Mcm4C3/C3 Atm+/− males neared statistical significance for decreased tumor latency (LRMCT P=0.0751; GBWT P=0.0729), and Mcm4C3/+ Atm+/− male tumor latency was similar to Mcm4C3/C3 alone (LRMCT P=0.472; GBWT P=0.4339) (Figure 1b, Supplementary Table 2).
Heterozygosity for Atm had a striking effect on the spectrum of tumors in mice bearing the Chaos3 allele. Whereas histiocytic sarcoma was prevalent in Mcm4C3/C3 mice of mixed strain background (41% in females; 60% in males), its incidence declined in Mcm4C3/C3 Atm+/− mice (≤5% in females and males). Meanwhile, lymphoma and other cancer types increased (FET P=0.0093; P=0.0001; Figure 1c). The tumor spectrum also differed between genotypes and gender. Nearly all females (98%) of the Mcm4C3/C3, Mcm4C3/C3 Atm+/−, and Mcm4C3/+ Atm+/−genotypes developed cancer by the end of the study, vs. 72% of males of the same genotypes (FET P=0.0001). In particular, Mcm4C3/+ Atm+/− females were far more susceptible to cancer than males (FET P=0.0223; Figure 1b, Figure S1). The incidence of mammary tumors was also high in females of these genotypes, but absent in males, influencing overall differences in tumor spectrum.
Chk2 deficiency impacts tumor latency in Chaos3 females and susceptibility in males
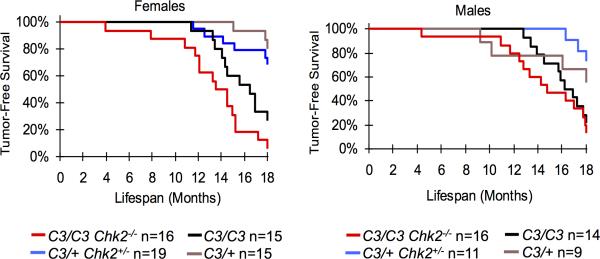
CHK2 is a phosphorylation target of ATM and serves as a downstream effector of the DSB checkpoint response (Figure 1a)42. In some circumstances, CHK2 can also be activated by ATR 43, 44. When activated, the CHK2 kinase can phosphorylate p53, protecting it from MDM2-catalyzed ubiquitination and degradation 42. Other targets include BRCA1, which is involved in HR repair 45,46. In sum, CHK2 activation can lead to DNA repair, cell cycle arrest, or apoptotic cell death. Unlike Atm or p53, several studies have shown that Chk2 null mice do not spontaneously develop tumors 47,48-51. However, Mcm4C3/C3 Chk2−/− females had decreased tumor latency compared to Mcm4C3/C3 alone in a mixed C3H x B6 background (LRMCT P=0.0189, GBWT P=0.027; Figure 2, Supplementary Table 2). Interestingly, although the overall tumor incidence was identical, the fraction of mammary tumors in Mcm4C3/C3 Chk2−/− females rose significantly from 15% to 50% (Supplementary Figure 2; FET P=.002). Mcm4C3/C3 Chk2−/− males did not have a statistically different latency compared to Mcm4C3/C3 alone, and their cancer incidence was similar to females of the same genotype (Figure 2). However, Mcm4C3/+ Chk2+/− males were more susceptible to cancer (73%) than Mcm4C3/+ controls (44%; Supplementary Figure 2).
Figure 2. Effects of Chk2 deficiency upon tumorigenesis in Chaos3 mice.
Kaplan-Meier graphs of the indicated genotypes and sexes are shown. Mcm4C3/C3 Chk2−/− female mice have significantly decreased time to tumor onset than Mcm4C3/C3 alone. C3 = Mcm4C3.
Hus1 deficiency has no impact on tumor latency or cancer susceptibility in Chaos3 mice
The study of ATR pathway genes in tumorigenesis is complicated by embryonic lethality that occurs in nulls for Atr, Chk1, the RAD9-RAD1-HUS1 (9-1-1) complex members Rad9a and Hus1, and the 9-1-1 clamp loader Rad17. The 9-1-1 complex is a PCNA-like clamp that loads onto damage sites and recruits the ATR activator TOPBP1 52. Mice with genetically reduced HUS1 levels are viable, normal in appearance, but are not tumor susceptible, and do not experience accelerated tumorigenesis in a p53-deficient background 53. Graded levels of Hus1 expression can be achieved using the following combinations of null (Hus1Δ1) and hypomorphic (Hus1neo) alleles: Hus1+/neo (71.4% of WT), Hus1Δ1/+ (43.5% of WT), and Hus1Δ1/neo (20.8% of WT) 53. We used these allele combinations to examine the effects of ATR pathway perturbation upon cancer latency and frequency in Mcm4C3/C3 mice. However, none of the Hus1 mutant genotypes had significantly different cancer susceptibility or latency compared to Mcm4C3/C3 Hus1+/+ mice (Figure 3a, Supplementary Figure 3, Supplementary Table 2).
Figure 3. Hus1 deficiency has no effect upon tumorigenesis or checkpoint signaling in Chaos3 mice and tumors.
(A) Chaos3 x Hus1 tumor latency. Mcm4C3/C3 x Hus1 mice do not have significantly different (see statistics in Supplementary Table 2) time to tumor onset than Mcm4C3/C3 alone. C3 = Mcm4C3. (B) Western blot analysis of Mcm4C3/C3 mammary tumors with a gradation of Hus1 hypomorphism. The genotypes are abbreviated as follows: “Δ” is a null allele (Hus1Δ1); “Neo” is a hypomorphic (Hus1neo) allele; “+” is the WT allele. For levels of HUS1 in these genotypes, see the text. Antibodies used are as indicated to the left of the panels. The results shown are from the same Western blot that was stripped and reprobed sequentially, following verification of effective stripping.
The lack of an effect upon cancer phenotypes led us to test whether the hypomorphic Hus1 genotypes actually impact checkpoint signaling in Mcm4C3/C3 mammary tumors. Consistent with previous genomic analyses of Mcm4C3/C3 mammary tumors showing that p53 deletions are infrequent in this model 32, p53 levels were robust in most of the 8 tumors tested by Western blotting (Figure 3b), likely reflecting checkpoint-mediated stabilization 54. There was no correlation between levels of p53 and four genotypes of Hus1 representing a gradation of HUS1 levels (see above). CHK1 activation, as indicated by phosphorylation of SER345 that is catalyzed by ATR in response to replication or genotoxic stress 55, roughly paralleled the p53 levels in this tumor set. These data indicate that Hus1 hypomorphism has little impact on ATR axis damage signalling in these tumors. Interestingly however, Mcm4C3/C3 Hus1Δ1/neo mice exhibited abnormal craniofacial features (not shown) similar to mice deficient for both Hus1 and Atm 56, suggesting that there is an impact of HUS1 deficiency in some non-tumorigenic cell types during development of Mcm4C3/C3 mice.
p21 deficiency exacerbates tumor frequency and onset in Chaos3 mice
p21 is a cyclin-dependent kinase inhibitor and downstream target of p53 that halts cell cycle progression when activated (Figure 1a). It functions by blocking the activity of cyclin-CDK complexes (CDK2 and CDC2), and can inhibit proliferating cell nuclear antigen (PCNA) and therefore DNA replication 57. Despite being a p53 target, mice lacking p21 are not cancer-prone as are p53 mutants 58. Mice homozygous for Chaos3 or the hypomorphic Mcm2 allele (Mcm2IresCreERT2) exhibit modestly elevated p53 phosphorylation and p21 expression. Furthermore, p53 mutation in either of these backgrounds increases embryonic lethality and accelerates cancer formation in survivors 23, 28. These results are indicative of important cellular roles for the downstream targets of checkpoint pathways in replication-deficient mice.
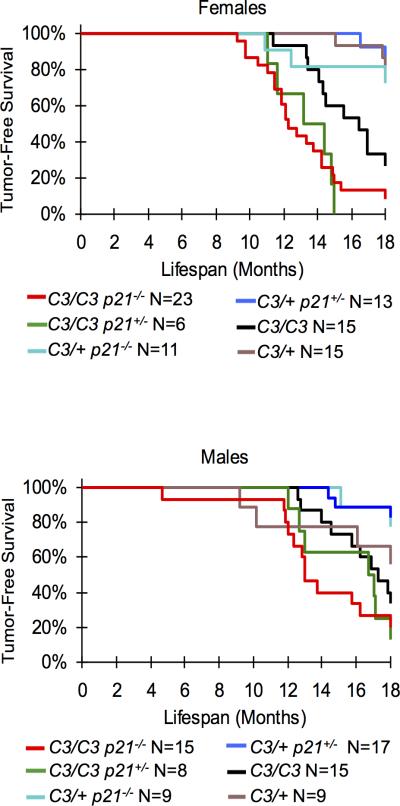
To explore if p53 signaling to p21 (Figure 1a) is important for tumor prevention in animals with intrinsic replication stress, the effects of p21 deficiency was examined in Chaos3 mice. While embryonic development of double mutant animals was not affected as are p53/Mcm4Chaos3 embryos 28, p21 nullizygosity significantly decreased time to tumor onset of Chaos3 males and females (Figure 4; Supplementary Table 2), with the predominant tumor class being histiocytic sarcomas in this mixed C3H × B6 background (Supplementary Figure 4). Mcm4C3/C3 p21+/− females, but not males, also had significantly decreased tumor latency compared to Mcm4C3/C3 alone (Figure 4; Supplementary Table 2). Finally, cancer susceptibility was elevated in Mcm4C3/+ p21−/− and Mcm4C3/+ p21+/− vs. Mcm4C3/+ females (55%, 42% and 21%, respectively; Supplementary Figure 4).
Figure 4. p21 deficiency impacts Chaos3 tumor latency in males and females and tumor susceptibility in females.
Kaplan-Meier graphs of the indicated genotypes and sexes are shown. Mcm4C3/C3 p21−/− male and female mice have significantly decreased time to tumor onset than Mcm4C3/C3 alone. Mcm4C3/C3 p21+/− females, but not males, also have significantly decreased tumor latency compared to Mcm4C3/C3 alone. See statistics in Supplementary Table 2. C3 = Mcm4C3.
DISCUSSION
Much is known about the molecular biology of the ATM and ATR pathways, their roles in responding to various types of DNA damage, and the impacts upon the cell cycle. However, most of this knowledge is based upon in vitro biochemical studies or experiments performed in cultured cells or in yeast. Regarding in vivo roles, mouse knockout models have been created for most genes in the ATM and ATR pathways, and phenotypes defined and compared to corresponding human diseases. Especially for the ATM pathway, these mouse models (and cells derived from them) have been exploited to characterize the types of DNA damage to which they primarily respond, such as DSBs. However, certain complications have limited studies on the effects of, and responses to, replication stress in vivo, despite the recognition that it is a major driver of genomic instability and tumorigenesis 59, 60. These complications include the embryonic lethality of null mutations in the Atr pathway, and the dearth of suitably relevant models of non-oncogene-associated replication stress.
Here, we utilized the Chaos3 mouse model to better understand the importance of DDR pathways in whole organisms with intrinsic replication stress, particularly with respect to carcinogenesis. This model is powerful and unique in that the replicative helicase mutation it bears (Mcm4Chaos3) is not so disruptive that development is affected. The mutation destabilizes the MCM2-7 hexamer but not its unwinding activity, causes a decrease in dormant replication origins, and triggers multiple fork recovery pathways. These defects ultimately lead to elevated chromosome breaks, chromosome segregation defects and tumorigenesis 19, 25, 26. Thus, there is opportunity to study the roles of both major DDR pathways (ATR and ATM) in cancer susceptibility without applying exogenous agents. Finally, the Chaos3 model does not involve artificial oncogene overexpression, the most commonly used strategy for inducing and studying replication stress in cancer 59.
Disruption of the ATM pathway via Atm or Chk2 mutation had the effect of exacerbating Chaos3 phenotypes. Most dramatic was that the Mcm4C3/C3 Atm−/− genotype caused semilethality that was traceable to retarded in utero growth. One interpretation of this result is that Chaos3 cells, which sustain elevated DSBs that may arise from collapsed and/or persistently stalled replication forks that fail to be compensated by nearby dormant origin firing (dormant origins are reduced in Chaos3 mice 25), accumulate a lethal level of persistent unrepaired DNA damage from the concurrent lack of DDR signaling. Stochastic factors or segregating background genetic variation may underlie the incomplete penetrance of lethality. Although early lymphoma onset in all Atm−/− animals obscured possible effects of Chaos3 upon other cancer susceptibilities, both Mcm4C3/C3 Atm+/− and Mcm4C3/+ Atm+/− mice exhibited decreased tumor latency and/or increased tumor susceptibility compared to controls (Mcm4C3/C3 and Mcm4C3/+, respectively). Heterozygosity for Atm alone does not markedly elevate cancer rates or decrease latency in mice 34, but it does render them sensitive to sublethal doses ionizing irradiation 61. Considering that Mcm4C3 heterozygotes have modestly elevated GIN (2-5 increase in erythrocyte micronuclei vs. 20 fold in homozygotes) but are not cancer prone 19, these data indicate that a synthetic phenotype results from the combination of either genetic (Chaos3 heterozygosity) or environmental (radiation) genomic stresses with a normally benign genetic reduction in ATM signaling. Similarly, heterozygosity for Chk2 also increased tumor incidence in Mcm4C3 heterozygotes. We consider these results as being supportive of the concept that heterozygosity of multiple key genes can drive carcinogenesis 62. Notably, there is some evidence that human ATM mutation carriers are at moderately elevated risk for breast and possibly other cancers (for example, see 63); it is unclear whether cancer outcome in these individuals is strictly an issue of penetrance or is modified by genetic background or environmental factors.
Chk2 deficiency also increased tumor incidence and decreased tumor latency in Chaos3 mice, although viability wasn't affected as with the Mcm4C3/C3 Atm−/− genotype. We interpret this to indicate that most cells from such animals do not retain a catastrophic level of unrepaired DSBs. The presence of ATM is predicted to allow initial localized responses to DSBs that may occur at collapsed forks, such as H2AX phosphorylation (γH2AX) and subsequent HR repair by RAD51 64, which may reduce the damage burden below the threshold of cellular lethality or compromised proliferation. It is also possible that in the absence of CHK2, ATM activates CHK1 to stimulate repair responses 65. Overall, both sets of experiments indicate that perturbation of the ATM pathway, which is involved primarily in the response to DSBs, increases cancer susceptibility in mice with intrinsic replication stress and elevated chromosomal instability/DSBs.
HUS1 was shown to be is critical for CHK1 phosphorylation in response to exogenous genotoxins 66, and genetic reduction of Hus1 expression was shown to increase genome instability and hypersensitivity to replication inhibitors but not cancer susceptibility 53. Using this hypomorphic Hus1 model for putative ATR pathway attenuation, we reasoned that Chaos3 mice might provide a sensitized system for uncovering possible roles of the ATR pathway in tumor suppression in mice with genetically predisposed replication stress. Notably, Chaos3 MEFs exhibit signs of ATR pathway activation in the form of modestly increased levels of RPA foci, RAD17 phosphorylation, and Chk1 phosphorylation (the latter in the B6 but not (B6 x C3H)F1 background 25, 28. However, overall tumor latency and susceptibility were not altered in Chaos3 mice deficient for Hus1. In contrast, depletion of Atr in mice has been shown to suppress oncogene-induced tumors that normally exhibit replication stress 67, 68. These observations contribute to the proposal that while ATR may suppress neoplastic transformation to some degree via its role in DNA damage responses, it may be required for subsequent survival and proliferation of tumors 68, 69. Interestingly, severe depletion of ATR in a human patient was associated with growth defects and genomic instability but not cancer 70. In light of those reports, we can offer two interpretations for our observations. One is that ~80% of HUS1 in Hus1Δ1/neo mice does not impact the levels of replication stress in Mcm4C3/C3 cells. Another is that HUS1 may have a more significant role in DNA repair activities distinct from checkpoint signaling 71, 72, a concept not inconsistent with findings that compound deficiency for Atm & Hus1 or Hus1 & p53 severely affects animal growth and mammary epithelial maintenance, respectively, without increasing tumorigenesis.
As mentioned earlier, the strong interaction between p53 and MCM deficiency (Chaos3 or McmIresCreIRCT2 homozygosity) demonstrated that intrinsic replication stresses ultimately trigger p53-dependent damage responses that preserve normal development and inhibit neoplastic transformation 23,28. A previous study suggested that the p21 upregulation observed in (C3HxB6)-Chaos3 mice was unlikely to contribute to tumor suppression because the mean tumor latency in Mcm4C3/C3 p21−/− was very similar to that of Mcm4C3/C3 p21+/− 28. However, that study did not include Mcm4C3/C3 animals as controls. We expanded that study to include both male and female mice and all the relevant control genotypes from related litters. The results indicate a role a tumor suppressive role of p21 in the Chaos3 model, but that it is probably relevant only in a subset of cells bearing a level of DNA damage that results in p53-mediated p21 transcription.
During the course of this project, a total of 687 detailed necropsies were performed (Supplementary Table 4). Overall, the results are consistent with previous studies showing that genetic MCM depletion causes extreme cancer predisposition, but that genetic background is the primary determinant of cancer type 19, 23-25. Because of this strong influence of strain background, possible Mcm- or checkpoint gene-specific alterations in tumor spectrum must be analyzed with caution. With this caveat, the shift towards mammary tumor susceptibility in the Chk2 -deficient Chaos3 mice of mixed background is notable. Although overall cancer rates were similar, the mammary tumor incidence in Mcm4C3/C3 Chk2−/− females (50%) was > 3 fold higher than that of Mcm4C3/C3 relatives (15%, consistent with that in true C3HxB6 F1s 25). Therefore, rather than a factor of genetic background, the increased mammary tumorigenesis may be attributable to Chk2 deficiency. Certain Chk2 alleles (not null alleles) are known to convey a 2-3 fold increased breast cancer risk 73. Since Chk2 deficiency alone has not been associated with cancer in mice, the Chaos3 mutation may bring out a susceptibility that is evident in longer-living humans.
In addition to genetic background effects, we found that tumor latency and susceptibility differed between genders in some of genotypes. Aside from cancers related to sexually dimorphic tissues such as mammary, ovary and prostate, differences in latency or frequency between sexes has been a longstanding puzzle. Differences are often hypothesized to be related to factors such as hormones, immune system differences, and differences in sex chromosome constitution 74. Here, we observed that females had an overall greater cancer susceptibility to Atm and p21 deficiency than males. Cancer incidence in Mcm4C3/+ p21+/− and Mcm4C3/+ Atm+/− females was double that of males of the same genotypes. Additionally, p21 nullizygosity increased the cancer incidence of Mcm4C3/+ females by 34%, but had no effect on males (Supplemental Fig 4). These results hint at a role for DNA repair pathways in sexual dimorphism in cancer susceptibility, which is not unprecedented in consideration of the consequences of BRCA1/2 deficiencies in female cancers. In humans, certain inherited Atm and p21 polymorphisms (ATM Ex1-81G>A, ATM D126E, and CDKN1A S31R) lead to decreased DDR response and efficiency, which is associated with increased risk of developing lung cancer in African American women 75. It is possible that further studies in mice can get at the root of cancer susceptibility gender differences and interactions with genetic background.
Overall, this study marks the importance of intact DDR pathways in responding to replication stress, providing protection from carcinogenesis when the DNA replication machinery is defective from birth. It remains unclear if lifelong exposure to exogenous sources of replication stress would benefit from the same DDR genes, but in vitro studies indicate this is likely to be so. Our findings also indicate that gender and genetic background significantly impacts cancer susceptibility and tumor latency when DNA replication integrity and DDR pathways are concurrently compromised. DDR pathways are being recognized as potential therapeutic targets in cancer treatment, since tumor cells can be hypersensitized to DNA damaging drugs when both overlapping pathways are inactivated or attenuated 76. With increasing use of personalized genomics, it may be possible to effectively characterize the status of a tumor's endogenous DDR, and exploit weaknesses in an effective and targeted manner.
MATERIALS AND METHODS
Mice
p21 mice (B6;129S2-Cdkn1atm1Tyj) were purchased from the Jackson Laboratory. Hus1 mutant mice (Hus1tm2Rsw , abbreviated as Hus1neo; Hus1tm1Led, abbreviated as Hus1Δ1) were obtained from R. Weiss 77, 78 as were Atm mutants (Atmtm1Led, abbreviated as Atm-) 36, and Chk2 (Chek2 tm1Mak , abbreviated as Chk2-) from Tak Mak 79. At the time of crossings, Chk2 and p21 mutants were congenic in C57BL/6J (B6), Atm was congenic in FvB, and the Hus1 animals were congenic in 129S6. Chaos3 C3HeB/FeJ (C3H) congenic animals were crossed to DDR mutants to generate double mutant animals that were of mixed genetic background. Progeny were genotyped as described in the original publications or as indicated by The Jackson Laboratory for those mice obtained from that source (http://jaxmice.jax.org). Double mutants and littermates of the same gender were aged to a terminal endpoint of eighteen months or until animals showed clinical signs of disease. Prism (GraphPad 5) statistical software was used to analyze survival curves and generate Kaplan-Meier plots.
MEF studies
Timed matings were conducted to collect embryos at embryonic days 12.5, 13.5, and 18.5. MEFs were generated, cultured, and cell proliferation assays performed as previously described 19.
Histopathology
Tumor samples were formalin-fixed and embedded in paraffin for sectioning and histological analysis. Slides were stained with hematoxylin and eosin (H&E) prior to histopathological evaluation.
Statistical analyses
The following tests of significance were performed and abbreviated as follows: LRMCT= Log-rank/Mantel-Cox Test; GBWT= Gehan-Breslow-Wilcoxon Test. LRMCT and GBWT are alternative methods that are applied to the survival curves; the latter gives more weight to deaths at earlier time points. The analysis was performed with Prism software (Graphpad). χ2 analysis was used to determine statistical significance of observed versus expected genotype ratios. FET was used to examine the significance of the association (contingency) between genotypes and gender to cancer susceptibility/frequency or subtype.
Western Blotting
Tissues were homogenized in T-PER (Pierce), plus complete EDTA-free proteinase inhibitor (Roche). Then, 40 ug of protein was subjected to electrophoresis on a 10% denaturing PAGE gel, transferred to a polyvinylidene difluoride membrane and blocked with 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBST). Membranes were incubated with the following antibodies at 1:1000 in 5% BSA in TBST overnight at 4 deg C: p53 Abcam #26 and CHK1 (SER345) Cell Signaling #2341. Beta-actin (Sigma #A1978) was employed at 1:10,000 in 5% BSA in TBST.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank R.S. Weiss and T. Mak for providing mice. We also thank R.S. Weiss and G. Balmus for critical feedback on the manuscript. This study was supported by NIH training grants IT32HDO57854 and 5T32GM007617, and Empire State Stem Cell Fund contract numbers C026442 and C024174 to J.C.S.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.TCGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warmerdam DO, Kanaar R. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat Res. 2009;704:2–11. doi: 10.1016/j.mrrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci U S A. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shechter D, Gautier J. MCM proteins and checkpoint kinases get together at the fork. Proc Natl Acad Sci U S A. 2004;101:10845–10846. doi: 10.1073/pnas.0404143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, et al. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 13.Arana ME, Kunkel TA. Mutator phenotypes due to DNA replication infidelity. Semin Cancer Biol. 2010;20:304–311. doi: 10.1016/j.semcancer.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayraghavan S, Schwacha A. The eukaryotic mcm2-7 replicative helicase. Subcell Biochem. 2012;62:113–134. doi: 10.1007/978-94-007-4572-8_7. [DOI] [PubMed] [Google Scholar]
- 19.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 20.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- 21.Bagley BN, Keane TM, Maklakova VI, Marshall JG, Lester RA, Cancel MM, et al. A dominantly acting murine allele of mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet. 2012;8:e1003034. doi: 10.1371/journal.pgen.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honeycutt KA, Chen Z, Koster MI, Miers M, Nuchtern J, Hicks J, et al. Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene. 2006;25:4027–4032. doi: 10.1038/sj.onc.1209435. [DOI] [PubMed] [Google Scholar]
- 23.Kunnev D, Rusiniak ME, Kudla A, Freeland A, Cady GK, Pruitt SC. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010;29:3630–3638. doi: 10.1038/onc.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang CH, Wallace MD, Abratte C, Southard T, Schimenti JC. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 2010;6:e1001110. doi: 10.1371/journal.pgen.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang CH, Yang D, Bai G, Freeland A, Pruitt SC, Schimenti JC. Post-transcriptional homeostasis and regulation of MCM2-7 in mammalian cells. Nucleic Acids Res. 2012;40:4914–4924. doi: 10.1093/nar/gks176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XC, Tye BK. Ploidy dictates repair pathway choice under DNA replication stress. Genetics. 2011;187:1031–1040. doi: 10.1534/genetics.110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawabata T, Yamaguchi S, Buske T, Luebben SW, Wallace M, Matise I, et al. A reduction of licensed origins reveals strain-specific replication dynamics in mice. Mamm Genome. 2011;22:506–517. doi: 10.1007/s00335-011-9333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443:13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- 31.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–235. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 32.Wallace MD, Pfefferle AD, Shen L, McNairn AJ, Cerami EG, Fallon BL, et al. Comparative oncogenomics implicates the Neurofibromin 1 gene (NF1) as a breast cancer driver. Genetics. 2012;192:385–396. doi: 10.1534/genetics.112.142802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatti S, Kozlov S, Farooqi AA, Naqi A, Lavin M, Khanna KK. ATM protein kinase: the linchpin of cellular defenses to stress. Cell Mol Life Sci. 2011;68:2977–3006. doi: 10.1007/s00018-011-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 35.Reiman A, Srinivasan V, Barone G, Last JI, Wootton LL, Davies EG, et al. Lymphoid tumours and breast cancer in ataxia telangiectasia; substantial protective effect of residual ATM kinase activity against childhood tumours. Br J Cancer. 2011;105:586–591. doi: 10.1038/bjc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci U S A. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 38.Spring K, Ahangari F, Scott SP, Waring P, Purdie DM, Chen PC, et al. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat Genet. 2002;32:185–190. doi: 10.1038/ng958. [DOI] [PubMed] [Google Scholar]
- 39.Mao JH, Wu D, DelRosario R, Castellanos A, Balmain A, Perez-Losada J. Atm heterozygosity does not increase tumor susceptibility to ionizing radiation alone or in a p53 heterozygous background. Oncogene. 2008;27:6596–6600. doi: 10.1038/onc.2008.280. [DOI] [PubMed] [Google Scholar]
- 40.Lu S, Shen K, Wang Y, Santner SJ, Chen J, Brooks SC, et al. Atm-haploinsufficiency enhances susceptibility to carcinogen-induced mammary tumors. Carcinogenesis. 2006;27:848–855. doi: 10.1093/carcin/bgi302. [DOI] [PubMed] [Google Scholar]
- 41.Karabinis ME, Larson D, Barlow C, Wynshaw-Boris A, Moser AR. Heterozygosity for a mutation in Brca1 or Atm does not increase susceptibility to ENU-induced mammary tumors in Apc(Min)/+ mice. Carcinogenesis. 2001;22:343–346. doi: 10.1093/carcin/22.2.343. [DOI] [PubMed] [Google Scholar]
- 42.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 43.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–6583. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 46.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 47.Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El GS, Pamidi A, Halaby MJ, Bohgaki M, Cardoso R, Li L, et al. Inactivation of chk2 and mus81 leads to impaired lymphocytes development, reduced genomic instability, and suppression of cancer. PLoS Genet. 2011;7:e1001385. doi: 10.1371/journal.pgen.1001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niida H, Murata K, Shimada M, Ogawa K, Ohta K, Suzuki K, et al. Cooperative functions of Chk1 and Chk2 reduce tumour susceptibility in vivo. Embo J. 2010;29:3558–3570. doi: 10.1038/emboj.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stracker TH, Couto SS, Cordon-Cardo C, Matos T, Petrini JH. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol Cell. 2008;31:21–32. doi: 10.1016/j.molcel.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemp M, Sancar A. DNA distress: just ring 9-1-1. Curr Biol. 2009;19:R733–734. doi: 10.1016/j.cub.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Levitt PS, Zhu M, Cassano A, Yazinski SA, Liu H, Darfler J, et al. Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol Cell Biol. 2007;27:2189–2201. doi: 10.1128/MCB.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 55.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balmus G, Zhu M, Mukherjee S, Lyndaker AM, Hume KR, Lee J, et al. Disease severity in a mouse model of ataxia telangiectasia is modulated by the DNA damage checkpoint gene Hus1. Hum Mol Genet. 2012;21:3408–3420. doi: 10.1093/hmg/dds173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 59.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 60.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 61.Barlow C, Eckhaus MA, Schaffer AA, Wynshaw-Boris A. Atm haploinsufficiency results in increased sensitivity to sublethal doses of ionizing radiation in mice. Nat Genet. 1999;21:359–360. doi: 10.1038/7684. [DOI] [PubMed] [Google Scholar]
- 62.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97:813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 64.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nature Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 65.Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, et al. Ataxiatelangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 66.Weiss RS, Matsuoka S, Elledge SJ, Leder P. Hus1 acts upstream of chk1 in a mammalian DNA damage response pathway. Curr Biol. 2002;12:73–77. doi: 10.1016/s0960-9822(01)00626-1. [DOI] [PubMed] [Google Scholar]
- 67.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J Clin Invest. 2012;122:241–252. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010;9:1249–1255. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mokrani-Benhelli H, Gaillard L, Biasutto P, Le GT, Touzot F, Vasquez N, et al. Primary microcephaly, impaired DNA replication, and genomic instability caused by compound heterozygous ATR mutations. Hum Mutat. 2013;34:374–384. doi: 10.1002/humu.22245. [DOI] [PubMed] [Google Scholar]
- 71.Lieberman HB, Bernstock JD, Broustas CG, Hopkins KM, Leloup C, Zhu A. The role of RAD9 in tumorigenesis. J Mol Cell Biol. 2011;3:39–43. doi: 10.1093/jmcb/mjq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 74.Dorak MT, Karpuzoglu E. Gender differences in cancer susceptibility: an inadequately addressed issue. Front Genet. 2012;3:268. doi: 10.3389/fgene.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng YL, Kosti O, Loffredo CA, Bowman E, Mechanic L, Perlmutter D, et al. Elevated lung cancer risk is associated with deficiencies in cell cycle checkpoints: genotype and phenotype analyses from a case-control study. Int J Cancer. 2010;126:2199–2210. doi: 10.1002/ijc.24771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 77.Levitt PS, Liu H, Manning C, Weiss RS. Conditional inactivation of the mouse Hus1 cell cycle checkpoint gene. Genomics. 2005;86:212–224. doi: 10.1016/j.ygeno.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 78.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 79.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.