Abstract
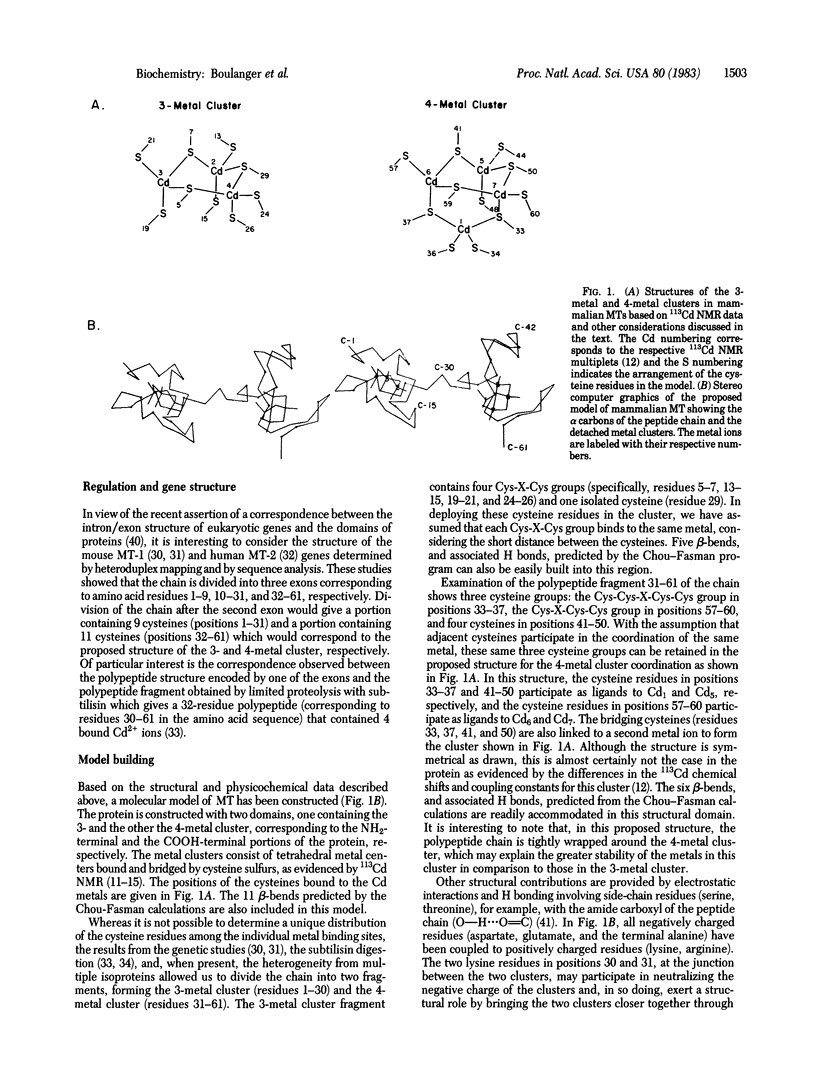
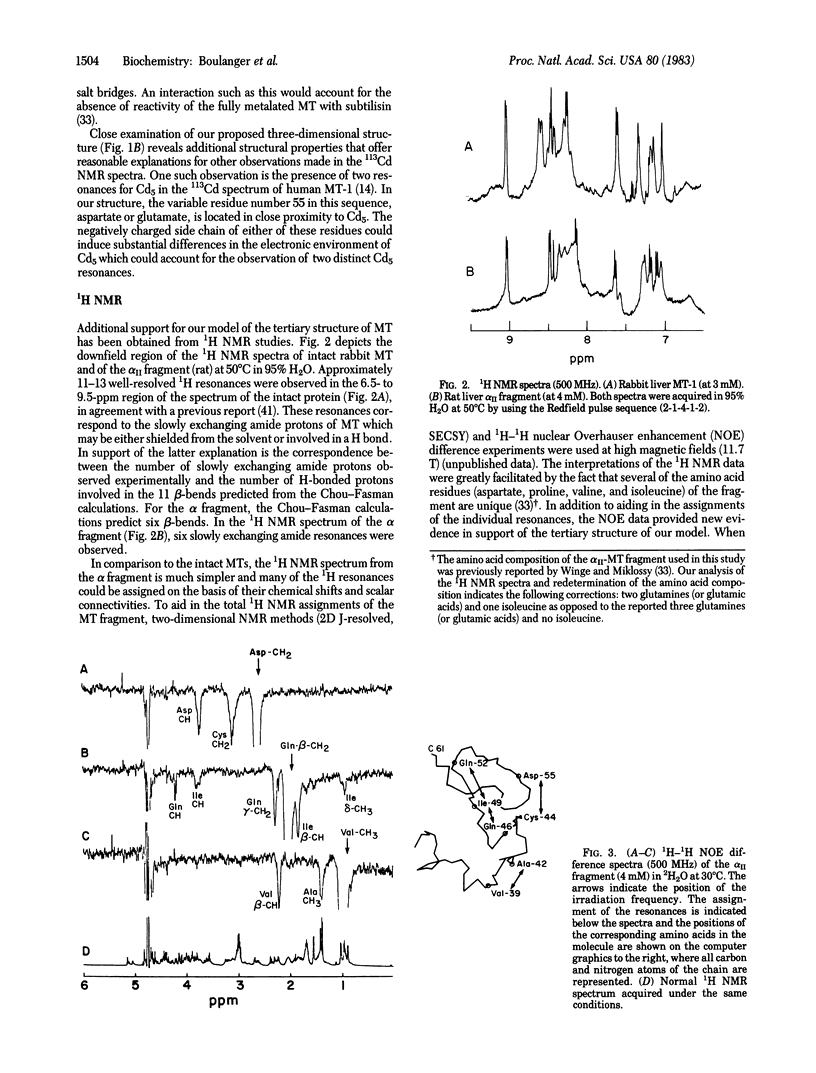
The results of physicochemical studies of mammalian metallothioneins are summarized and used to propose a model of the protein. The primary structures of all mammalian metallothioneins are very homologous; there are 38 invariant residues and 20 of them are cysteines. The results of UV and CD optical studies indicated that all 20 cysteines are involved in the ligation of 7 mol of metal per mol of metallothionein and that the protein does not contain any alpha-helix structure. A theoretical analysis by the Chou-Fasman method has predicted 11 beta-bends, each one involving at least one cysteine residue. The most significant structural data, provided by 113Cd NMR, demonstrated that the 7 mol of bound Cd2+ are arranged in two separate metal clusters, one containing four metal ions and the other containing three, with all Cd2+ tetrahedrally coordinated to cysteine thiolate ligands. The 11 cysteine residues of the carboxyl-terminal portion of the metallothionein chain (residues 30-61) are ligated to the 4-metal cluster as shown by 113Cd NMR of this enzymatically cleaved fragment. The remaining cysteine residues from the amino-terminal polypeptide portion (residues 1-29) form the 3-metal cluster. Such a division of the chain is consistent with the presence of an intron in the mouse metallothionein-1 gene corresponding to residue 32 in the polypeptide chain. A two-domain molecular model has been constructed based on an analysis of all the available data and is described in detail. The accuracy of this model was tested by 1H NMR at 500 MHz and the data are in agreement with our proposed structure.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulanger Y., Armitage I. M. 113Cd nmr study of the metal cluster structure of human liver metallothionein. J Inorg Biochem. 1982 Oct;17(2):147–153. doi: 10.1016/s0162-0134(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Boulanger Y., Armitage I. M., Miklossy K. A., Winge D. R. 113Cd NMR study of a metallothionein fragment. Evidence for a two-domain structure. J Biol Chem. 1982 Nov 25;257(22):13717–13719. [PubMed] [Google Scholar]
- Briggs R. W., Armitage I. M. Evidence for site-selective metal binding in calf liver metallothionein. J Biol Chem. 1982 Feb 10;257(3):1259–1262. [PubMed] [Google Scholar]
- Cherian M. G., Goyer R. A. Methallothioneins and their role in the metabolism and toxicity of metals. Life Sci. 1978 Jul 3;23(1):1–9. doi: 10.1016/0024-3205(78)90317-x. [DOI] [PubMed] [Google Scholar]
- Durnam D. M., Perrin F., Gannon F., Palmiter R. D. Isolation and characterization of the mouse metallothionein-I gene. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6511–6515. doi: 10.1073/pnas.77.11.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin A. M., Peterson G., Brady F. O. Cd-metallothionein-a test object for dark-field studies of protein structure. Ultramicroscopy. 1977 Aug;2(4):389–395. doi: 10.1016/s0304-3991(76)92252-x. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Glanville N., Durnam D. M., Palmiter R. D. Structure of mouse metallothionein-I gene and its mRNA. Nature. 1981 Jul 16;292(5820):267–269. doi: 10.1038/292267a0. [DOI] [PubMed] [Google Scholar]
- Hennessey J. P., Jr, Johnson W. C., Jr Information content in the circular dichroism of proteins. Biochemistry. 1981 Mar 3;20(5):1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- Huang I. Y., Kimura M., Hata A., Tsunoo H., Yoshida A. Complete amino acid sequence of mouse liver metallothionein-II. J Biochem. 1981 Jun;89(6):1839–1845. doi: 10.1093/oxfordjournals.jbchem.a133385. [DOI] [PubMed] [Google Scholar]
- Huang I. Y., Yoshida A. Mouse liver metallothioneins. Complete amino acid sequence of metallothionein-I. J Biol Chem. 1977 Nov 25;252(22):8217–8221. [PubMed] [Google Scholar]
- Johnson D. R., Foulkes E. C. On the proposed role of metallothionein in the transport of cadmium. Environ Res. 1980 Apr;21(2):360–365. doi: 10.1016/0013-9351(80)90038-9. [DOI] [PubMed] [Google Scholar]
- KAGI J. H., VALLEE B. L. Metallothionein: a cadmium and zinc-containign protein from equine renal cortex. II. Physico-chemical properties. J Biol Chem. 1961 Sep;236:2435–2442. [PubMed] [Google Scholar]
- Karin M., Richards R. I. Human metallothionein genes--primary structure of the metallothionein-II gene and a related processed gene. Nature. 1982 Oct 28;299(5886):797–802. doi: 10.1038/299797a0. [DOI] [PubMed] [Google Scholar]
- Kissling M. M., Kägi H. R. Primary structure of human hepatic metallothionein. FEBS Lett. 1977 Oct 15;82(2):247–250. doi: 10.1016/0014-5793(77)80594-2. [DOI] [PubMed] [Google Scholar]
- Kojima Y., Berger C., Vallee B. L., Kägi J. H. Amino-acid sequence of equine renal metallothionein-1B. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3413–3417. doi: 10.1073/pnas.73.10.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi J. H., Himmelhoch S. R., Whanger P. D., Bethune J. L., Vallee B. L. Equine hepatic and renal metallothioneins. Purification, molecular weight, amino acid composition, and metal content. J Biol Chem. 1974 Jun 10;249(11):3537–3542. [PubMed] [Google Scholar]
- Lerch K. Amino-acid sequence of copper-metallothionein from Neurospora crassa. Experientia Suppl. 1979;34:173–179. doi: 10.1007/978-3-0348-6493-0_9. [DOI] [PubMed] [Google Scholar]
- Lerch K., Ammer D., Olafson R. W. Amino acid sequence of crab metallothionein. FEBS Lett. 1981 Apr 20;126(2):165–168. doi: 10.1016/0014-5793(81)80232-3. [DOI] [PubMed] [Google Scholar]
- Ohi S., Cardenosa G., Pine R., Huang P. C. Cadmium-induced accumulation of metallothionein messenger RNA in rat liver. J Biol Chem. 1981 Mar 10;256(5):2180–2184. [PubMed] [Google Scholar]
- Otvos J. D., Armitage I. M. Structure of the metal clusters in rabbit liver metallothionein. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7094–7098. doi: 10.1073/pnas.77.12.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp H., Weser U. Circular dichroism of metallothioneins. A structural approach. Biochim Biophys Acta. 1978 Mar 28;533(1):209–226. doi: 10.1016/0005-2795(78)90565-2. [DOI] [PubMed] [Google Scholar]
- Squibb K. S., Cousins R. J., Feldman S. L. Control of zinc-thionein synthesis in rat liver. Biochem J. 1977 Apr 15;164(1):223–228. doi: 10.1042/bj1640223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. T., Yamamura M. Changes of metal contents and isometallothionein levels in rat tissues after cadmium loading. Biochem Pharmacol. 1980 Sep 15;29(18):2407–2412. doi: 10.1016/0006-2952(80)90342-1. [DOI] [PubMed] [Google Scholar]
- Vasák M., Galdes A., Hill H. A., Kägi J. H., Bremner I., Young B. W. Investigation of the structure of metallothioneins by proton nuclear magnetic resonance spectroscopy. Biochemistry. 1980 Feb 5;19(3):416–425. doi: 10.1021/bi00544a003. [DOI] [PubMed] [Google Scholar]
- Vasák M., Kägi J. H., Hill H. A. Zinc(II), cadmium(II), and mercury(II) thiolate transitions in metallothionein. Biochemistry. 1981 May 12;20(10):2852–2856. doi: 10.1021/bi00513a022. [DOI] [PubMed] [Google Scholar]
- Winge D. R., Miklossy K. A. Domain nature of metallothionein. J Biol Chem. 1982 Apr 10;257(7):3471–3476. [PubMed] [Google Scholar]
- Winge D. R., Premakumar R., Rajagopalan K. V. Metal-induced formation of metallothionein in rat liver. Arch Biochem Biophys. 1975 Sep;170(1):242–252. doi: 10.1016/0003-9861(75)90115-0. [DOI] [PubMed] [Google Scholar]


