Abstract
Mesenchymal stem cells (MSCs) display multipotent characteristics that make them ideal for potential therapeutic applications. MSCs are typically cultured as monolayers on tissue culture plastic, but there is increasing evidence suggesting that they may lose their multipotency over time in vitro and eventually cease to retain any resemblance to in vivo resident MSCs. Three-dimensional (3D) culture systems that more closely recapitulate the physiological environment of MSCs and other cell types are increasingly explored for their capacity to support and maintain the cell phenotypes. In much of our own work, we have utilized fibrin, a natural protein-based material that serves as the provisional extracellular matrix during wound healing. Fibrin has proven to be useful in numerous tissue engineering applications and has been used clinically as a hemostatic material. Its rapid self-assembly driven by thrombin-mediated alteration of fibrinogen makes fibrin an attractive 3D substrate, in which cells can adhere, spread, proliferate, and undergo complex morphogenetic programs. However, there is a significant need for simple cost-effective methods to safely retrieve cells encapsulated within fibrin hydrogels to perform additional analyses or use the cells for therapy. Here, we present a safe and efficient protocol for the isolation of MSCs from 3D fibrin gels. The key ingredient of our successful extraction method is nattokinase, a serine protease of the subtilisin family that has a strong fibrinolytic activity. Our data show that MSCs recovered from 3D fibrin gels using nattokinase are not only viable but also retain their proliferative and multilineage potentials. Demonstrated for MSCs, this method can be readily adapted to retrieve any other cell type from 3D fibrin gel constructs for various applications, including expansion, bioassays, and in vivo implantation.
Introduction
Bone marrow stromal cells, commonly referred to as mesenchymal stem cells (MSCs), are nonhematopoietic cells found in the adult bone marrow that possess multipotent characteristics. MSCs have the ability to differentiate into multiple lineages, including osteogenic, adipogenic, and chondrogenic phenotypes. Due to their high degree of plasticity and relative ease of isolation from many tissues,1–5 MSCs have been explored in numerous clinical trials for tissue engineering and regenerative medicine applications. Despite the apparent therapeutic potential of MSCs, most likely through trophic factor secretion,6 current understanding of the intrinsic and extrinsic components of the microenvironment that regulate their activity in vivo remains incomplete. Fundamental knowledge regarding these components is desirable, not only to better understand MSC biology but also to improve the translational potential of these cells.
The current dogma is that developing physiologically relevant artificial models capable of instructing stem cells will require a more accurate recapitulation of their native niche.7,8 In an attempt to reconstruct the stem cell microenvironment that more closely mimics in vivo conditions, many investigators are exploring the use of three-dimensional (3D) culture systems. Recent studies suggest that MSCs maintained in two-dimensional (2D) culture systems gradually lose their proliferative potential, colony-forming efficiency, and differentiation capacity with time.9–11 While the evidence that 3D culture methods provide a cellular environment more consistent with that in vivo is persuasive,12–17 there is still a need for optimized culture models for large-scale long-term expansion of stem cells with uniform properties that are capable of differentiating into selected mature cell types with high efficiency and purity.18 Furthermore, the development of efficient methods to safely extract these cells from 3D tissue culture is important, both to meet the high cell volumes required for therapeutic applications and to characterize how cells grown in 3D models are regulated by various components of an artificial niche.
In this particular study, we focused on MSC encapsulation within fibrin, in part, because fibrin is a widely used material,19 which has been shown to promote cell survival and proliferation both in vitro and in vivo.20–23 There is compelling evidence that fibrin supports the delivery of stem cells, such as bone marrow mononuclear cells24,25 and human MSCs,20,26,27 and stimulates the MSC differentiation toward osteogenic and chondrogenic differentiation.27–29 In our own work, we have used fibrin extensively as an extracellular matrix analog capable of supporting capillary morphogenesis in vitro30–33 and neovascularization in vivo.31,34 We have also shown that cocultures of MSCs and endothelial cells in 3D fibrin hydrogels readily form pericyte-invested capillary networks,32,35,36 which have prompted our efforts to better understand how the perivascular location of MSCs may influence their phenotype.35 However, our efforts were limited, in part, by the lack of a simple yet effective method to safely recover and characterize MSCs residing within the fibrin hydrogels.
Recovery of cells from collagen hydrogels and collagen-based tissues can readily be achieved using collagenase, but no comparably simple method to retrieve viable cells from 3D fibrin culture models exists, to the best of our knowledge. In most cases, commonly used proteolytic enzymes, including trypsin and collagenase, have been used for primary cell isolation from a variety of tissue types.37–40 However, when used to dissolve fibrin for in vitro models, these enzymes do not yield a single-cell suspension effectively. Furthermore, longer incubation times with these enzymes required for dissolving the gels may damage the cells harvested for subcultivation or other studies. Previous studies have used 3D fibrin gels as biomimetic substrates for the isolation of stem cells residing in various tissues.41,42 Using urokinase, cells outgrown from these tissues were isolated by selective degradation of the 3D fibrin gels. In this article, we used nattokinase, a bacillus-derived serine protease that is known for its potent fibrinolytic activity,43–45 to recover encapsulated MSCs from 3D fibrin gels. Compared with other fibrinolytic enzymes, such as urokinase and plasmin, nattokinase is reportedly more efficient in degrading fibrin gels.44 We demonstrated that nattokinase yields significantly higher MSC recovery compared with other proteolytic enzymes, including trypsin and TrypLE. In addition, we found that this enzyme-mediated recovery is not harmful, as assessed by the cellular proliferation and viability in 3D culture. Finally, using our extraction protocol, we showed that cells recovered from 3D hydrogels were capable of differentiating into osteogenic and adipogenic lineages. This extraction method is an effective system that could potentially be used to safely and efficiently harvest a variety of cell types from 3D fibrin cultures for subsequent use in numerous applications, including expansion, bioassays, and in vivo implantation.
Materials and Methods
Cell culture
Human bone marrow-derived MSCs were obtained from a commercial source (Lonza) at passage 2. As part of the manufacturer's quality control, these MSCs were tested for purity by flow cytometry and for their ability to differentiate into osteogenic, chondrogenic, and adipogenic lineages. Cells are positive for the cell surface markers CD105, CD166, CD29 (integrin β1), and CD44 and negative for CD14, CD34, and CD45. MSCs were maintained in the high glucose (4.5 g/L) Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). All cultures were incubated at 37°C and 5% CO2. Media were changed every 2 days. MSCs were routinely expanded in 2D cultures and harvested with 0.05% trypsin–ethylenediaminetetraacetic acid (EDTA) (Invitrogen). Cells were used before passage 8 for differentiation experiments and before passage 11 for viability and cell extraction experiments. For 2D controls to the 3D experiments described below, MSCs were cultured for 1, 7, or 14 days. We identify these times as preculture, indicating the culture time before harvesting and subsequent analysis.
Construction of the 3D culture model
MSCs were encapsulated within 3D fibrin gels through methods similar to those used previously to create a 3D coculture model of capillary morphogenesis.32 In brief, 5×104 MSCs were mixed within a 2.5, 5, or 10 mg/mL fibrinogen solution (Sigma-Aldrich; Lot No. 069K7636v, 65%–85% protein). Five hundred microliters of this solution containing MSCs was combined with 10 μL of thrombin (50 U/mL; Sigma) in a single well of a 12-well plate to make one gel construct. This process was repeated until the desired number of gels was constructed. Constructs were left undisturbed for 5 min to allow partial gelation before incubating for an additional 25 min at 37°C and 5% CO2. Gels were then cultured in the DMEM supplemented with 10% FBS. Media were changed every 2 days. Cells were retrieved from 3D fibrin gels on days 1, 7, and 14 postassembly. We identify the time of culture in 3D fibrin gels as preculture, indicating the culture time before retrieval and subsequent analysis.
Retrieving viable cells from 3D fibrin gel constructs
MSCs embedded in 3D fibrin gels were recovered using one of three methods: our novel recovery technique (Fig. 1) involving the fibrinolytic enzyme nattokinase or the methods involving 0.05% trypsin-EDTA (Gibco) or 100% TrypLE (Invitrogen). In our new method, a fibrinolytic solution was prepared by dissolving 50 FU/mL (fibrin degradation units) of nattokinase (NSK-SD; Japan Bio Science Laboratory Co., Ltd) in phosphate-buffered saline (PBS) containing 1 mM EDTA (Fisher Scientific). The gels were washed with PBS before dislodging them from the well siding using a small spatula. The gels were subsequently dissolved by adding 500 μL of the fibrinolytic solution and incubating at 37°C for 30 min (for the 2.5 mg/mL fibrin gels) or 60 min (for the 5 and 10 mg/mL fibrin gels). These incubation times were determined empirically based on the observations of gel dissolution. Upon dissolution, the contents of each well were collected and centrifuged. Cells were then washed with cold PBS before subsequent procedures. The same dissolution conditions, as described above, were implemented in the methods involving trypsin-EDTA or TrypLE.
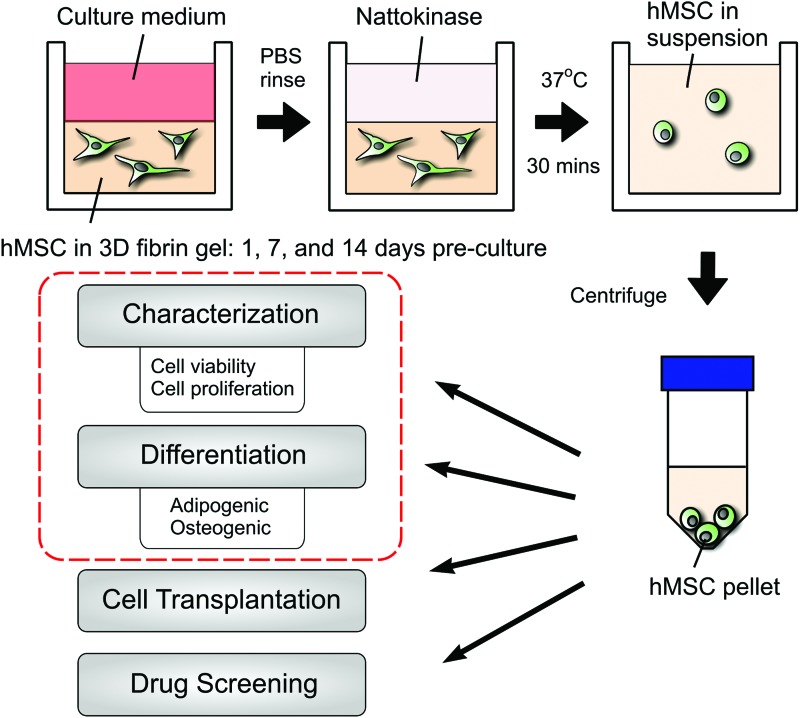
FIG. 1.
Illustration depicting the method by which mesenchymal stem cells (MSCs) were extracted from three-dimensional (3D) fibrin gels. In this study, we developed a simple new method for efficiently retrieving MSCs encapsulated within 3D fibrin hydrogels. Cells were cultured within the fibrin gels for 1, 7, or 14 days before retrieval. The key component of the extraction process is a fibrinolytic enzyme, nattokinase. Treatment of the cell-seeded hydrogels with nattokinase for 30 min (longer incubations required for more concentrated gels) results in a single-cell suspension, which can be collected and concentrated through centrifugation and then subsequently used in additional assays or for different applications. Color images available online at www.liebertpub.com/tec
Quantitative polymerase chain reaction
The multilineage potential of MSCs recovered following fibrinolysis was determined, in part, through quantitative polymerase chain reaction (qPCR) to assess the expression of genes associated with osteogenic and adipogenic differentiation. In brief, MSCs cultured for up to 14 days were either retrieved from 3D fibrin gels using our novel nattokinase-based recovery method or collected from 2D cultures through standard trypsinization. Harvested cells were then subjected to standard adipogenic or osteogenic induction protocols (described below) in 2D culture for 7 and 21 days, respectively.46–50 Total RNA was isolated from cells using the SV Total RNA Isolation System (Promega). The RNA concentration and the purity of each sample were determined by A260/A280 absorptions using a Nanodrop ND-1000 (Thermo Scientific) spectrometer. Equal amounts of total RNA from each sample were used to create the first-strand cDNA using the ImProm-II Reverse Transcription System (Promega). The PCR amplification was performed with the KAPA SYBR® Fast Universal Master Mix (Kapa Biosystems) on a 7500 Fast Real-Time PCR System (Applied Biosystems) in a final volume of 20 μL using cycling parameters (3 min, 95°C; 3 s, 95°C; 20 s, 60°C with the latter two steps repeated for 40 times). Each reaction was performed in triplicate, and the ΔΔCT method was used for the gene expression analysis.51 The gene encoding for peptidylprolyl isomerase (PPIA) was used as the housekeeping gene as it has been shown to have the most stable expression levels of a variety of housekeeping gene candidates under different conditions.52 The primer sequences of the genes for qPCR are provided in Table 1.
Table 1.
Primer Sequences Designed by Primer-Blast and Used for Quantitative Polymerase Chain Reaction
| Gene | Sense primer | Antisense primer |
|---|---|---|
| BGLAP | 5′-AGGCACCCTTCTTTCCTCTTC-3′ | 5′-TTCCTCTTCTGGAGTTTATTTGGGA-3′ |
| CEBPA | 5′-ATGCAAACTCACCGCTCCAAT-3′ | 5′-GAGGCAGGAAACCTCCAAATAAA-3′ |
| PPARG | 5′-ATTACGAAGACATTCCATTCACAAG-3′ | 5′-CTCAGAATAATAAGGTGGAGATGC-3′ |
| PPIA | 5′-GTCTTGTGTGTTGTCTGGTTA-3′ | 5′-ATGTTTGATGTTTATTTCCACCTTG-3′ |
| RUNX2 | 5′-CAGAAGGGAGGAGATGTGTGTA-3′ | 5′-TTGCTAATGCTTCGTGTTTCCA-3′ |
Quantification of cell viability through fluorescent-activated cell sorting
To assess the viability, cells were retrieved from 3D cultures 6 h after initial cell seeding by incubating the gels in the nattokinase fibrinolytic solution, as described above. For these assays, the 3D fibrin gels were incubated in the fibrinolytic solution for 90 min. Cells were then resuspended in ice-cold PBS, pelleted by centrifugation at 2000 rpm at 4°C for 5 min, and then incubated with a 3 μM solution of propidium iodide (PI; Invitrogen) in PBS (pH 7.2) for 15 min at room temperature. Samples were then washed twice and resuspended in 2% FBS in PBS for flow cytometry analysis. Unstained cell suspensions were prepared in parallel as control samples.
Multilineage differentiation protocols
After preculture on 2D substrates or in 3D fibrin gels, MSCs were tested for their differentiation capacity. For adipogenic differentiation, MSCs retrieved from 2D cultures or extracted from a 3D fibrin gel were reseeded at 20,000 cells/cm2 in a 24-well plate for functional assays or in a 6-well plate for gene expression assays through qPCR. Cells were maintained in either adipogenic growth media (AGM, a control), consisting of αMEM (Minimum Essential Medium, alpha modification; Gibco), 10% FBS, 1% penticillin/streptomycin (CellGro), and 5 mg/mL gentamicin (Gibco), or adipogenic induction media (AIM), consisting of AGM, 1 μM dexamethasone (Sigma-Aldrich), 0.5 mM 3-isobutyl-1-methylxanthine (Acros Organics), 10 μg/mL insulin (Gibco), and 0.2 mM indomethacin (Sigma-Aldrich).48,53
For osteogenic differentiation, MSCs precultured on 2D substrates or in 3D fibrin gels for up to 14 days were retrieved and subsequently reseeded at 5000 cells/cm2 in a 24-well plate for functional assays or in a 6-well plate for qPCR. Cells were maintained in osteogenic growth media (OGM), consisting of αMEM (Gibco), 20% FBS, 2 mM L-glutamine (CellGro), 1% penticillin/streptomycin, and 5 mg/mL gentamicin, or in osteogenic base media (OBM), consisting of OGM, 10 mM β-glycerol phosphate (Sigma-Aldrich), and 50 μg/mL L-ascorbic acid (Fisher Scientific). After 14 days in OBM, cells were cultured in osteogenic mineralization media [OMM, containing OBM+100 nM dexamethasone (Sigma-Aldrich)], as previously reported.48,53 For simplicity, OBM and OMM will be referred to as osteogenic induction media (OIM).
Oil Red O staining, imaging, and quantification
Adipogenic differentiation was assessed, in part, by staining cultures with Oil Red O and quantifying, as described above.48,54 Briefly, a 12.2 mM stock solution of Oil Red O dye (Sigma) was dissolved in isopropanol. Cells were fixed in 4% paraformaldehyde at 4°C for 30 min after 7 and 14 days of culture in the AIM. Cells were then rinsed in PBS at least twice. Stock Oil Red O solution was added to PBS at a ratio of 3:2 to create the working solution. The working solution was filtered with a 0.22-μm filter (Millipore) before use. Each well was immersed in the Oil Red O working solution for 20 min. After staining, each well was quickly rinsed 3×in a 60/40 isopropanol/PBS solution to remove excess Oil Red O. The wells were then rinsed 2×in PBS and imaged on an Olympus microscope IX81 equipped with a DP25 color camera. After imaging, 4% IGEPAL-CA630 (Sigma) in isopropanol was added to each well and protected from light for 15 min. Each well was then analyzed with a Thermo Scientific Multiskan Spectrum spectrophotometer at 520 nm to determine the absorbance of each well. The absorbance of Oil Red O was normalized to the total cell number in each well as determined by nuclei counting using DAPI staining.55 Three images per condition were analyzed to determine the number of cells per well. Four wells per condition were used to quantify the levels of Oil Red O.
von Kossa staining
Cells were rinsed in PBS 2×and then fixed in 4% paraformaldehyde at 4°C for 30 min after 14 and 21 days in the OIM. After fixation, cells were rinsed in double distilled (DD) water 3×and then immersed in 5% silver nitrate (Sigma) and subjected to ultraviolet (UV) light (∼365 nm) for 40 min. After UV exposure, cells were rinsed 3×in DD water. The cells were then rinsed in sodium thiosulfate (Sigma) for 3 min and rinsed in DD water 3×. Images were taken on an Olympus IX81 with a DP25 color camera.
Calcium quantification
The calcium content in osteogenic cultures was quantified using the ortho-cresolphthalein complexone (OCPC) method, as previously described.49,50 Cells were washed in PBS twice before incubation in 1 mL of 1 N acetic acid overnight. The OCPC solution was prepared by adding OCPC to DD water with 1 N potassium hydroxide (KOH) and 1 N acetic acid. The dissolved solutions (10 μL per replicate) were then mixed with a working solution (300 μL per replicate) of the OCPC solution and ethanolamine/boric acid/8-hydroxyquinoline buffer [all from Sigma, except KOH (Acros)]. The absorbance values were recorded using a Thermo Scientific Multiskan Spectrum spectrophotometer at 570 nm. The calcium values were quantified by a standard curve from 0 to 150 μg/mL. Samples and standards were assayed in triplicate.
Statistical analyses
Statistical analyses were carried out using GraphPad Prism software. Data are reported as means±standard deviations. All statistical comparisons were made by performing a one-way analysis of variance, followed by Tukey's multiple comparison tests to judge significance between two data sets at a time. P values<0.05 were considered statistically significant. Statistics for qPCR were performed on ΔΔCT values.
Results
Nattokinase efficiently degrades 3D fibrin gels without damaging cells
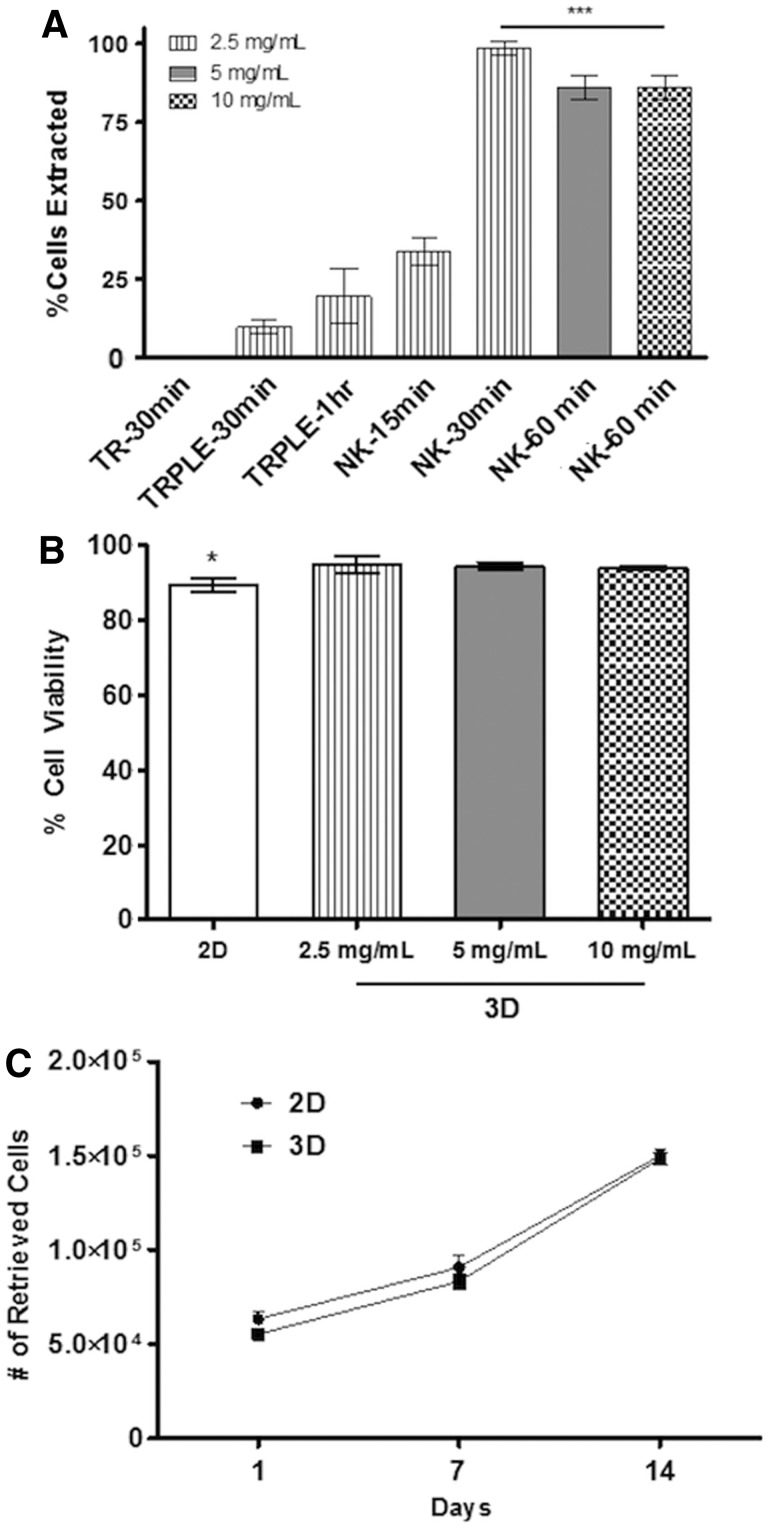
In this study, we developed and applied a new method (Fig. 1) to recover cells encapsulated within 3D fibrin hydrogels based on nattokinase, a powerful fibrinolytic enzyme that is mostly known for its blood-thinning effects. To validate the method, we first quantified the percentage of cells extracted from 3D fibrin gels (Fig. 2A). Six hours after initial cell encapsulation, fibrin gels were degraded using trypsin, TrypLE, or our nattokinase solution. A 30-min incubation in the nattokinase solution enabled nearly 100% recovery of the cells entrapped in 2.5 mg/mL fibrin gels. By comparison, a significantly lower percentage of the encapsulated MSCs were retrieved from the gels using either trypsin or TrypLE. A 60-min incubation with nattokinase was optimal for the more concentrated 5 and 10 mg/mL fibrin gels, resulting in an efficient cell retrieval comparable to that attained with nattokinase in lower concentration gels and significantly better than that attained with 60-min incubations with TrypLE (Fig. 2A). Furthermore, the quantification of PI staining through flow cytometry revealed similar levels of viability when comparing cells recovered from 2.5 mg/mL 3D fibrin gels (cultured for 14 days) digested using nattokinase to those recovered from 2D cultures (Fig. 2B). The viability of the cells was maintained even when the cell-seeded gels were incubated in the nattokinase solution for 90 min (Fig. 2B), but anecdotally, we observed no changes in the viability after incubation times up to 2 h (data not shown). In addition, we also cultured the MSCs within 2.5 mg/mL of 3D fibrin gels for up to 14 days and quantified the number of cells retrieved by nattokinase to assess their proliferation rates. Data showed that MSCs proliferated at comparable rates in both 2D and 3D (Fig. 2C). Looking across time points, these data also suggest that our nattokinase-based method is effective for recovering cells encapsulated within 3D fibrin gels across a range of cell densities.
FIG. 2.
Enzyme-assisted extraction of MSCs from two-dimensional (2D) and 3D cultures. (A) Nattokinase (NK) yielded a significantly higher percentage of MSCs extracted from 3D fibrin gels relative to the other proteolytic enzymes [trypsin (TR) and TrypLE (TRPLE)]. To quantify the percentage of cells recovered, MSC-seeded fibrin gels were dissolved 6 h after initial cell seeding (50K cells/gel). For 2.5 mg/mL fibrin gels, cells were efficiently retrieved after a 30-min incubation in the nattokinase-based fibrinolytic solution; 5 and 10 mg/mL gels were incubated for 60 min. Retrieved cells were pooled together from a total of three gels for each technical sample. *** indicates that all of the groups under the line are significantly different from those groups not under the line (p≤0.001). (B) After 14 days of preculture in 3D fibrin gels, nearly 100% of the MSCs extracted from nattokinase were viable, as quantified by propidium iodide staining and flow cytometry. (C) After 1, 7, or 14 days of preculture, MSCs were retrieved from 3D fibrin gels (using nattokinase) or harvested from 2D cultures (using trypsin). The number of cells retrieved from the cultures was comparable at all time points for both culture conditions.
MSCs retrieved from 3D fibrin gels maintain their adipogenic potential
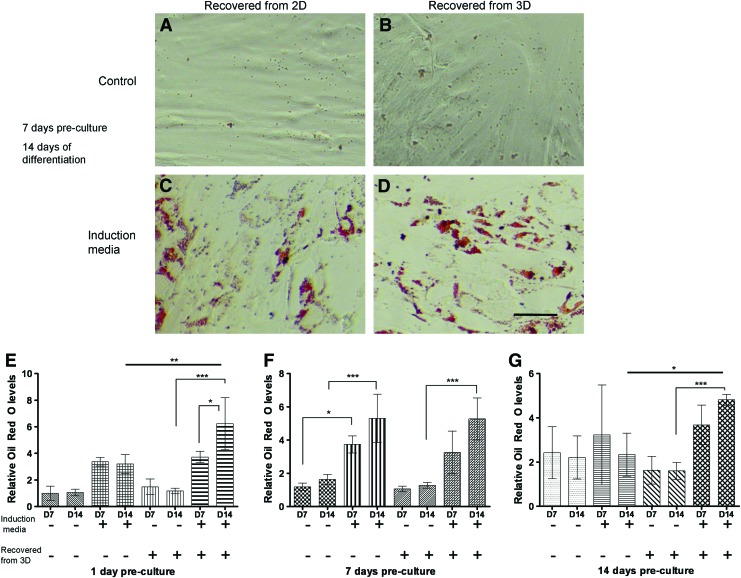
To assess the adipogenic differentiation potential of MSCs precultured for 1, 7, or 14 days in fibrin gels and subsequently retrieved by nattokinase, harvested cells were grown in media with various factors known to induce adipogenic differentiation followed by staining for the presence of lipid deposits with Oil Red O. Lipid deposits were detected 7 and 14 days after induction (Fig. 3C, D). Quantitative assessment of Oil Red O levels (Fig. 3E–G) showed that MSCs retrieved from 3D fibrin gels by nattokinase were readily induced to form lipid droplets in the presence of soluble adipogenic supplements, with the levels of Oil Red O comparable to those in cells cultured exclusively in 2D. These data suggest that nattokinase extraction of MSCs from fibrin does not diminish their ability to differentiate into adipocytes.
FIG. 3.
MSCs maintain the potential to become adipogenic after extraction from 3D fibrin gels. Micrographs represent MSCs grown on 2D tissue culture polystyrene (A, C) or in 3D fibrin gels (B, D) for 7 days of preculture and then extracted and maintained in growth media (A, B) or differentiated in adipogenic media (C, D) for 14 additional days in 2D culture. Cells were stained using the Oil Red O method. Scale bar represents 200 μm. (E, F, G) MSCs were grown for periods of 1 (E), 7 (F), or 14 days (G) in either 2D or 3D environments, extracted using trypsin (2D) or nattokinase (3D) and subsequently replated in 2D cultures. These cultures were then subjected to either growth media or adipogenic induction media for up to 14 days. Relative Oil Red O levels were generated by dividing the measured values first by the number of cells in each well and then normalized to the baseline levels expressed by MSCs cultured in 2D growth media after 1 day of preculture (i.e., the first data point on the bar graph in E). These data show that the presence of soluble adipogenic supplements and prolonged culture times in these supplements generally enhance adipogenic differentiation of the MSCs, as expected. They also show that MSCs retrieved from 3D fibrin gels using nattokinase have no apparent deficits in adipogenesis. *p≤0.05, **p≤0.01, ***p≤0.001 for statistical significance. Color images available online at www.liebertpub.com/tec
MSCs retrieved from 3D fibrin gels maintain their osteogenic potential
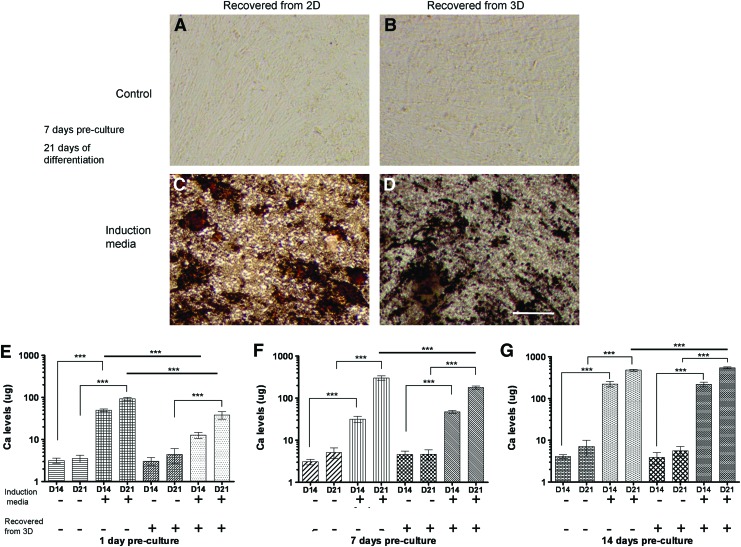
To assess the osteogenic differentiation potential of MSCs precultured for up to 14 days in fibrin gels and subsequently extracted by nattokinase, harvested cells were cultured in the OIM and compared to those grown exclusively on tissue culture polystyrene (TCPS) as a control. Mineral deposition was visualized by the common von Kossa phosphate staining protocol. MSCs differentiated in the OIM after growth on 2D TCPS or recovered from 3D fibrin gels stained positive for phosphates after 14 and 21 days (Fig. 4C, D). In parallel, the amounts of calcium deposited by the MSCs were quantified by the OCPC method. Cells cultured in the OIM showed elevated calcium levels (compared to noninduced controls), regardless of whether or not they were grown exclusively in 2D or were first extracted from 3D fibrin gels by nattokinase (Fig. 4E–G). Specifically, cells precultured for 14 days in 3D fibrin gels, recovered with nattokinase, and then differentiated for 14 additional days showed equivalent calcium levels compared to cells grown on a 2D surface [∼220 μg]. Collectively, these data qualitatively and quantitatively suggest that nattokinase extraction of MSCs from fibrin does not reduce their osteogenic differentiation potential.
FIG. 4.
MSCs maintain the potential to become osteogenic after extraction from 3D fibrin gels. Micrographs represent MSCs grown on 2D TCPS (A, C) or in 3D fibrin gels (B, D) for 7 days of preculture and then extracted and maintained in growth media (A, B) or differentiated in osteogenic media (C, D) for 21 additional days in 2D culture. Cells were stained using the von Kossa method. Scale bar represents 200 μm. (E, F, G) MSCs were grown for periods of 1 (E), 7 (F), or 14 days (G) in either 2D or 3D environments, extracted using trypsin (2D) or nattokinase (3D) and subsequently replated in 2D cultures. These cultures were then subjected to either growth media or osteogenic induction media for up to 21 days. Total calcium levels were then quantified as an indication of osteogenic differentiation as described in the “Materials and Methods” section. These data show that the presence of soluble osteogenic supplements and prolonged culture times in these supplements generally enhance osteogenic differentiation of the MSCs, as expected. They also show that MSCs retrieved from 3D fibrin gels using nattokinase have no apparent deficits in osteogenesis. ***p≤0.001 for statistical significance. Color images available online at www.liebertpub.com/tec
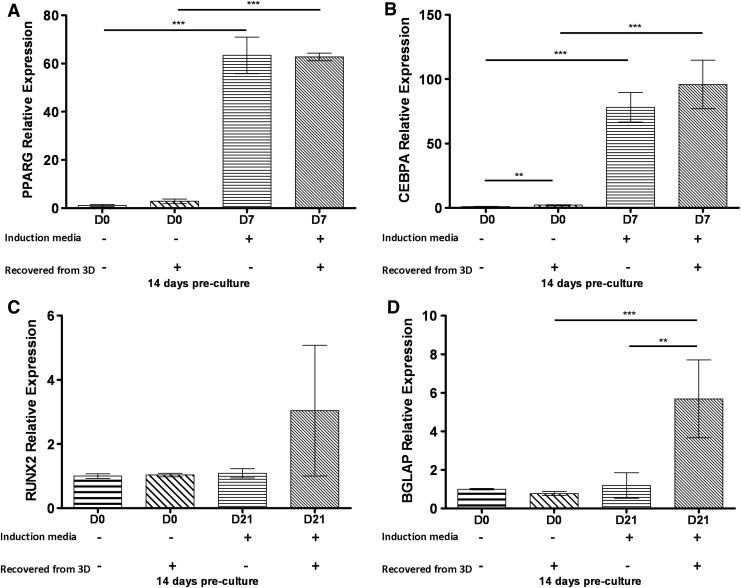
MSCs retrieved from 3D fibrin gels express genes associated with osteogenic and adipogenic lineages
Finally, we quantified the expression of several genes associated with adipogenic and osteogenic differentiation to further assess how well MSCs precultured for 1, 7, and 14 days and subsequently extracted from 3D fibrin gels by nattokinase sustain their multipotency. Specifically, for Oil Red O, von Kossa, and calcium assays described above, MSCs were first grown on 2D TCPS or within 3D fibrin gels for up to 14 days, recovered, and subjected to the appropriate induction media favorable for differentiation. Gene expression analysis was performed on cells after 7 and 21 days in adipogenic- and osteogenic-specific culture conditions, respectively. Similar levels of PPARγ and CEBPα were detected in cells cultured in adipogenic media for 7 days (Fig. 5A, B), regardless of whether they had first been cultured in 3D fibrin gels and recovered with nattokinase or cultured exclusively on 2D TCPS. However, control cultures grown in the baseline medium did not show adipogenic differentiation. Likewise, qPCR analysis confirmed that MSCs retrieved from 3D fibrin gels by nattokinase were also able to upregulate the gene expression levels of Runx2 and BGLAP in response to osteogenic inductive media (Fig. 5C, D). These findings collectively suggest that nattokinase extraction of MSCs from 3D fibrin gels does not negatively impact their ability to express key genes associated with adipogenic and osteogenic differentiation.
FIG. 5.
Quantitative polymerase chain reaction (qPCR) analysis of adipogenic and osteogenic marker gene expression levels in MSCs retrieved from culture conditions. MSCs were recovered from 2D (by trypsin) and 3D cultures (by nattokinase) after 14 days of preculture and then subjected to either adipogenic or osteogenic induction media for either an additional 7 or 21 days, respectively. Total RNA was extracted from the cells and subjected to qPCR analysis to assess the expression levels of (A) PPARγ, (B) CEBPα, (C) Runx2, and (D) BGLAP. Statistics were performed on ΔΔCT values and are indicated as shown (**p≤0.01, ***p≤0.001). Collectively, these data showed that MSCs precultured in 3D fibrin gels and subjected to nattokinase extraction were capable of subsequently expressing genes consistent with osteogenic and adipogenic differentiation potentials.
Discussion
In an effort to recapitulate the structural and functional characteristics of in vivo microenvironments, significant emphasis in the tissue engineering and biomaterials communities has been placed on the development of the 3D cell culture systems.56 However, analytical assays and tools commonly used to assess cell phenotypes in 2D cultures are typically more complicated in 3D and, generally, require that cells first be retrieved from the 3D environment. The recovery of viable cells from 3D cultures has been limited by the lack of suitable methods to retrieve encapsulated cells. In our own previous work, we have extensively used fibrin to investigate capillary morphogenesis in vitro and neovascularization in vivo.32,35,57 However, despite our experience with fibrin, we lacked a simple yet effective method to recover cells encapsulated within fibrin hydrogels for further characterization. Here, we have presented a safe and efficient protocol for the isolation of cells from 3D fibrin gels based on the strong fibrinolytic enzyme, nattokinase, a serine protease of the subtilisin family that has a strong fibrinolytic activity.
To validate this method, we first assessed the efficiency of nattokinase for retrieving cells from fibrin gels compared to other commonly used proteolytic enzymes, trypsin (another serine protease)58 and TrypLE™ (a recombinant fungal trypsin-like protease).59,60 Using MSCs as a model cell type, we showed that nattokinase was >4×more efficient than TrypLE for cell extraction (Fig. 2A). The number and viability of MSCs extracted from fibrin by nattokinase were also nearly equivalent to cells harvested from 2D tissue culture plastic by trypsin (Fig. 2B, C), suggesting that MSCs proliferate to a comparable extent in 3D fibrin as they do on 2D polystyrene and that nattokinase does not compromise their plasma membranes.
We next investigated the effects of cell extraction using nattokinase on the multilineage potential of MSCs harvested from 3D fibrin gels. Traditional adipogenic and osteogenic differentiation assays were performed in 2D cultures using MSCs that were first propagated within and retrieved from 3D fibrin gels and compared to cells that were grown and then differentiated entirely in 2D. The analysis of Oil Red O levels, an indicator of lipid formation, showed that MSCs extracted from 3D fibrin gels through nattokinase extraction were equally capable of adipogenic differentiation as controls from 2D cultures (Fig. 3). Furthermore, qPCR analyses of the genes encoding for PPARγ, a key regulator of adipogenesis, and CEBPα, a positive feedback loop regulator of PPARγ expression,46,61,62 suggested that nattokinase extraction after 14 days of preculture in 3D fibrin gels followed by 7 days of exposure to adipogenic differentiation media did not alter the adipogenic potential of MSCs (Fig. 5A, B). Similarly, von Kossa staining and the quantification of calcium levels revealed qualitatively and quantitatively that nattokinase did not alter the ability of MSCs to synthesize a matrix capable of mineralization (Fig. 4). Quantitative PCR analysis confirmed that cells extracted from fibrin using nattokinase were capable of osteogenic gene expression. The expression of BGLAP (the gene encoding for osteocalcin) in cells extracted from 3D was elevated with respect to cells grown on 2D surfaces (Fig. 5D), suggesting that priming the cells for a period of time in 3D fibrin gels before induction may, in fact, enhance the osteogenic phenotype. Differences in Runx2 gene expression, an early marker of osteogenic differentiation,63 showed similar trends (Fig. 5C), but were not significantly different in MSCs induced down an osteogenic lineage for 21 days preceded by 14 days of growth in 3D fibrin gels relative to cells cultured exclusively in 2D. Collectively, these data illustrate that nattokinase extraction does not diminish the potential of MSCs to undergo subsequent adipogenesis or osteogenesis or to express genes characteristic of these two phenotypes. Although we did not explicitly subject the retrieved MSCs to a chondrogenic differentiation protocol, we anticipate that the MSCs recovered through this method would indeed be capable of undergoing chondrogenesis. Since many studies have explored the ability of fibrin-based materials to support the formation of cartilaginous tissues both in vitro and in vivo,64–67 the method we have developed here may enable other researchers to retrieve MSCs undergoing chondrogenesis to better understand the process.
In the human body, fibrinolysis is achieved mainly by the serine protease plasmin68 and can also be achieved by matrix metalloproteinases in certain circumstances.69,70 Plasmin is generated by enzymatic cleavage of plasminogen, either by the urokinase plasminogen activator (uPA) or by the tissue plasminogen activator (tPA). The activation by uPA requires initial binding of uPA to the cell membrane-anchored uPA receptor, thereby sequestering plasmin activation and proteolysis to the immediate vicinity of the cell surface. Conversely, the activation by tPA does not require prior binding to a cell surface receptor and results in a global activation of plasmin. Given these mechanisms, a reasonable enzyme to extract cells from 3D fibrin gels would be plasmin or the activators of plasmin, uPA or tPA. In fact, prior studies have used purified urokinase to isolate stromal cells from 3D fibrin gel cultures.41,42 In those studies, the authors dissolved their fibrin gels in a solution consisting of the medium containing serum (the source of the plasminogen) and 5000 units of urokinase. However, we did not compare these enzymes side-by-side in this study primarily due to their high cost relative to nattokinase. Achieving the comparable levels of fibrinolytic activity by purified urokinase would be ∼300×more expensive than nattokinase, whereas plasmin would cost nearly 42,000×more. Other possible enzymes were also considered, including proteinase K, collagenase, and Accutase; however, since none of these are specific for fibrin,71–73 we reasoned that they would not be as efficient.
Trypsin is commonly used to passage cells during 2D cell culture, but our data show that retrieving cells from 3D fibrin gels using trypsin is very inefficient, even when the gels are exposed to the enzyme for 30 min. As prolonged exposure to trypsin causes an upregulation in proteins that regulate apoptosis,74 we concluded that incubation times longer than 30 min would be undesirable. We speculate that the increased efficiency of cell retrieval and the high degree of cell viability achieved with 30 min of nattokinase are likely due to its high affinity and fibrinolytic specificity to cross-linked fibrin.43–45 Although we are unaware of direct comparisons of the relative affinities of nattokinase and trypsin to fibrin, trypsin has a markedly lower affinity to fibrin compared to plasmin.75 Furthermore, a previous report suggested that nattokinase has a higher affinity to cross-linked fibrin than plasmin.76 Thus, it is reasonable to infer that nattokinase also has a higher affinity to fibrin compared to trypsin.
MSCs from bone marrow and a variety of other adult tissues are already the focus of numerous human clinical trials77,78 and have shown enormous promise in preclinical studies to facilitate bone regeneration,79 promote tissue neovascularization,80–82 and reduce inflammation.83 Much of their therapeutic benefits seem to be related to their trophic effects, that is, through the secretion of numerous growth factors.83 In the case of bone marrow, MSCs are relatively rare cells (∼0.01% of the nucleated cells from a low-density Percoll gradient84) and are typically isolated based on their adherent properties.84 Comparatively, a large number of cells (∼107) are needed for therapeutic applications, in part, because the number of cells that actually engraft within target tissues may be quite low.85 As a result, MSCs are typically expanded using standard 2D cell cultures. However, it has previously been shown that culturing MSCs on 2D surfaces, over time, diminishes the expression levels of surface markers commonly associated with MSCs (e.g., VCAM-1, ICAM-1, and CD157)86 and adversely affects their proliferation and telomere length.9,10 A quasi-3D fibrin culture, where cells were grown on fibrin gels, retained multipotentiality of MSCs,87 but it is unclear if a protocol consisting entirely of 3D culture would be even better in terms of maintaining MSC multipotency. With the simple enzymatic method to digest fibrin without harming the cells that we have described here, the possibility now exists that MSCs can be cultured exclusively in 3D from the time of harvest (or perhaps, the first passage, to exploit their adherent properties to isolate them from other cell types) to the time of therapeutic application. Furthermore, cultures of multiple cell types could now theoretically be grown in 3D fibrin gels, extracted by nattokinase, and subsequently sorted through fluorescent-activated cell sorting for subsequent analyses or applications. In our own work, we expect that this methodology will enable our efforts to better understand cross talk between MSCs and endothelial cells in the perivascular niche.35
Conclusion
Retrieving viable cells with high efficiency from 3D environments is nontrivial. We described here a simple yet effective method to harvest MSCs encapsulated within 3D fibrin gels using a powerful fibrinolytic enzyme, nattokinase. Our data show that MSCs recovered from 3D fibrin gels using nattokinase are not only viable but also retain their proliferative and multilineage potential. Demonstrated for MSCs, this method will likely be useful to also retrieve other cell types from 3D fibrin gels for subsequent applications, including expansion, bioassays, and in vivo injection.
Acknowledgments
We thank Mr. Kazuya Ogasawara from Japan Bio Science Laboratory Co., Ltd., Oita, Japan, for the kind gift of the nattokinase (NSK-SD) extract used in this study. We also acknowledge Dr. Mohamed El-Sayed, Dr. Scott Medina, and Dr. Rameshwar Rao for equipment access and assay technical advice. Financial support for this study was provided by the U.S. National Institutes of Health (R01-HL085339 and R21-DE021537 to A.J.P.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Rao M.S., and Mattson M.P.Stem cells and aging: expanding the possibilities. Mech Ageing Dev 122,713, 2001 [DOI] [PubMed] [Google Scholar]
- 2.Sarugaser R., Ennis J., Stanford W.L., and Davies J.E.Isolation, propagation, and characterization of human umbilical cord perivascular cells (HUCPVCs). Methods Mol Biol 482,269, 2009 [DOI] [PubMed] [Google Scholar]
- 3.De Coppi P., Bartsch G., Jr., Siddiqui M.M., Xu T., Santos C.C., Perin L., Mostoslavsky G., Serre A.C., Snyder E.Y., Yoo J.J., Furth M.E., Soker S., and Atala A.Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol 25,100, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Yen B.L., Chang C.J., Liu K.J., Chen Y.C., Hu H.I., Bai C.H., and Yen M.L.Brief report—human embryonic stem cell-derived mesenchymal progenitors possess strong immunosuppressive effects toward natural killer cells as well as T lymphocytes. Stem Cells 27,451, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Yen M.L., Hou C.H., Peng K.Y., Tseng P.C., Jiang S.S., Shun C.T., Chen Y.C., and Kuo M.L.Efficient derivation & concise gene expression profiling of human embryonic stem cell-derived mesenchymal progenitors (EMPs). Cell Transplant 20,1529, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Caplan A.I., and Correa D.The MSC: an injury drugstore. Cell Stem Cell 9,11, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutolf M.P., and Blau H.M.Artificial stem cell niches. Adv Mater 21,3255, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert P.M., and Blau H.M.Engineering a stem cell house into a home. Stem Cell Res Ther 2,3, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banfi A., Muraglia A., Dozin B., Mastrogiacomo M., Cancedda R., and Quarto R.Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp Hematol 28,707, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Baxter M.A., Wynn R.F., Jowitt S.N., Wraith J.E., Fairbairn L.J., and Bellantuono I.Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells 22,675, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Reiser J., Zhang X.Y., Hemenway C.S., Mondal D., Pradhan L., and La Russa V.F.Potential of mesenchymal stem cells in gene therapy approaches for inherited and acquired diseases. Expert Opin Biol Ther 5,1571, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., and Frenette P.S.Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466,829, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markway B.D., Tan G.K., Brooke G., Hudson J.E., Cooper-White J.J., and Doran M.R.Enhanced chondrogenic differentiation of human bone marrow-derived mesenchymal stem cells in low oxygen environment micropellet cultures. Cell Transplant 19,29, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Wang W., Itaka K., Ohba S., Nishiyama N., Chung U.I., Yamasaki Y., and Kataoka K.3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells. Biomaterials 30,2705, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Jahn K., Richards R.G., Archer C.W., and Stoddart M.J.Pellet culture model for human primary osteoblasts. Eur Cell Mater 20,149, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Ungrin M.D., Joshi C., Nica A., Bauwens C., and Zandstra P.W.Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One 3,e1565, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook M.M., Futrega K., Osiecki M., Kabiri M., Kul B., Rice A., Atkinson K., Brooke G., and Doran M.Micromarrows—three-dimensional coculture of hematopoietic stem cells and mesenchymal stromal cells. Tissue Eng Part C Methods 18,319, 2012 [DOI] [PubMed] [Google Scholar]
- 18.King J.A., and Miller W.M.Bioreactor development for stem cell expansion and controlled differentiation. Curr Opin Chem Biol 11,394, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breen A., O'Brien T., and Pandit A.Fibrin as a delivery system for therapeutic drugs and biomolecules. Tissue Eng Part B Rev 15,201, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Ho W., Tawil B., Dunn J.C., and Wu B.M.The behavior of human mesenchymal stem cells in 3D fibrin clots: dependence on fibrinogen concentration and clot structure. Tissue Eng 12,1587, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Christman K.L., Vardanian A.J., Fang Q., Sievers R.E., Fok H.H., and Lee R.J.Injectable fibrin scaffold improves cell transplant survival, reduces infarct expansion, and induces neovasculature formation in ischemic myocardium. J Am Coll Cardiol 44,654, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Syedain Z.H., Bjork J., Sando L., and Tranquillo R.T.Controlled compaction with ruthenium-catalyzed photochemical cross-linking of fibrin-based engineered connective tissue. Biomaterials 30,6695, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang N.F., Lam A., Fang Q., Sievers R.E., Li S., and Lee R.J.Bone marrow-derived mesenchymal stem cells in fibrin augment angiogenesis in the chronically infarcted myocardium. Regen Med 4,527, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu J.H., Kim I.K., Cho S.W., Cho M.C., Hwang K.K., Piao H., Piao S., Lim S.H., Hong Y.S., Choi C.Y., Yoo K.J., and Kim B.S.Implantation of bone marrow mononuclear cells using injectable fibrin matrix enhances neovascularization in infarcted myocardium. Biomaterials 26,319, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Ruger B.M., Breuss J., Hollemann D., Yanagida G., Fischer M.B., Mosberger I., Chott A., Lang I., Davis P.F., Hocker P., and Dettke M.Vascular morphogenesis by adult bone marrow progenitor cells in three-dimensional fibrin matrices. Differentiation 76,772, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Bensaid W., Triffitt J.T., Blanchat C., Oudina K., Sedel L., and Petite H.A biodegradable fibrin scaffold for mesenchymal stem cell transplantation. Biomaterials 24,2497, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Catelas I., Sese N., Wu B.M., Dunn J.C., Helgerson S., and Tawil B.Human mesenchymal stem cell proliferation and osteogenic differentiation in fibrin gels in vitro. Tissue Eng 12,2385, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Dickhut A., Gottwald E., Steck E., Heisel C., and Richter W.Chondrogenesis of mesenchymal stem cells in gel-like biomaterials in vitro and in vivo. Front Biosci 13,4517, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Baumgartner L., Arnhold S., Brixius K., Addicks K., and Bloch W.Human mesenchymal stem cells: influence of oxygen pressure on proliferation and chondrogenic differentiation in fibrin glue in vitro. J Biomed Mater Res A 93,930, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Ghajar C.M., Blevins K.S., Hughes C.C., George S.C., and Putnam A.J.Mesenchymal stem cells enhance angiogenesis in mechanically viable prevascularized tissues via early matrix metalloproteinase upregulation. Tissue Eng 12,2875, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Ghajar C.M., Chen X., Harris J.W., Suresh V., Hughes C.C., Jeon N.L., Putnam A.J., and George S.C.The effect of matrix density on the regulation of 3-D capillary morphogenesis. Biophys J 94,1930, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghajar C.M., Kachgal S., Kniazeva E., Mori H., Costes S.V., George S.C., and Putnam A.J.Mesenchymal cells stimulate capillary morphogenesis via distinct proteolytic mechanisms. Exp Cell Res 316,813, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kachgal S., and Putnam A.J.Mesenchymal stem cells from adipose and bone marrow promote angiogenesis via distinct cytokine and protease expression mechanisms. Angiogenesis 14,47, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kniazeva E., Kachgal S., and Putnam A.J.Effects of extracellular matrix density and mesenchymal stem cells on neovascularization in vivo. Tissue Eng Part A 17,905, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrion B., Huang C.P., Ghajar C.M., Kachgal S., Kniazeva E., Jeon N.L., and Putnam A.J.Recreating the perivascular niche ex vivo using a microfluidic approach. Biotechnol Bioeng 107,1020, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kachgal S., Carrion B., Janson I.A., and Putnam A.J.Bone marrow stromal cells stimulate an angiogenic program that requires endothelial MT1-MMP. J Cell Physiol 227,3546, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kossack N., Meneses J., Shefi S., Nguyen H.N., Chavez S., Nicholas C., Gromoll J., Turek P.J., and Reijo-Pera R.A.Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells 27,138, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun N., Panetta N.J., Gupta D.M., Wilson K.D., Lee A., Jia F., Hu S., Cherry A.M., Robbins R.C., Longaker M.T., and Wu J.C.Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A 106,15720, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang G.T., Yamaza T., Shea L.D., Djouad F., Kuhn N.Z., Tuan R.S., and Shi S.Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng Part A 16,605, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farias V.A., Linares-Fernandez J.L., Penalver J.L., Paya Colmenero J.A., Ferron G.O., Duran E.L., Fernandez R.M., Olivares E.G., O'Valle F., Puertas A., Oliver F.J., and Ruiz de Almodovar J.M.Human umbilical cord stromal stem cell express CD10 and exert contractile properties. Placenta 32,86, 2011 [DOI] [PubMed] [Google Scholar]
- 41.Yang Y.I., Kim H.I., Shelby J., Choi M.Y., Jang S.H., Kim J.T., Jang W.H., Choi C.S., and Cheong S.H.Fibrin matrix-supported three-dimensional organ culture of adipose tissue for selective outgrowth, expansion, and isolation of adipose-derived stem cells. Acta Biomater 7,4109, 2011 [DOI] [PubMed] [Google Scholar]
- 42.Choi M.Y., Kim H.I., Yang Y.I., Kim J.T., Jang S.H., Park C.M., Jang W.H., Youn Y.C., Cheong S.H., Choi C.S., Kim D.K., and Lee S.J.The isolation and in situ identification of MSCs residing in loose connective tissues using a niche-preserving organ culture system. Biomaterials 33,4469, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Urano T., Ihara H., Umemura K., Suzuki Y., Oike M., Akita S., Tsukamoto Y., Suzuki I., and Takada A.The profibrinolytic enzyme subtilisin NAT purified from Bacillus subtilis cleaves and inactivates plasminogen activator inhibitor type 1. J Biol Chem 276,24690, 2001 [DOI] [PubMed] [Google Scholar]
- 44.Sumi H., Hamada H., Tsushima H., Mihara H., and Muraki H.A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 43,1110, 1987 [DOI] [PubMed] [Google Scholar]
- 45.Fujita M., Nomura K., Hong K., Ito Y., Asada A., and Nishimuro S.Purification and characterization of a strong fibrinolytic enzyme (nattokinase) in the vegetable cheese natto, a popular soybean fermented food in Japan. Biochem Biophys Res Commun 197,1340, 1993 [DOI] [PubMed] [Google Scholar]
- 46.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., and Chen C.S.Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6,483, 2004 [DOI] [PubMed] [Google Scholar]
- 47.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., and Marshak D.R.Multilineage potential of adult human mesenchymal stem cells. Science 284,143, 1999 [DOI] [PubMed] [Google Scholar]
- 48.Prockop D.J.P., Donald G.; Bunnell B.A., ed. Mesenchymal Stem Cells: Methods and Protocols. Totowa, NJ: Humana Press, 2008 [Google Scholar]
- 49.Rao R.R., Jiao A., Kohn D.H., and Stegemann J.P.Exogenous mineralization of cell-seeded and unseeded collagen-chitosan hydrogels using modified culture medium (vol 8, pg 1560, 2012). Acta Biomater 8,2417, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ter Brugge P.J., and Jansen J.A.In vitro osteogenic differentiation of rat bone marrow cells subcultured with and without dexamethasone. Tissue Eng 8,321, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Schmittgen T.D., and Livak K.J.Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3,1101, 2008 [DOI] [PubMed] [Google Scholar]
- 52.Fink T., Lund P., Pilgaard L., Rasmussen J.G., Duroux M., and Zachar V.Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol 9,98, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krause U., Seckinger A., and Gregory C.A.Assays of osteogenic differentiation by cultured human mesenchymal stem cells. Methods Mol Biol 698,215, 2011 [DOI] [PubMed] [Google Scholar]
- 54.Stacey D.H., Hanson S.E., Lahvis G., Gutowski K.A., and Masters K.S.In vitro adipogenic differentiation of preadipocytes varies with differentiation stimulus, culture dimensionality, and scaffold composition. Tissue Eng Part A 15,3389, 2009 [DOI] [PubMed] [Google Scholar]
- 55.Tarnowski B.I., Sens D.A., Nicholson J.H., Hazenmartin D.J., Garvin A.J., and Sens M.A.Automatic quantitation of cell-growth and determination of mitotic index using DAPI nuclear staining. Pediatr Pathol 13,249, 1993 [DOI] [PubMed] [Google Scholar]
- 56.Lutolf M.P., Gilbert P.M., and Blau H.M.Designing materials to direct stem-cell fate. Nature 462,433, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grainger S.J., and Putnam A.J.Assessing the permeability of engineered capillary networks in a 3D culture. PLoS One 6,e22086, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kocholaty W., Ellis W.W., and Jensen H.Activation of plasminogen by trypsin and plasmin. Blood 7,882, 1952 [PubMed] [Google Scholar]
- 59.T'Joen V., Declercq H., and Cornelissen M.Expansion of human embryonic stem cells: a comparative study. Cell Prolif 44,462, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marley M.S.D., Givens M.D., Galik P.K., Riddell K.P., Looney C.R., and Stringfellow D.A.Efficacy of a recombinant trypsin product against bovine herpesvirus I associated with in vivo- and in vitro-derived bovine embryos. Theriogenology 69,746, 2008 [DOI] [PubMed] [Google Scholar]
- 61.Kanematsu D., Shofuda T., Yamamoto A., Ban C., Ueda T., Yamasaki M., and Kanemura Y.Isolation and cellular properties of mesenchymal cells derived from the decidua of human term placenta. Differentiation 82,77, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Kim W.K., Meliton V., Amantea C.M., Hahn T.J., and Parhami F.20(S)-Hydroxycholesterol inhibits PPAR gamma expression and adipogenic differentiation of bone marrow stromal cells through a hedgehog-dependent mechanism. J Bone Miner Res 22,1711, 2007 [DOI] [PubMed] [Google Scholar]
- 63.Psaltis P.J., Paton S., See F., Arthur A., Martin S., Itescu S., Worthley S.G., Gronthos S., and Zannettino A.C.W.Enrichment for STRO-1 expression enhances the cardiovascular paracrine activity of human bone marrow-derived mesenchymal cell populations. J Cell Physiol 223,530, 2010 [DOI] [PubMed] [Google Scholar]
- 64.Ho S.T., Cool S.M., Hui J.H., and Hutmacher D.W.The influence of fibrin based hydrogels on the chondrogenic differentiation of human bone marrow stromal cells. Biomaterials 31,38, 2010 [DOI] [PubMed] [Google Scholar]
- 65.Jung S.N., Rhie J.W., Kwon H., Jun Y.J., Seo J.W., Yoo G., Oh D.Y., Ahn S.T., Woo J., and Oh J.In vivo cartilage formation using chondrogenic-differentiated human adipose-derived mesenchymal stem cells mixed with fibrin glue. J Craniofac Surg 21,468, 2010 [DOI] [PubMed] [Google Scholar]
- 66.Wu X., Ren J., and Li J.Fibrin glue as the cell-delivery vehicle for mesenchymal stromal cells in regenerative medicine. Cytotherapy 14,555, 2012 [DOI] [PubMed] [Google Scholar]
- 67.Diederichs S., Baral K., Tanner M., and Richter W.Interplay between local versus soluble transforming growth factor-beta and fibrin scaffolds: role of cells and impact on human mesenchymal stem cell chondrogenesis. Tissue Eng Part A 18,1140, 2012 [DOI] [PubMed] [Google Scholar]
- 68.Lijnen H.R.Elements of the fibrinolytic system. Ann N Y Acad Sci 936,226, 2001 [DOI] [PubMed] [Google Scholar]
- 69.Hiraoka N., Allen E., Apel I.J., Gyetko M.R., and Weiss S.J.Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell 95,365, 1998 [DOI] [PubMed] [Google Scholar]
- 70.Hotary K.B., Yana I., Sabeh F., Li X.Y., Holmbeck K., Birkedal-Hansen H., Allen E.D., Hiraoka N., and Weiss S.J.Matrix metalloproteinases (MMPs) regulate fibrin-invasive activity via MT1-MMP-dependent and -independent processes. J Exp Med 195,295, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brendlerschwaab S.Y., Schmezer P., Liegibel U., Weber S., Michalek K., Tompa A., and Poolzobel B.L.Cells of different tissues for in-vitro and in-vivo studies in toxicology—compilation of isolation methods. Toxicol In Vitro 8,1285, 1994 [DOI] [PubMed] [Google Scholar]
- 72.MacCoss M.J., Wu C.C., and Yates J.R.Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal Chem 74,5593, 2002 [DOI] [PubMed] [Google Scholar]
- 73.Shao L.J., Feng W., Sun Y., Bai H., Liu J., Currie C., Kim J.J., Gama R., Wang Z., Qian Z.J., Liaw L., and Wu W.S.Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res 19,296, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang H.L., Hsing H.W., Lai T.C., Chen Y.W., Lee T.R., Chan H.T., Lyu P.C., Wu C.L., Lu Y.C., Lin S.T., Lin C.W., Lai C.H., Chang H.T., Chou H.C., and Chan H.L.Trypsin-induced proteome alteration during cell subculture in mammalian cells. J Biomed Sci 17,36, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolev K., Tenekedjiev K., Komorowicz E., and Machovich R.Functional evaluation of the structural features of proteases and their substrate in fibrin surface degradation. J Biol Chem 272,13666, 1997 [DOI] [PubMed] [Google Scholar]
- 76.Fujita M., Ito Y., Hong K., and Nishimuro S.Characterization of nattokinase-degraded products from human fibrinogen or cross-linked fibrin. Fibrinolysis 9,157, 1995 [Google Scholar]
- 77.Giordano A., Galderisi U., and Marino I.R.From the laboratory bench to the patient's bedside: an update on clinical trials with mesenchymal stem cells. J Cell Physiol 211,27, 2007 [DOI] [PubMed] [Google Scholar]
- 78.Wagner J., Kean T., Young R., Dennis J.E., and Caplan A.I.Optimizing mesenchymal stem cell-based therapeutics. Curr Opin Biotechnol 20,531, 2009 [DOI] [PubMed] [Google Scholar]
- 79.Simmons C.A., Alsberg E., Hsiong S., Kim W.J., and Mooney D.J.Dual growth factor delivery and controlled scaffold degradation enhance in vivo bone formation by transplanted bone marrow stromal cells. Bone 35,562, 2004 [DOI] [PubMed] [Google Scholar]
- 80.Nagaya N., Kangawa K., Itoh T., Iwase T., Murakami S., Miyahara Y., Fujii T., Uematsu M., Ohgushi H., Yamagishi M., Tokudome T., Mori H., Miyatake K., and Kitamura S.Transplantation of mesenchymal stem cells improves cardiac function in a rat model of dilated cardiomyopathy. Circulation 112,1128, 2005 [DOI] [PubMed] [Google Scholar]
- 81.Silva G.V., Litovsky S., Assad J.A., Sousa A.L., Martin B.J., Vela D., Coulter S.C., Lin J., Ober J., Vaughn W.K., Branco R.V., Oliveira E.M., He R., Geng Y.J., Willerson J.T., and Perin E.C.Mesenchymal stem cells differentiate into an endothelial phenotype, enhance vascular density, and improve heart function in a canine chronic ischemia model. Circulation 111,150, 2005 [DOI] [PubMed] [Google Scholar]
- 82.Kinnaird T., Stabile E., Burnett M.S., Shou M., Lee C.W., Barr S., Fuchs S., and Epstein S.E.Local delivery of marrow-derived stromal cells augments collateral perfusion through paracrine mechanisms. Circulation 109,1543, 2004 [DOI] [PubMed] [Google Scholar]
- 83.Caplan A.I.Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 213,341, 2007 [DOI] [PubMed] [Google Scholar]
- 84.Bruder S.P., Jaiswal N., and Haynesworth S.E.Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation. J Cell Biochem 64,278, 1997 [DOI] [PubMed] [Google Scholar]
- 85.Parekkadan B., and Milwid J.M.Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng 12,87, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Honczarenko M., Le Y., Swierkowski M., Ghiran I., Glodek A.M., and Silberstein L.E.Human bone marrow stromal cells express a distinct set of biologically functional chemokine receptors. Stem Cells 24,1030, 2006 [DOI] [PubMed] [Google Scholar]
- 87.Colley H., McArthur S.L., Stolzing A., and Scutt A.Culture on fibrin matrices maintains the colony-forming capacity and osteoblastic differentiation of mesenchymal stem cells. Biomed Mater 7,2012 [DOI] [PubMed] [Google Scholar]