Abstract
Flaviviral RNA-dependent RNA polymerases (RdRps) initiate replication of the single-stranded RNA genome in the absence of a primer. The template sequence 5′-CU-3′ at the 3′-end of the flaviviral genome is highly conserved. Surprisingly, flaviviral RdRps require high concentrations of the second incoming nucleotide GTP to catalyze de novo template-dependent RNA synthesis. We show that GTP stimulates de novo RNA synthesis by RdRp from Japanese encephalitis virus (jRdRp) also. Crystal structures of jRdRp complexed with GTP and ATP provide a basis for specific recognition of GTP. Comparison of the jRdRpGTP structure with other viral RdRp-GTP structures shows that GTP binds jRdRp in a novel conformation. Apo-jRdRp structure suggests that the conserved motif F of jRdRp occupies multiple conformations in absence of GTP. Motif F becomes ordered on GTP binding and occludes the nucleotide triphosphate entry tunnel. Mutational analysis of key residues that interact with GTP evinces that the jRdRpGTP structure represents a novel pre-initiation state. Also, binding studies show that GTP binding reduces affinity of RdRp for RNA, but the presence of the catalytic Mn2+ ion abolishes this inhibition. Collectively, these observations suggest that the observed pre-initiation state may serve as a checkpoint to prevent erroneous template-independent RNA synthesis by jRdRp during initiation.
INTRODUCTION
Japanese encephalitis virus (JEV) is a positive-strand RNA virus that belongs to the genus Flavivirus of the family Flaviviridae. Other members of this genus include dengue (DENV), West Nile (WNV), yellow fever and tick-borne encephalitis viruses (1). JEV is a neurotropic virus and the leading cause of viral encephalitis. The disease caused by this virus is endemic in China, South and South-east Asia. Conservative estimates state that ∼50 000 cases are reported annually, one-third of which are fatal and around half result in permanent neurological damage in survivors (2). JEV is thought to be responsible for hundreds of fatalities—mostly children—that occur every year in northern India due to viral encephalitis. Although vaccines are available, a rigorously tested effective low-cost vaccine that will afford long-term protection against infection is still lacking. As in the case of other pathogenic flaviviruses, therapeutic agents to combat JEV infection and to prevent disease progression are not available.
The JEV genome is represented by a single-stranded RNA (ssRNA) molecule ∼11-kb long, with a 5′ cap (m7G5′ppp5′A) and no polyadenylated tail. This viral genome codes for three structural (C, prM and E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins (3). The primary translation product of the viral genome is a 370-kDa polyprotein, which is processed by a host signalase and viral protease to yield structural and non-structural proteins.
NS5 is the largest (∼105 kDa) and the most conserved protein among the members of the Flavivirus genus (1,4). The N-terminal region of this protein (∼30 kDa) houses a methyltransferase activity, which is responsible for adding the methyl group to the 5′ RNA cap structure (5,6). Toward the C-terminus is present the RNA-dependent RNA polymerase (RdRp) module (∼70 kDa) that is critical for replication of the viral genome (7,8). NS5 associates strongly with NS3 and along with other viral proteins, host factors and genomic RNA “forms” a replication complex (RC) (9,10). Initially, the negative strand is synthesized to give rise to a double-stranded replicative form (dsRF) within the membrane-associated RC. The RC is localized inside double-layered membrane vesicles, presumably to protect the dsRF from host anti-viral factors (11,12). The RF is then used as a template to generate replicative intermediates. These replicative intermediates ultimately resolve to give rise to single-stranded genomic RNA and dsRF on completion of one round of synthesis. Thus, the synthesis of progeny RNA occurs in a semi-conservative and asymmetric manner on recycling dsRF (13). In addition to its core function in viral genome replication, NS5 has been implicated in regulating expression of host genes involved in anti-viral responses (4,14).
Unlike replicative DNA polymerases, viral RdRps use diverse strategies to initiate RNA synthesis (15). The polymerase of Picornaviridae family uses a protein primer (16), while influenza virus of the Orthomyxoviridae family can use an unusual cap snatching method to prime replication (17). For members of the families Flaviviridae, Cystoviridae (phi6 bacteriophage) and Reoviridae (rotaviruses), the RdRp enzyme can initiate RNA synthesis at the template termini de novo without the need for any primer (15). Once a phosphodiester bond is formed between the first two nucleotides, the enzyme switches from initiation to elongation mode and mediates template-dependent extension of the dinucleotide primer (18).
In the case of DENV RdRp, a high concentration of GTP is required for initiation of synthesis of RNA (18–20). The presence of the CU sequence at the 3′-end of the genomic RNA implies that GTP should be the second incoming nucleotide. The CU sequence is conserved in the genomes of most flaviviruses including JEV, at 3′-ends of both positive and negative strands. The requirement for higher concentration of GTP is also true for RdRp from distant viruses like hepatitis C virus (HCV; genus Hepacivirus; family Flaviviridae) (21), classical swine fever virus (genus Pestivirus; family Flaviviridae) (22) and phi6 bacteriophage (family Cystoviridae) (23). It should be mentioned here that the genomes of these viruses exhibit distinct terminal dinucleotide sequences that can be different for the negative and positive strands.
We show here that JEV RdRp (jRdRp) requires high concentration of GTP for initiation, but not for elongation. We have determined two crystal structures of jRdRp in complex with GTP (2.4 Å) and ATP (2.3 Å). In addition, we have obtained a structure of RdRp in the apo-state (3.65 Å). The structures of the jRdRpGTP and jRdRpATP complexes provide a basis for specific recognition of GTP by this enzyme. Additionally, these structures showed that the conserved motif F undergoes substantial ordering and occludes the nucleotide triphosphate (NTP) entry tunnel on GTP binding. Comparison of the jRdRpGTP structure with that of other viral RdRp-NTP complexes showed that the GTP molecule is bound in a novel conformation. A number of point mutants of jRdRp were prepared on the basis of the comparison of the jRdRpGTP structure with that of full initiation complex of RdRp from the bacteriophage phi6. These mutants were assessed for their ability to catalyze initiation and elongation of RNA synthesis. These experiments showed that jRdRpGTP structure represents a novel pre-initiation state of genome replication. Additionally, it was seen that GTP inhibits binding of jRdRp to RNA and that the presence of Mn2+ ions abolishes this inhibitory effect. Also, this inhibitory effect of GTP on RNA binding is lost or reduced in a mutant of RdRp that shows reduced initiation activity. Taken together, these studies allow formulation of a possible sequence of events during initiation that prevents non-templated and wasteful RNA synthesis that could be erroneous.
MATERIALS AND METHODS
Cloning, expression and purification of the jRdRp enzyme
The gene segment corresponding to the RdRp module (corresponding to amino acid 272–905 of the NS5 protein) was amplified from pGEX6P1 plasmid containing the NS5 gene of JEV strain P20778 (GenBank accession number AF080251). The amplified product was purified and ligated into the pGEX6P1 that would yield a fusion polypeptide with a GST tag at the N-terminus connected to the RdRp module by a linker with the PreScission protease site. Sequencing was carried out for a few clones and one without any mutations was selected for expression and purification of the RdRp protein. For making site-specific mutations in the jRdRp enzyme, QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies) was used. Primers were designed, and mutants were prepared according to manufacturer’s instructions.
The GST–jRdRp fusion protein was expressed in Escherichia coli C41(DE3) cells. Five liters of LB medium (100 μg/ml ampicillin) was inoculated with 25 ml of starter culture and incubated at 37°C at 200 rpm. When the optical density at 600 nm reached 0.6–0.8, isopropyl 1-thio-β-d-galactopyranoside was added to the culture at a final concentration of 0.25 mM and was incubated for 18 h at 18°C. Cells were harvested by centrifugation, resuspended in 25 mM Tris-Cl buffer, pH 8.0 (4°C), 500 mM NaCl, 5% glycerol, 2 mM dithiothreitol (DTT), 0.01% IGEPAL CA-630 and 1 mM phenyl methyl sulfonyl fluoride and then frozen in −80°C until use. Cells were lysed by sonication, and the lysate was clarified by centrifugation at 17 000 rpm for 45 min at 4°C. Glutathione-sepharose resin pre-equilibrated with buffer A (25 mM Tris-Cl buffer, pH 8.0, 250 mM NaCl, 5% glycerol and 2 mM DTT) was incubated with the clarified supernatant for 3 h. The beads were then washed with buffer B (25 mM Tris-Cl buffer, pH 8.0, 750 mM NaCl, 5% glycerol and 2 mM DTT) to remove non-specifically bound proteins, and then again with buffer A. The resin was then incubated with PreScission protease overnight to cleave the GST tag. jRdRp protein was eluted with buffer A, concentrated and further purified by size exclusion chromatography using a Superdex-200 column equilibrated with buffer C (25 mM Tris-Cl, pH 7.4, 250 mM NaCl, 2% glycerol and 2 mM DTT). The presence of jRdRp in the major peak was confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis, and the purified jRdRp was concentrated to 8–12 mg/ml and stored at −80°C.
Primer-free initiation assay
Primer-free de novo RNA synthesis was assessed by tracking the incorporation of 6-carboxy Fluorescein (6FAM)-labeled UTP in the presence of a template. The incorporation will occur only if a dinucleotide primer is formed and extended by the RdRp enzyme. Hence, in the absence of productive initiation, addition of labeled UTP to the growing primer will not occur. This method avoids the use of radioactive NTPs, which constitute a significant health and environmental hazard (24). The 13-nt RNA template (20 pmole) (5′-UAAUUCCCCCCCU-3′) was mixed with jRdRp protein (10 pmole) along with ATP (10 μM), CTP (10 μM), FAM-UTP (10 μM) and GTP (100 μM) in a 20-μl reaction mix. The buffer used for initiation was 50 mM Tris-Cl, pH 8.0 (at 30°C), 1 mM DTT, 5% glycerol and 0.05 mg/ml bovine serum albumin. To assess the role of GTP and ATP in initiation, reactions were carried out with varying concentrations of these two NTPs with the concentration of the other NTPs maintained at 10 μM. The concentration of divalent ion Mn2+ was 2.5 mM in the reaction mix. The reaction mixes were incubated at 30°C for 3 h, and the reactions were stopped by adding 10 µl of a solution containing 20 mM ethylenediaminetetraacetic acid and 80% formamide. The samples were heated for 5 min at 98°C followed by cooling on ice for 5 min. Next, the reaction products were resolved on a 20% polyacrylamide gel containing 8 M urea that was prepared and run in 1× Tris-borate + ethylenediaminetetraacetic acid buffer. Resolved products on the gel were observed by excitation at 488 nm, and the bands were visualized and recorded using Pharos Imager (BioRad Corp.). The intensities of the observed bands were quantified using Quantity one, 1D analysis software. The initiation activity for each reaction mixture was quantified by calculating the cumulative intensity of all the product bands in the corresponding lane on the gel.
A similar protocol was used to assess the effect of site-specific mutations in the jRdRp enzyme on primer-free initiation. For estimating relative activity of mutants, the activity of wild-type (wt) was taken as 100%.
Primer extension assay
To probe the elongation activity of jRdRp, a primer (5′-GUUCACACAGAUAAACUUCU-3′) with a 6-FAM labeled at the 5′-end was used as an alternative to radioactive labelling (24). RNA template (5′-AGAUCCUGCCCCCCCCCCCAAGAAGUUUAUCUGUGUGAAC-3′) and the labeled primer were mixed in equimolar quantity in 1× TE buffer and annealed by heating at 90°C for 5 min followed by cooling to room temperature (25°C). The annealed RNA template-primer duplex (20 picomole) was incubated with jRdRp (10 picomole) in 20 μl of 50 mM Tris-Cl, pH 8.0 (37°C), 1 mM DTT, 5% glycerol and 0.05 mg/ml bovine serum albumin. All four NTPs were added to the reaction volume to a final concentration of 100 μM, and the catalytic ion Mn2+ (MnCl2) was maintained at concentration of 2.5 mM. The reaction mixtures were incubated at 37°C for 3 h, and the samples were then processed using a protocol similar to that for initiation assay. The intensity of the unextended primer in each lane was quantified using Quantity one, 1D analysis software. The values obtained for each reaction mixture were divided by that obtained for a sample wherein reaction cannot occur to calculate the fraction of total primer that was extended by jRdRp. To quantify the elongation activity of different mutants of jRdRp relative to wt enzyme, the fraction of total primer extended by wt RdRp was taken as 100%.
Crystallization
jRdRp (0.08 mM) was incubated with GTP/ATP (5 mM) for 30 min before setting crystallization trials. After screening and optimization, the best crystals were obtained in 10–18% wt/vol PEG 4000, 100 mM Tris-Cl buffer, pH 8.0 (at 25°C), and 7% v/v 1-propanol by hanging drop method. The size of the crystals was improved by macroseeding. For data collection, the crystals were cryoprotected by soaking for 1 min in reservoir solutions containing 5, 10, 15 and 20% glycerol, respectively, and then flash frozen in liquid nitrogen.
Crystals of apo-jRdRp (0.096 mM) were obtained in 10–15% PEG 5000 mono methyl ether, 100 mM Tris-Cl, pH 8.0 (at 25°C), and 200 mM NaSCN. The crystals were cryoprotected using a protocol similar to that for crystals of jRdRpGTP.
Structure determination
X-ray diffraction data were collected at the BM14 beamline of ESRF, and a single crystal was used for data collection. Data were indexed, integrated, scaled and merged using HKL2000 (25).
The jRdRpGTP structure was determined by molecular replacement (MR) using MolRep and Phaser programs in CCP4i suite (26). The structure of apo- RdRp from WNV (2HFZ) and DENV (2J7W) was used as search models (27,28). For successful MR, search models corresponding to the palm, fingers and thumb domains of 2HFZ were prepared. WNV RdRp provided the best solutions for the thumb and palm domains with PHASER, and the collated coordinates were fixed during a MOLREP run to obtain a solution for the fingers domain. The composite solution was then subjected to rigid body refinement followed by B-factor and positional refinement using CNS (29). Subsequently, electron density maps (2Fo-Fc, Fo-Fc and composite simulated annealing omit map) were calculated and showed good fit between chemical model and electron density. These maps showed clear electron density corresponding to the GTP molecule near the active site. GTP was placed in this electron density, the sequence of 2HFZ was slowly changed to that of jRdRp and every change in the model was subjected to crystallographic refinement. After several rounds of model building and refinement, the automatic water picking procedure of CNS Package were used to place water molecules. This was followed by further refinement, and on convergence of the Rfree value, Translation/Libration/Screw (TLS) refinement was carried using Refmac program in CCP4i (30).
The jRdRpATP structure was determined by MR with the jRdRpGTP structure as a search model. Before MR, the coordinates corresponding to GTP and water molecules were removed from the search model. MR with Phaser yielded a clear solution, and after a round of rigid body and b-factor refinement, electron density maps were calculated. These maps showed density for the ATP molecule in the active site, but the density for base moiety was broken. ATP was placed in this density, and this was followed by model building, refinement and placement of water molecules iteratively until the Rfree value converged. Finally, TLS refinement was carried out using the Refmac program in CCP4i.
The apo-jRdRp structure was determined by MR using the jRdRpGTP structure as a search model in Phaser. Before MR, the coordinates corresponding to GTP and water molecules were removed from the search model. Refinement was carried out in PHENIX with NCS constraints until convergence of the Rfree value (31). Data collection and refinement statistics for jRdRpGTP, jRdRpATP and apo jRdRp are provided in Table 1. Coordinates and structure factors for the three structures have been deposited with the Protein Data Bank with the following accession codes: 4HDG (jRdRpGTP), 4HDH (jRdRpATP) and 4MTP (apo- jRdRp).
Table 1.
Data collection and refinement statistics
| jRdRpGTP | jRdRpATP | Apo jRdRp | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.95373 | 0.95373 | 0.95373 |
| Space group | P21212 | P21212 | P212121 |
| Unit cell dimensions–a, b, c (Å) | 144.6, 86.6, 112.7 | 144.8, 86.5, 112.8 | 119.5, 174.0,182.7 |
| Angles–α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50–2.38 | 50–2.28 | 56.78–3.65 |
| Rsym | 8.4 (47.3) | 7.0 (43.5) | 15.5 (56.9) |
| I/δI | 17.0 (2.7) | 21.2 (2.8) | 6.7 (2.4) |
| Completeness (%) | 99.73 (99.50) | 99.4 (97.4) | 99.6 (99.9) |
| Redundancy | 5.3 (5.3) | 5.0 (4.0) | 4.8 (4.8) |
| Number of reflections | 57 470 | 65 396 | 428 66 |
| Refinement | |||
| Rcrysta/Rfreeb | 22.0/27.6 | 22.4/27.0 | 23.4/27.3 |
| Number of atoms | |||
| Protein | 9838 | 9830 | 18 702 |
| Nucleotide | 64 | 62 | |
| Zn2+ ion | 4 | 4 | 8 |
| Water | 455 | 422 | |
| Average B-factors (Å2) | |||
| Protein | 36.6 | 38.3 | 47.1 |
| Nucleotide triphosphate | 67.0 | 64.1 | |
| Zn2+ ion | 38.6 | 36.6 | 44.9 |
| Water | 36.6 | 38.1 | |
| R.M.S.D | |||
| Bond lengths (Å) | 0.017 | 0.019 | 0.002 |
| Bond angles (°) | 1.587 | 1.567 | 0.64 |
Values in brackets refer to the highest resolution shell.
aRcryst = 
bRfree was calculated with 7% of reflections excluded from the whole refinement procedure for jRdRpGTP and jRdRpATP. For apo jRdRp, 5% of reflections were excluded.
Fluorescence anisotropy measurements to monitor binding of RdRp and RNA
Fluorescence anisotropy was measured using a SpectraMax M5 microplate reader (Molecular Devices). jRdRp and ssRNA (5′- XAUAAACUUCU-3′, where X is 6FAM label) were mixed and equilibrated for 20 min at room temperature in a binding buffer (50 mM Tris-Cl, pH 8.0, at 25°C, 5% glycerol, 1 mM DTT). RNA concentration was maintained at 5 nM, and jRdRp concentration was varied from 0 to 4000 nM. When added, Mn2+ ion was present in 100-fold molar excess of the protein. To study the effect of GTP on jRdRp–ssRNA binding, the mixture of jRdRp (1 µM) and FR10 (5 nM) was incubated with varying amounts of GTP. The ratio of the concentration of jRdRp to that of GTP was varied from 1:0.1 to 1:400. Fraction of RNA bound (FSB) was calculated using the following equation (32),
where AOBS is the observed anisotropy for a particular ratio of jRdRp and labeled RNA. AB and AF denote anisotropies of bound and free-labeled RNA, respectively. Q is the ratio of fluorescence intensities of bound and free-labeled RNA measured in the experiment. To determine the apparent dissociation constants (Kd), fraction of RNA bound (FSB) was plotted as a function of total RdRp concentration (Rt), and the data were fit to the following Hill equation using non-linear regression in Origin 8 software, where n is the average number of interacting sites:
RESULTS
GTP and initiation of RNA synthesis by jRdRp
The two nucleotides CU at the 3′-ends of positive and negative strands are conserved in the genomes of most flaviviruses and, therefore, could be essential for initiation. Additionally, RdRp molecules are known to go through rounds of abortive initiation before switching to elongation mode (20). To test the role of GTP in initiation, we designed a template RNA (5′-UAAUUCCCCCCCU-3′) that retains the conserved dinucleotide CU at the 3′-end and will not loop back to prime RNA synthesis. For this template, we observed maximum RNA synthesis at high GTP concentrations (>100 µM) with the concentration of other nucleotides maintained at 10 µM (Figure 1A and B). At high concentrations of GTP, a number of products corresponding to different lengths of RNA were synthesized giving rise to a ladder-like pattern on the gel (Figure 1A). The ladder-like pattern is probably due to polymerase slippage caused by the stretch of consecutive cytosines in the template, a phenomenon seen routinely in the case of DNA-dependent DNA polymerases (33). Additionally, RdRp molecules are known to go through rounds of abortive initiation before switching to elongation mode and this may heighten the effect of polymerase slippage (20). Unlike the case of GTP, there was no comparable increase in the amount of initiation products with increasing concentrations of ATP (Figure 1B and C).
Figure 1.
Role of GTP in initiation and elongation by jRdRp. The results of initiation assay in the absence of primer with varying GTP (A) and ATP (B) concentrations are displayed. For the initiation assay, 6FAM-labeled UTP was used to visualize the reaction products. Lane 1 in (A) shows the migration of molecular markers, and the bands corresponding to 5, 12, 15 and 20mer ssRNA labeled with 6-FAM at the 3' end are marked. Lane 2 corresponds to the reaction mix with the catalytic mutant of jRdRp, D668A. The wedge and the corresponding values show the increasing concentration of GTP (A) or ATP (B) in the reaction mix. (C) Graph showing the cumulative intensity of initiation products obtained in the presence of increasing concentrations of GTP and ATP. Error bars represent the standard deviation (independent assays, n = 3). (D) Primer extension assay with the jRdRp enzyme. The primer is labeled with fluorophore 6FAM at 5' end as indicated by the cartoon. Lane 1 is reaction mix without Mn2+ and Lane 2 is reaction mix with the catalytic mutant, D668A RdRp. The maximally extended primer is marked by an arrow. GTP concentration varies across the lanes as indicated by the wedge. ATP, CTP and UTP were maintained at identical concentrations of 100 µM, each. (E) Graph showing the fraction of total primer extended for different amounts of GTP and ATP. Error bars represent the standard deviation (independent assays, n = 3).
It was seen that Mn2+ ions were necessary for initiation, and no products were formed in the presence of Mg2+ ions. The requirement for Mn2+ was also observed in earlier reports on biochemical characterization of recombinant jRdRp (34,35). However, for replication complexes isolated from membranes of cells infected with JEV, it has been shown that in addition to low concentrations of Mn2+ (0.05–0.4 mM), higher concentrations of Mg2+ (2–10 mM) can also support RNA synthesis (36).
We also assessed the effect of varying concentrations of GTP on elongation of a labeled primer annealed to a template. The conditions for primer extension were standardized, and optimal activity was obtained in the presence of Mn2+ ions at 37°C. In the substrate used for the primer extension assay, the label is present at the 5′-end of the primer and the ladder-like pattern observed in the gel appears due to dissociation of the polymerase after a few rounds of replication. There was no significant qualitative or quantitative difference in the products formed in the presence of different concentrations of GTP or ATP (other NTPs were maintained at a concentration of 100 µM) (Figure 1D and E). Taken together, these experiments show that high concentration of GTP aids initiation but is not necessary for elongation.
Structure of the RdRp enzyme in jRdRpGTP and jRdRpATP
The two complexes jRdRpGTP and jRdRpATP crystallized in the space group P21212 with the cell constants of a = 144.6 Å, b = 86.6 Å, c = 112.7 Å and α = β = γ = 90° (Table 1). The jRdRpGTP complex was refined to a final Rfree of 27.6% and Rcryst of 22.0% (2.4 Å resolution). In the case of jRdRpATP, the final Rfree and Rcryst are 27.0% and 22.4%, respectively (2.3 Å resolution). The asymmetric unit contains two representatives of the jRdRpNTP complex. Each complex contains one molecule of jRdRp with residues from 274 to 315 and 321 to 889 (the regions 316–320 and 890–905 are disordered) (Figure 2A), one molecule of GTP/ATP, water molecules and two Zn2+ ions. Each Zn2+ ion is present at a distinct location away from the active site (Supplementary Figures S1 and S2). These locations are different from the ones observed for the catalytic ion (Mn2+/Mg2+) in structures of ternary complexes of other viral RdRps. Zn2+ ions in jRdRp are present at the same location as in the case of DENV RdRp (2J7W) and WNV RdRp (2HFZ).
Figure 2.
Structure of the jRdRpGTP complex. The finger, thumb and palm domains are colored yellow, orange and cyan, respectively. (A) The alpha-helices and beta-strands are numbered from the N-terminal of RdRp construct. The side chains of the active site residues (D668, D669 and D536) along with GTP are shown in stick representation. (B) The simulated annealing omit map (Fo-Fc) for GTP is displayed at a contour of σ = 3.0.
The presence and conformation of the bound GTP was confirmed through calculation of a simulated annealing Fo–Fc omit map (Figure 2B). The structure of the two RdRp molecules in the asymmetric unit is nearly identical and can be superimposed with a root-mean-square deviation (RMSD) of 0.3 Å (612 Cα pairs). The jRdRp shares a high sequence identity with DENV RdRp (66%) and WNV RdRp (84%) (Supplementary Figure S3) (27,28). The structure of RdRp adopts the right-handed topology and shows the presence of the three domains characteristic of replicative DNA and RNA polymerases, namely, the palm (368–387, 498–545, 602–716), fingers (274–367, 388–497, 546–601) and thumb (717–889) (Figure 2A). The palm domain houses the catalytic residues D668, D669 and D536. A detailed description of the jRdRp structure is provided in the Supplementary Information.
Motif F and GTP binding
The motif F in the jRdRpGTP structure presented here is ordered unlike the case of apo- structures of DENV and WNV reported before (Figure 2 and Supplementary Figure S2). On superimposition of the structure of jRdRpGTP with that of available structures of flaviviral RdRps, RMSDs of 2.7 Å, 3.8 Å and 2.1 Å were obtained with the structures of DENV apo-RdRp (562 Cα pairs; 2J7U), WNV apo-RdRp (571 Cα pairs; 2HFZ.pdb) and DENV apo-RdRp soaked with GTP (562 Cα pairs; 2J7W), respectively (28). If the motif F substructure and the loop stretching from residues 407–423 (sometimes referred to as motif G) are removed from jRdRpGTP structure, then these RMSD values come down to 1.6 Å, 1.8 Å and 1.5 Å (37). Motif F can be divided into three subparts—Motif F1: M457 to K463; F2: K469 to A470; and F3: K471 to I476. This motif is part of the longest stretch of amino acids, which is highly conserved among Flaviviruses (454–490) (Supplementary Figure S3).
The structure of the jRdRpGTP complex shows that motif F adopts a beta-hairpin substructure that is oriented toward the active site center (Figure 3A and Supplementary Figure S4A). The jRdRpATP complex also has a similar motif F substructure. The region just beyond the motif F in DENV (474–479) and WNV (475–480) forms the first turn of a helix (α10) and this helix is oriented in a direction perpendicular to the active site loop (Supplementary Figures S4B and S4C). In the case of jRdRp, the equivalent stretch (475–480) adopts an extended conformation and is part of the beta-hairpin substructure adopted by motif F.
Figure 3.
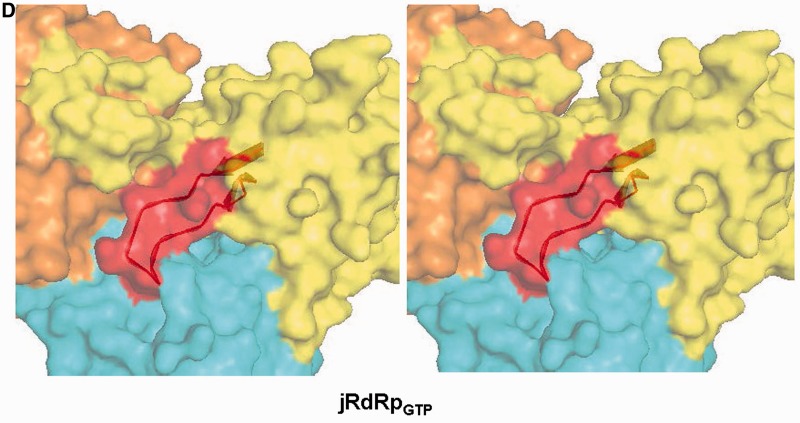
Motif F in structures of jRdRp. (A–C) Stereo images of the G loop (green) and the motif F (red) substructure of jRdRpGTP, apo- jRdRp (molecule D) and apo- NS5 from JEV (4K6M), respectively. Residues 398–526 are shown. In apo- jRdRp, the motif F and G-loop are disordered (B). In apo- NS5 (C) and jRdRpGTP structure (A), the motif F forms a beta-stranded substructure, but the loop connecting the two strands adopts distinct structures. (D) Stereo image of surface representation of the jRdRp showing the NTP tunnel in the jRdRpGTP structure. The polymerase surface corresponding to palm, fingers and thumb is colored cyan, yellow and orange, respectively. The RNA binding groove is on backside of the displayed surface. Motif F (red–ribbon and surface) occludes the NTP tunnel.
To ascertain whether motif F becomes ordered on binding GTP/ATP, we determined the structure of jRdRp in its apo-state (3.65 Å). jRdRp was crystallized in the space group P212121 with the cell constants of a = 119.5 Å, b = 174.0 Å, c = 182.7 Å and α = β = γ = 90° (Table 1). The complex was refined to a final Rfree of 27.3% and Rcryst of 23.4%. There are four molecules of RdRp in the asymmetric unit, namely, molecules A, B, C and D. In molecules A and B, motif F is ordered and has a beta-hairpin substructure similar to that in jRdRpGTP structure (Supplementary Figures S5A and S5B). In molecules A and B, the residues E466 and F467, which are at the tip of motif F loop, form interactions with thumb residues of molecules B and A, respectively. These interactions stabilize motif F in a conformation similar to that observed in the jRdRpGTP structure. In the case of molecule C, the beta-strands of the motif F are ordered, whereas the electron density for the loop (residues 460–475) connecting the two strands is weak (Supplementary Figure S5C). For molecule D, the entire motif F is disordered (Figure 3B). The average real space correlation for motif F substructure (residue 453–480) is 0.90, 0.89 and 0.78, respectively, for molecules A, B and C in apo-RdRp structure. Unlike for molecules A and B, there is no polypeptide chain adjacent to the motif F loop of molecules C and D, and consequently, the interactions formed by E466 and F467 are absent. Because there is no external stabilization of motif F in the case of molecules C and D, the disorder exhibited by the motif F in these molecules would be a closer representation of their state in solution.
The structure of JEV NS5 in the apo-state was recently reported (38). This structure (4K6M) showed an ordered motif F in a different conformation than that observed in the jRdRpGTP structure (Figure 3C). In NS5, the tip of motif F forms interactions with the Mtase domain interaction stabilizing motif F in the observed conformation (38). Additionally, the authors suggest that—as in the case of NS5 from DENV—the Mtase domain can adopt different relative orientations with respect to the RdRp domain, and therefore the observed stabilization of the motif F would be transient (39). Overall, the observations made here and the data available for JEV apo-NS5, WNV and DENV RdRp suggest that motif F substructure exhibits conformational heterogeneity in flaviviruses and can adopt multiple structures in absence of interaction with NTP.
Motif F in RdRp (apo- or ligand-bound form) from HCV, bovine viral diarrhea virus, norovirus, foot-and-mouth disease virus, poliovirus and human immunodeficiency virus -1 reverse transcriptase adopts a similar beta-stranded substructure (40) (Supplementary Figure S6). However, in the jRdRp structure, the hairpin loop within the motif F substructure points downward and obstructs the tunnel formed between the fingers and thumb domain through which NTP enters into the active site (Figure 3D). The length and composition of motif F in these enzymes is different from that of flaviviral RdRps and it does not occlude the NTP entry tunnel. Additionally, in the case of apo- jRdRp (molecules C and D) and apo- NS5 from JEV, the NTP entry tunnel is not blocked by motif F.
Interactions of jRdRp with GTP
We observed clear electron density for GTP, and in the final refined structure, the average B-factor of the nucleotide is 67 Å2 (Figure 2B and Table 1). To obtain the structure of DENV in complex with GTP, Yap and colleagues soaked apo-RdRp crystals with GTP (28). In the consequent structure, only the triphosphate moiety of the GTP could be modeled. However, in the jRdRpGTP structure, the entire GTP molecule could be placed in the observed electron density.
In the jRdRpGTP structure, the triphosphate moiety of GTP interacts with the side chains of basic residues from motif F (R460, K463, K471 and R474) and from motif E (R734 and R742) (Figure 4A and B). In a recent report, Iglesias and colleagues have shown for DENV that the residues K463 and K462 are responsible for specifically selecting viral template for initiation (41). These residues are part of motif F and K463 does interact with GTP. The residues of the priming loop (S799 side chain hydroxyl group, main chain atoms of W800 and S801) also interact with the triphosphate and sugar moieties of GTP. The conformation of the priming loop in jRdRpGTP is nearly identical to that in DENV RdRp structures (apo or GTP soaked) as this stretch superimposes with an RMSD of 0.4 Å (26 Cα atoms). In the case of apo-RdRp from WNV, the RMSD for superimposition of the priming loop with that of jRdRp is 1.6 Å (26 Cα atoms). Four residues from motif E—C714, R734, N716 and Y771—form a network of water-mediated hydrogen bonds with the gamma phosphate of GTP. From motif C, the catalytic residue D668 interacts with 2′O of the ribose sugar. D668 forms a water-mediated hydrogen bond with the N3 of the guanine base in one of the molecules in the asymmetric subunit.
Figure 4.
jRdRp-GTP interactions. The finger, thumb and palm domains are colored yellow, orange and cyan, respectively. GTP and the interacting amino acid residue are shown in stick representation. The water molecules responsible for water-mediated interactions between jRdRp and GTP are depicted in the form of red spheres. (A) The residues that interact with the triphosphate and sugar moieties of GTP are displayed. (B) The residues that form interactions with the guanine base and ribose sugar of GTP displayed. The base-specific hydrogen bonds formed between D541 and S604 with the N2 and O6 atoms of guanine are highlighted by dotted lines.
The carboxyl group of D541 (motif A) interacts with N1 (2.7 Å) and N2 (3.0 Å) of the guanine base. Also, the O6 atom of guanine base makes a H-bond (3.1 Å) with the hydroxyl group of S604 (motif B). These interactions with the N2 and O6 atoms are specific for guanine base and are not possible with the adenine base of ATP. Almost all the aforementioned residues that interact with GTP are identical in different flaviviral RdRps. The residues S799 and N716 are unique to jRdRp and replaced by threonine and histidine, respectively, in other flaviviruses (Supplementary Figure S3). Overall, the GTP molecule is stabilized in the jRdRp active site through interactions with residues of the A, B, C, E and F motifs and that of the priming loop.
jRdRpATP
The backbone structure of jRdRp when bound to GTP or ATP is nearly identical (Supplementary Figure S7A), and the two structures superimpose with an RMSD value of 0.2 (612 Cα pairs). The electron density map (simulated annealing Fo–Fc omit map) for ATP in the jRdRpATP structure was not as clear as the one observed for GTP and exhibited only partial density for the base moiety (Figure 5A). This observation suggests that the base moiety of ATP is not as well-stabilized as in the case of GTP and exhibits rotational flexibility. Comparison of the conformation of GTP and ATP in the jRdRpATP and jRdRpGTP complexes shows that the triphosphate region of ATP and GTP aligns well, but the sugar and base moieties adopt distinct conformations (Figure 5B). The residues of jRdRp that interact with the triphosphate moiety in the jRdRpATP and jRdRpGTP structures are identical and attain the same conformation. However, the residues of jRdRp that are proximal to the sugar moiety of ATP (C714, S715 and S801) are distinct from that seen in case of jRdRpGTP. Owing to weak electron density for the adenine base, it is difficult to be confident about the interactions formed between the base atoms and residues of jRdRp.
Figure 5.
Conformation of ATP in jRdRpATP. (A) The simulated annealing omit map for ATP is displayed at a contour of σ = 3.0. (B) The bound conformations of ATP and GTP are compared by superimposing the jRdRpGTP and jRdRpATP structures (RMSD = 0.2 Å, 612 C-α pairs).
The interactions formed between residues of motif F and the triphosphate moiety of ATP are similar to that seen in the case of jRdRpGTP. Consequently, the motif F in the jRdRpATP structure is ordered and exhibits a conformation similar to that seen in the case of jRdRpGTP. As in the case of jRdRpATP, the motif F occludes the NTP entry tunnel in the jRdRpATP structure also (Supplementary Figure S7B).
Diversity in conformation and binding sites of NTPs
The location and conformation of bound GTP was compared with NTP in other available RdRp–NTP complexes (Supplementary Figure S8). In the available structure of a flavivirus (DENV) RdRp in complex with GTP (2J7W.pdb), the triphosphate moiety could be modeled, but this moiety refined with high B-factors (>100 Å2). In most of the available structures of RdRp-NTP complexes, the NTP ligand refines to high B-factors and/or <1 occupancy. The triphosphate moiety shows some overlap with that of GTP in the jRdRpGTP structure (Supplementary Figure S8A) (28). In the RdRp-GTP structure (1S49.pdb) of bovine viral diarrhea virus (family: Flaviviridae; genus: Pestivirus), the base and sugar of GTP points to a different direction on comparison (Supplementary Figure S8B) (42). In the case of HCV (family: Flaviviridae; genus: Hepacivirus), multiple structures of RdRp-GTP or RdRp-UTP complex are available (43–45). In these structures, GTP and UTP bind at slightly different locations in distinct conformations that show no overlap with GTP in jRdRpGTP (Supplementary Figure S8C–S8G).
The structures of RdRp in complex with ATP (1HI1.pdb) and GTP (2JLG.pdb) for the phi6 bacteriophage (family: Cystoviridae; genus: Cystovirus) are also available (46,47). For 2JLG, GTP attains different conformations in the three RdRp molecules in the asymmetric unit. In one of the RdRp molecules in the asymmetric unit, two molecules of GTP are bound in a location and conformation similar to that observed in the full initiation complex for this enzyme (1UVN). In the other two molecules, the orientation of GTP (and that of ATP in 1HI1.pdb) is anti-parallel to the GTP conformation in jRdRpGTP (Supplementary Figure S4H). For Rotavirus (family: Reoviridae; genus: Rotavirus) RdRp the GTP molecule (2R7X.pdb) attains an inverted orientation at a location slightly displaced compared with GTP in jRdRpGTP (Supplementary Figure S4I). The GTP molecule is present in a location that overlaps with the priming loop in jRdRpGTP. In the RdRp–NTP structures from Poliovirus (family: Picornaviridae), foot and mouth disease virus (family: Picornaviridae) and norovirus (family: Caliciviridae), the conformation of the bound NTP is similar to that of the incoming nucleotide during the elongation phase (48–50). The binding site and orientation of the bound NTPs in these structures does not overlap with that of GTP in jRdRpGTP (Supplementary Figure S8J–S8L).
Overall, the GTP binding-site location and conformation in jRdRpGTP was different from that for NTP in other known structures of viral RdRp–NTP complexes. Thus, GTP binds in the active site of jRdRp in a novel orientation.
Functional relevance of jRdRpGTP structure
The structures of the initiation state of RdRp molecule from phi6 bacteriophage (1UVN.pdb) and reovirus (1N1H.pdb) are available (46,51). These structures contain the RdRp molecule bound to an RNA template, the first two 3′ nucleotides of which form base pairs with two NTPs of the nascent primer in the presence of two catalytic ions. The jRdRp structure aligns better with the phi6 RdRp structure than the reovirus RdRp enzyme. The jRdRpGTP and phi6 RdRpRNA:GTP:GTP structures were aligned through superimposition of the palm domains. As Figure 6 shows, although the palm domain and the active site motif aligns well, the GTP molecule in the jRdRpGTP structure are oriented almost perpendicular to the first two nucleotides (GTPs) present base paired to RNA in the phi6 RdRpRNA:GTP:GTP complex. We identified four residues—D541, R734, R742 and S799—that interact with the GTP in jRdRp, but the equivalent residues (D329, N317, N324 and N626, respectively) do not exhibit functional interaction with template or nucleotides in the phi6 RdRpRNA:GTP:GTP complex. The residues R734 and R742 are part of a substructure spanning from residues 733–745 in jRdRpGTP. The alignment of jRdRpGTP with phi6 RdRpRNA:GTP:GTP shows that this region will have to move away from the active site to accommodate the first incoming nucleotide of the nascent primer.
Figure 6.
Comparison of the structures of jRdRpGTP complex with that of phi6 RdRpRNA:GTP:GTP (A) The superimposition of the two structures is displayed and the C-alpha trace of jRdRp and phi6 RdRp (1UVN.pdb) is colored magenta and green, respectively. The RNA template (colored according to element with carbon atoms in yellow) and two GTP molecules (green) from phi6 RdRpRNA:GTP:GTP are displayed in stick representation. GTP in the case of jRdRpGTP is colored slate blue. (B) Superposition of the nucleotide and the RNA template is shown separately. The comparison shows that the GTP molecule in jRdRp is present in a unique conformation in a distinct binding site. In the full initiation complex of jRdRp, GTPj is expected to reorient and adopt the same conformation as GTPP2 for productive catalysis to occur. Also, ATP bound to jRdRp may reorient to adopt the conformation exhibited by GTPP1.
To ascertain whether the jRdRpGTP structure is functionally relevant, the residues D541, R734, R742 and S799 were mutated to alanine. The residue S799 was also mutated to tyrosine. We also generated single and double alanine mutants of S604 and D541—the two residues that interact with the Watson–Crick edge of the guanine base of GTP. All the mutants could be purified in soluble form and were assessed for their ability to initiate RNA synthesis in comparison with the wt enzyme. If the observed conformation of GTP in the jRdRpGTP structure represents a critical step during initiation, these mutations should perturb de novo synthesis without affecting elongation. We observed almost complete abolition of initiation for the D541A, S604A, R734A and R742A mutants (Figure 7A and B).
Figure 7.
Initiation and elongation activity of mutant and wt jRdRp. The results of primer-free initiation assay (A, B) and primer extension assay (C, D) for different mutants and wt jRdRp are displayed. The level of initiation and extension activity of different mutants of jRdRp relative to wt enzyme are shown in (B) and (D), respectively. Error bars represent the standard deviation (independent assays, n = 3). For the initiation assays (A), 6FAM-labeled UTP was used. Lane 1 in (A) shows the migration of molecular markers and the bands corresponding to 5, 12, 15 and 20mer ssRNA labeled with 6-FAM at the 3'-end are marked. For extension assays, a 20mer primer strand with a 6FAM label at the 5'-end was annealed to a complementary 40mer template strand, as indicated by the cartoon in (C). To monitor the extension reaction, a reaction mix without Mn2+ was loaded in Lane 1 of the gel displayed in (C) to visualize the unextended primer. Lane 2 in both (A) and (C) correspond to the reaction mix with the catalytic mutant of jRdRp, D668A. For the extension assay, the bands corresponding to maximally extended primer (40 nt) in (C) are marked by an arrow.
In the case of S799A and S799Y mutants, the ability to initiate RNA synthesis was enhanced with respect to the wt enzyme (Figure 7A and B). The side chain hydroxyl of S799 residue makes H-bond with the main chain nitrogen of S801 and with the side chain hydroxyl of S804, both of which will be disrupted if S799 is mutated to alanine (Supplementary Figure S9). S799, S801 and S804 residues are part of the priming loop, and this loop has to move out of the active site for the double-stranded product RNA to extrude out from the enzyme. It is possible that the priming loop structure is more mobile in the absence of interactions contributed by S799 or due to steric interactions formed by the aromatic ring of tyrosine. As a result, it might be easier for the product RNA to extrude, leading to the observed higher activity.
Primer extension assays showed that the ability of wt jRdRp, R734A, R742A, S799A and S799Y to catalyze elongation was almost identical (Figure 7C and D). In the case of the D541A and S604A mutants, the elongation activity was marginally reduced. For the double mutant S604A-D541A, there was no detectable primer extension activity. From these assays, it is evident that the mutations have a much more pronounced effect on initiation than on elongation. Therefore, the observed bound conformation of GTP is mechanistically relevant.
GTP binding reduces the affinity of jRdRp for RNA in the absence of Mn2+
We assessed binding of jRdRp to a fluorescently labeled (5′ 6FAM) ssRNA. The strength of this association was estimated by measuring change in fluorescence anisotropy values on titrating jRdRp against a fixed amount (5 nM) of 10mer ssRNA. The apparent dissociation constant (KdApp) was calculated from the binding curves (Figure 8A) assuming a single binding site (see Methods). The KdApp value for jRdRp–ssRNA interaction is 39.3 ± 2.3 nM (Hill slope, n = 1.46). Pre-incubation of jRdRp at different concentrations with a 10-fold molar excess of GTP or ATP led to a substantial drop in affinity of jRdRp for RNA (Figure 8A, Supplementary Figure S10 and Table 2). In the presence of GTP and ATP, there was marginal change in anisotropy with increasing concentrations of RdRp, and the data cannot be used to fit a binding isotherm and derive binding constants (Figure 8A). Also, for a fixed concentration of jRdRp and RNA, increasing amounts of GTP led to gradual decrease in the amount of anisotropy, suggesting that the presence of GTP promoted dissociation of RNA and jRdRp (Figure 8B). As a control, GTP was titrated in the presence of labeled RNA and in the absence of RdRp, and no significant change in fluorescence intensity was observed (Supplementary Figure S11).
Figure 8.
Affinity of jRdRp for RNA in the presence and absence of nucleotide. (A) Binding curves for interaction of labeled (6FAM at the 5'-end) ssRNA with RdRp in the absence and presence of different combinations of NTP and Mn2+. (B) The graph displays the effect of increasing concentrations of GTP (in the absence of Mn2+) on the association of jRdRp and labeled ssRNA. For both (A) and (B), the error bars represent the standard deviation (independent experiment, n = 3).
Table 2.
Dissociation constants for the interaction of jRdRp with RNA in the presence/absence of NTPs and Mn2+
| ssRNA |
|||
|---|---|---|---|
| – | Addition of GTP along with Mn2+ | Addition of ATP along with Mn2+ | |
| KdApp (nM) | 39.3 ± 2.3 | 33.9 ± 0.6 | 40.4 ± 2.1 |
| Hill slope, n | 1.46 ± 0.07 | 1.12 ± 0.02 | 1.30 ± 0.05 |
| R2 value | 0.9857 | 0.9954 | 0.9962 |
The goodness of fit for the fitted curves displayed in Supplementary Figure S10 is shown as R2 values.
The binding affinity of jRdRp toward RNA in the presence of GTP/ATP was restored when Mn2+ ion was also included in the reaction buffer (Figure 8A). In the presence of Mn2+, the KdApp values for jRdRp-ssRNA-GTP-Mn2+ and jRdRp-ssRNA-ATP-Mn2+ are 33.9 nM (Hill slope n = 1.12) and 40.4 nM (Hill slope n = 1.30), respectively (Table 2). These observations show that the presence of GTP and ATP inhibits binding of jRdRp to RNA, and the presence of divalent manganese ions abolishes this antagonistic effect. It was seen that this inhibitory effect of GTP/ATP on RNA binding was lost in the case of the R734A mutant (Supplementary Figure S12). This observation is consistent with the inference that binding of GTP/ATP in the active site of jRdRp inhibits RNA binding in the absence of the catalytic ion.
DISCUSSION
The RdRp molecule from the phi6 bacteriophage is one of the best characterized with regard to de novo RNA synthesis. This enzyme also exhibits a requirement of high GTP concentration for optimal de novo synthesis. The first two template nucleotides in three different segments (M, S and L) of the negative-stranded phi6 genome are CC, CC and CU, respectively. Also, the first two template nucleotides in the complementary positive strand for all three segments will be UC. Hence, the first or second or both incoming nucleotides will be GTP during any initiation event for replication of the phi6 genome.
In the case of phi6 RdRp, it has been shown that Mg2+ is the divalent ion required for catalysis. According to the mechanism presented by Wright and colleagues, the full catalytically competent initiation state can be formed in diverse ways either by (i) binding of template followed by binding of the two NTPs (GTPs) with Mg2+ or (ii) NTP binding in a site that is not compatible with catalysis followed by reorientation to base pair with template RNA and the subsequent entry of another NTP with Mg2+ ions or (iii) both NTPs (at sites that are compatible with catalysis) with Mg2+ ions (catalytic ion) followed by entry of template RNA (52).
In the vast majority of known flaviviruses, the terminal dinucleotide sequence CU is conserved at the 3′-ends of both positive and negative strands of the dsRF. Therefore, the requirement for high concentrations of GTP is surprising because the first incoming nucleotide would be ATP. Assuming a similar final initiation state complex in the case of flaviviruses as seen for phi6, the data presented in this study suggest the following sequence of events during initiation in flaviviruses: (i) Apo-RdRp wherein motif F can adopt a multitude of conformations (Supplementary Figure S13A). (ii) RdRp binds GTP to form pre-initiation complex with concomitant ordering of motif F. This motif now blocks the NTP tunnel. In the observed conformation, GTP binds in a site that overlaps with the predicted RNA binding region. Hence, GTP binding will prevent entry of RNA into the active site (Supplementary Figure S13B). (iii) RdRp binds RNA in the presence of Mn2+; concomitantly, the GTP nucleotide reorients to base pair with C, the second nucleotide in the template. In the case of polymerases, the triphosphate moiety of the incoming nucleotide is closely involved in the octahedral coordination of the catalytic ion along with the catalytic residues (53). Hence, it is possible that the presence of Mn2+ may trigger reorientation of GTP so that its triphosphate moiety can coordinate the catalytic ion along with the catalytic residues D668 and D536. The reoriented GTP probably attains a conformation wherein it will not interfere with RNA binding and the template RNA can enter the active site of jRdRp. The GTP molecule can now base pair with the second 3′ nucleotide C of the template. Therefore, addition of Mn2+ could abolish the inhibitory effect of GTP toward RNA binding to jRdRp. Because the residues of the motif F interact with the triphosphate moiety of GTP, the reorientation of GTP in an anti-clockwise direction will lever the outward movement of motif F. It is also possible that this reorientation will lead to a dissociation of GTP and motif F. In either case, the motif F will no longer occlude the NTP tunnel (Supplementary Figure S13C). (iv) ATP diffuses through the now open NTP tunnel and base pairs with the first nucleotide U to form the full initiation complex (Supplementary Figure S13D). Because all the components are in place now, productive catalysis to form the first phosphodiester bond can occur, leading to the synthesis of a dinucleotide primer that can be extended.
In the case of the jRdRpATP complex, ATP will reorient to pair with the first nucleotide on template RNA (Supplementary Figures S14A and S14B). Base pairing of ATP with the first nucleotide U will result in significantly less reorientation—especially for the triphosphate moiety—as compared with that in the case of GTP base pairing with the second template nucleotide (Supplementary Figure S14C). Therefore, it is possible that motif F conformation will not undergo a drastic change, and the NTP entry tunnel will still be occluded. Consequently, GTP cannot diffuse in and pair with the second nucleotide of the template. Hence, the jRdRpATP represents a dead-end complex that cannot lead to productive initiation, and this inference is supported by biochemical data (Figure 1). Overall, the second incoming nucleotide GTP has to bind first to ensure that on pairing with the second nucleotide in template RNA in the presence of Mn2+ ions, the motif F moves to open the NTP entry tunnel and allow diffusion of the first incoming nucleotide into the active site to form a full initiation complex that will lead to productive catalysis.
The proposed mechanism would prevent stabilization of two nucleotides in the RdRp active site in the absence of template RNA and thus prevent non-templated RNA synthesis. GTP binding ensures that the two nucleotides bind temporally before and after RNA binding. The proposed chain of events would avoid adventitious and wasteful RNA synthesis that could be erroneous, and thus prevent the appearance of mutations in the conserved terminal sequences.
A recent study on NS5 from DENV suggests that this enzyme has the ability to synthesize AG dinucleotide in the absence of template and primer, and the authors propose that this ability enforces conservation of the terminal sequences (54). However, the amount of dinucleotide product obtained in the presence of template was substantially higher. Hence, the mechanism of initiation proposed above could be the preferred pathway for de novo template RNA synthesis by flaviviral RdRp enzymes. Also, it is possible that RdRp enzymes from different flaviviruses use distinct pathways for initiation.
The results presented here for JEV RdRp raise the possibility that chemical inhibitors that perturb GTP binding can compromise initiation of RNA synthesis and thus reduce viral titer. Overall, the study presented here provides a basis for the specific recognition of the initiator nucleotide GTP by a flaviviral RdRp enzyme and allows formulation of a possible mechanism of initiation that prevents non-templated RNA synthesis. The observed pre-initiation state and the proposed mechanism of initiation could be the target of therapeutic intervention to combat flaviviral infections.
ACCESSION NUMBERS
4HDG 4HDH 4MTP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Department of Atomic Energy (Government of India); Ramanujan fellowship from the Department of Science and Technology (Government of India) (to D.T.N.); Data collection at the BM14 beamline of ESRF (Grenoble, France) was funded by the BM14 project- a collaboration between DBT (Government of India), EMBL and ESRF. Funding for open access charge: Ramanujan fellowship from the Department of Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Taruni Roy and Manjula Devi Rangappa for help with cloning, expression and purification of the RdRp enzyme. D.T.N. thanks the X-ray diffraction facility located in the Molecular Biophysics Unit of the Indian Institute of Science (funded by Department of Biotechnology [DBT] and Department of Science and Technology [DST], Government of India) for facilitating screening and data collection. D.T.N. acknowledges the help rendered by Dr Hassan Belrhali (ESRF) and Dr Babu Manjashetty during data collection at the BM-14 beamline of ESRF.
REFERENCES
- 1.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The Viruses and Their Replication. 5th edn. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 2.Ghosh D, Basu A. Japanese encephalitis-a pathological and clinical perspective. PLoS Negl. Trop. Dis. 2009;3:e437. doi: 10.1371/journal.pntd.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 4.Davidson AD. New insights into flavivirus nonstructural protein 5. Adv. Virus Res. 2009;74:41–101. doi: 10.1016/S0065-3527(09)74002-3. [DOI] [PubMed] [Google Scholar]
- 5.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 8.Khromykh AA, Kenney MT, Westaway EG. trans-Complementation of flavivirus RNA polymerase gene NS5 by using kunjin virus replicon-expressing BHK cells. J. Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapoor M, Zhang LW, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between Ns3 and Ns5 proteins of dengue virus type-2 in the putative RNA replicase is linked to differential phosphorylation of Ns5. J. Biol. Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 10.Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 2006;80:5451–5464. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 12.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu PW, Westaway EG. Replication strategy of kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 14.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 15.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 16.Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 17.Hagen M, Tiley L, Chung TD, Krystal M. The role of template-primer interactions in cleavage and initiation by the influenza virus polymerase. J. Gen. Virol. 1995;76:603–611. doi: 10.1099/0022-1317-76-3-603. [DOI] [PubMed] [Google Scholar]
- 18.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 19.Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J. Virol. 2003;77:8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff MP, Canard B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology. 2006;351:145–158. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 21.Ranjith-Kumar CT, Gutshall L, Kim MJ, Sarisky RT, Kao CC. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 2002;76:12526–12536. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi GH, Zhang CY, Cao S, Wu HX, Wang Y. De novo RNA synthesis by a recombinant classical swine fever virus RNA-dependent RNA polymerase. Eur. J. Biochem. 2003;270:4952–4961. doi: 10.1046/j.1432-1033.2003.03897.x. [DOI] [PubMed] [Google Scholar]
- 23.Makeyev EV, Bamford DH. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballal R, Cheema A, Ahmad W, Rosen EM, Saha T. Fluorescent oligonucleotides can serve as suitable alternatives to radiolabeled oligonucleotides. J. Biomol. Tech. 2009;20:190–194. [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta. Crystallogr. D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 27.Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, et al. Crystal structure of the RNA polymerase domain of the west nile virus non-structural protein 5. J. Biol. Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 28.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roehrl MH, Wang JY, Wagner G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 33.Lovett ST. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004;52:1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim YG, Yoo JS, Kim JH, Kim CM, Oh JW. Biochemical characterization of a recombinant Japanese encephalitis virus RNA-dependent RNA polymerase. BMC Mol. Biol. 2007;8:59. doi: 10.1186/1471-2199-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu F, Hasebe F, Inoue S, Mathenge EG, Morita K. Identification and characterization of RNA-dependent RNA polymerase activity in recombinant Japanese encephalitis virus NS5 protein. Arch. Virol. 2007;152:1859–1869. doi: 10.1007/s00705-007-1007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchil PD, Satchidanandam V. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology. 2003;307:358–371. doi: 10.1016/s0042-6822(02)00130-7. [DOI] [PubMed] [Google Scholar]
- 37.Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu G, Gong P. Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9:e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bussetta C, Choi KH. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry. 2012;51:5921–5931. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J. Virol. 2011;85:5745–5756. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi KH, Groarke JM, Young DC, Kuhn RJ, Smith JL, Pevear DC, Rossmann MG. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl Acad. Sci. USA. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Farrell D, Trowbridge R, Rowlands D, Jager J. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 2003;326:1025–1035. doi: 10.1016/s0022-2836(02)01439-0. [DOI] [PubMed] [Google Scholar]
- 44.Bressanelli S, Tomei L, Rey FA, De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, Mahias K, Rey FA, Astier-Gin T, Bressanelli S. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J. Biol. Chem. 2010;285:32906–32918. doi: 10.1074/jbc.M110.151316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 47.Poranen MM, Salgado PS, Koivunen MR, Wright S, Bamford DH, Stuart DI, Grimes JM. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl Acad. Sci. USA. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamyatkin DF, Parra F, Machin A, Grochulski P, Ng KK. Binding of 2′-amino-2′-deoxycytidine-5′-triphosphate to norovirus polymerase induces rearrangement of the active site. J. Mol. Biol. 2009;390:10–16. doi: 10.1016/j.jmb.2009.04.069. [DOI] [PubMed] [Google Scholar]
- 51.Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Wright S, Poranen MM, Bamford DH, Stuart DI, Grimes JM. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus varphi6 from de novo initiation to elongation. J. Virol. 2012;86:2837–2849. doi: 10.1128/JVI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 54.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur JJ, Canard B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012;8:e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.