Continuous signaling of CXCR4 in MCF-7 cells results in epithelial-to-mesenchymal transition (EMT), up-regulation of metastasis-associated cytokines, cell migration, and metastasis. The EMT phenotype was reversed in 3D rBM with combined inhibition of CXCR4 and CXCR2 together or in combination with MEK or PI3K, supporting development for combinational therapy treatment in breast cancer.
Abstract
Aberrant expression of CXCR4 in human breast cancer correlates with metastasis to tissues secreting CXCL12. To understand the mechanism by which CXCR4 mediates breast cancer metastasis, MCF-7 breast carcinoma cells were transduced to express wild-type CXCR4 (CXCR4WT) or constitutively active CXCR4 (CXCR4ΔCTD) and analyzed in two-dimensional (2D) cultures, three-dimensional reconstituted basement membrane (3D rBM) cultures, and mice using intravital imaging. Two-dimensional cultures of MCF-7 CXCR4ΔCTD cells, but not CXCR4WT, exhibited an epithelial-to-mesenchymal transition (EMT) characterized by up-regulation of zinc finger E box–binding homeobox 1, loss of E-cadherin, up-regulation of cadherin 11, p120 isoform switching, activation of extracellular signal-regulated kinase 1/2, and matrix metalloproteinase-2. In contrast to the 2D environment, MCF-7 CXCR4WT cells cultured in 3D rBM exhibited an EMT phenotype, accompanied by expression of CXCR2, CXCR7, CXCL1, CXCL8, CCL2, interleukin-6, and granulocyte–macrophage colony stimulating factor. Dual inhibition of CXCR2 with CXCR4, or inhibition of either receptor with inhibitors of mitogen-activated protein kinase 1 or phosphatidylinositol 3-kinase, reversed the aggressive phenotype of MCF-7 CXCR4-expressing or MDA-MB-231 cells in 3D rBM. Intravital imaging of CXCR4-expressing MCF-7 cells revealed that tumor cells migrate toward blood vessels and metastasize to lymph nodes. Thus CXCR4 can drive EMT along with an up-regulation of chemokine receptors and cytokines important in cell migration, lymphatic invasion, and tumor metastasis.
INTRODUCTION
Chemokines provide directional cues for leukocytes during migration and tissue colonization and also contribute to tumor cell metastasis. CXC chemokine receptor 4 (CXCR4), a G protein–coupled receptor that selectively binds CXC ligand 12 (CXCL12, also known as SDF-1 α), has been widely studied in breast cancer metastasis. Studies show that aberrant expression of CXCR4 by breast cancer cells facilitates metastasis to organs that secrete CXCL12, including the lung, liver, bone marrow (Muller et al., 2001; Zlotnik et al., 2011), and axillary lymph nodes (Kato et al., 2003; Kang et al., 2005b; Su et al., 2006; Klevesath et al., 2013). Although a role for CXCR4 in the growth of a primary breast carcinoma lesion is not clear, enhanced expression of CXCR4 in the early stages of breast cancer suggests a role for CXCR4 and its signaling axis in the progression to metastasis. Moreover, inhibition of CXCR4 signaling in metastatic, triple-negative MDA-MB-231 breast cancer cells reduced breast cancer metastases to the lung and lymph nodes in vivo (Liang et al., 2004, 2005). Multiple kinases contribute to CXCL12-CXCR4–induced cell migration, such as mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K), and focal adhesion kinase (Fernandis et al., 2004; Zhao et al., 2008).
Of importance, expression of CXCR4 is up-regulated in human atypical ductal hyperplasia and human ductal carcinoma in situ, correlating with poor prognosis (Schmid et al., 2004; Kang et al., 2005a; Rhodes et al., 2011b). Tumors frequently overexpress chemokines and their receptors, and this has been correlated with shortened relapse-free survival (Bieche et al., 2007; Rhodes et al., 2011a, b). High levels of CXCR4 expression in both estrogen receptor (ER)-negative and ER-positive patients predicted poorer survival (Rhodes et al., 2011b), and the CXCL12–CXCR4 axis was highly effective in enhancing cell growth and tumorigenesis (Muller et al., 2001; Ali et al., 2007; Raman et al., 2007; Zlotnik et al., 2011). Relapse of ER-negative breast cancers is associated with a Src-responsive signature (Zhang et al., 2009), which increases the survival of indolent tumor cells in the bone marrow, and this survival was mediated by CXCL12 activation of the AKT/PKB survival pathway, thus increasing resistance to TRAIL death signals (Zhang et al., 2009).
CXCR4-activating mutations involving the carboxyl-terminus have been described in warts, hypogammaglobulinemia, recurrent bacterial infection, and myelokathexis (WHIM) syndrome (Gulino, 2003; Hernandez et al., 2003; Diaz, 2005), which results in enhanced neutrophil chemotaxis (Gulino et al., 2004; Kawai et al., 2005) and a lack of desensitization after ligand stimulation (Gulino et al., 2004; Balabanian et al., 2005b). Overexpression or mutation of chemokine receptors can yield oncogene-like functions. In human cancers, CXCR4 point mutations have been identified in clinical cases of medulloblastoma (Schuller et al., 2005), as well as in colon cancer and melanoma (Ierano et al., 2009). Carboxyl-terminal truncation of CXCR4 results in an inability to desensitize the receptor after ligand stimulation, producing a constitutively active receptor (Gulino et al., 2004; Balabanian et al., 2005b; Kawai et al., 2005; Liu et al., 2012). Expression of CXCR4 with truncation of the last 35 carboxyl-terminal residues of CXCR4 amino acids (CXCR4ΔCTD) in breast cancer cells results in impaired receptor desensitization, enhanced motility, epithelial-to-mesenchymal transition (EMT), enhanced estrogen-independent growth, and metastasis in vivo (Ueda et al., 2006; Rhodes et al., 2011b).
Although correlations between CXCR4 expression and metastasis were established (Rhodes et al., 2011b; Zlotnik et al., 2011), the mechanism by which CXCR4 signaling effects both morphological and behavioral changes that are fundamental to breast cancer invasion and metastasis has not been characterized. It has been shown that recruitment of inflammatory cells to tumor sites by interaction of chemokines with chemokine receptors favors metastasis (Yang et al., 2008).
Here we show that CXCR4WT (wild type) or CXCR4∆CTD in poorly metastatic MCF-7 cells results in 1) up-regulation of genes and signaling pathways important in EMT; 2) up-regulation of factors involved in cell migration, including CXCR2 and its ligands, as well as CXCR7; and 3) secretion of cytokines interleukin-6 (IL-6), CCL2, and granulocyte–macrophage colony stimulating factor (GM-CSF), which are important in tumor metastasis. In addition, using in vivo intravital imaging, we demonstrate that MCF-7 CXCR4WT cells migrate in single-cell streams toward blood vessels, whereas MCF-7 CXCR4∆CTD cells migrate as single cells toward blood vessels in the mammary gland, which correlates with increased metastasis to lymph nodes and lungs in athymic nude mice.
RESULTS
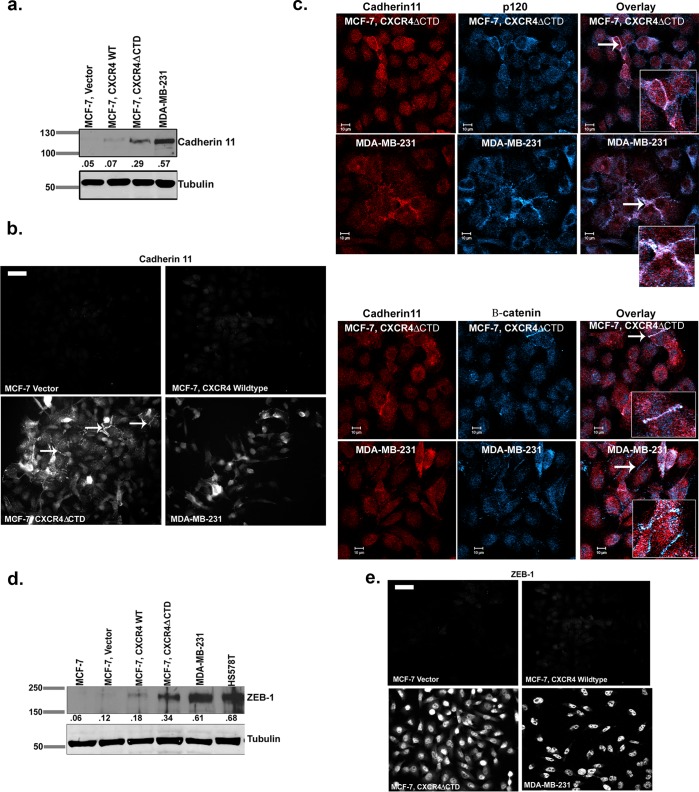
Expression of CXCR4ΔCTD in MCF-7 cells results in a switch from E-cadherin to cadherin 11, p120 isoform switching, and up-regulation of zinc finger E box–binding homeobox 1
To determine how activation of CXCR4 affected cell–cell adhesions in breast cancer cells with differing invasive properties, we expressed wild-type CXCR4 (CXCR4WT) or constitutively active CXCR4 (CXCR4ΔCTD) in MCF-7 breast cancer cells. MCF-7 vector and MCF-7 CXCR4WT cells displayed abundant cell–cell contacts and cobblestone morphology characterized by E-cadherin expression and localization to cell borders (Supplemental Figure S1, a–c). In contrast, MCF-7 CXCR4ΔCTD cells exhibited mesenchymal morphology with loss of cell contacts associated with loss of E-cadherin (Supplemental Figure S1, a–c). In contrast to E-cadherin, the mesenchymal cadherins N-cadherin and cadherin 11 were not detected in MCF-7 vector or MCF-7 CXCR4WT cells (Supplemental Figure S1b and Figure 1a). However, cadherin 11, but not N-cadherin, was expressed in MCF-7 CXCR4ΔCTD cells (Figure 1a and Supplemental Figure S1b). We used the CXCR4+, cadherin 11+, metastatic breast cancer cell line MDA-MB-231 as a control for monitoring the EMT phenotype in our assays. Consistent with these cadherin expression patterns, we found that the cadherin-associated signaling protein β-catenin was membrane localized in MCF-7 vector and MCF-7 CXCR4WT cells but was largely cytoplasmic in MCF-7 CXCR4ΔCTD cells (Supplemental Figure S1c). Western blot analysis detected two distinct β-catenin bands consistent with phosphorylated (upper band) and unphosphorylated β-catenin (Supplemental Figure S1b). Furthermore, the cadherin-associated protein p120 was primarily cytoplasmic in MCF-7 CXCR4ΔCTD cells, as compared with membrane localization of p120 in MCF-7 vector and MCF-7 CXCR4 WT cells (Supplemental Figure S1c). Analysis of p120 isoforms by Western blot revealed that the motility-associated p120 isoform 1 (Yanagisawa and Anastasiadis, 2006) was expressed in MCF-7 CXCR4ΔCTD cells, whereas the noninvasive p120 isoform 2 was expressed in MCF-7 vector and MCF-7 CXCR4WT cells (Supplemental Figure S1d). Immunofluorescence analysis of cadherin 11 in MCF-7 CXCR4ΔCTD cells showed localization at sites of cell–cell contact (white arrows, Figure 1b) and it colocalized with p120 and β-catenin at sites of cell–cell contact in MCF-7 CXCR4ΔCTD and MDA-MB-231 cells (Figure 1c). Expression of vimentin, an intermediate filament frequently expressed in mesenchymal cells, was up-regulated in both MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells compared with vector control (Supplemental Figure S1, e and f).
FIGURE 1:
Expression of CXCR4ΔCTD in MCF-7 breast carcinoma cells results in up-regulation of cadherin 11 and ZEB-1. (a) Western blot of cadherin 11 and tubulin. Densitometric scans from triplicate assays were quantitated and normalized to the loading control (tubulin). (b) Immunofluorescence staining of cadherin 11. Arrows indicate cadherin 11 localization at cell–cell contacts. Bars, 150 μm. (c) Immunofluorescence staining for colocalization of cadherin 11 and p120 and cadherin 11 and β-catenin in MCF-7 CXCR4ΔCTD and MDA-MB-231 cells. Cells were stained with mouse monoclonal anti-cadherin 11, rabbit polyclonal anti-p120, and rabbit polyclonal β-catenin antibodies and incubated with species-specific Cy3- and Cy5-conjugated secondary antibodies. Overlay images are pseudocolored; red is cadherin 11, and blue is p120 or β-catenin. Image represents a single z-section of 0.28 μm. Insets, enlarged 2× from original images. (d) Western blot of ZEB-1. Densitometric scans from triplicate assays were quantitated and normalized to the loading control (tubulin). (e) Immunofluorescence staining of ZEB-1. Bars, 150 μm.
Decreased expression of E-cadherin is frequently seen in EMT due to altered expression of transcription factors zinc finger E box–binding homeobox 1 (ZEB-1), ZEB-2, snail, twist, and slug (Yang and Weinberg, 2008). We failed to detect ZEB-2, snail, twist, and slug by quantitative real-time PCR (qRT-PCR), as their expression was below detection limits. However the protein expression of ZEB-1, a transcriptional repressor of E-cadherin, was up-regulated in MCF-7 CXCR4ΔCTD cells compared with MCF-7 vector cells (Figure 1d). Immunofluorescence showed increased nuclear localization of ZEB-1 in MCF-7 CXCR4ΔCTD cells (Figure 1e; MDA-MB-231 is a control cell line for ZEB-1). This is consistent with the idea that constitutive CXCR4 signaling maintains breast tumor cells in a more mesenchymal, single-cell, motile state through increased ZEB-1 expression.
CXCR4 signaling confers an invasive phenotype to MCF-7 cells in vitro and ex vivo
We previously showed that expression of CXCR4ΔCTD in MCF-7 cells was associated with constitutive CXCR4 activity, increased motility, and chemotaxis in the absence of CXCL12 in wound closure and chemokinesis assays (Ueda et al., 2006) and increased metastasis in vivo (Rhodes et al., 2011b). We used a Matrigel invasion assay to determine whether the CXCR4-specific inhibitor AMD3100 would impede the invasive behavior in vitro. To show that CXCR4-expressing cells respond to CXCL12 stimulation, we examined the invasiveness of cells in the presence of CXCL12 (100 ng/ml) using the Transwell (Matrigel) invasion assay with MDA-MB-231 cells as a positive control. MCF-7 CXCR4WT cells (overexpressing CXCR4) were highly invasive in response to CXCL12 (average of 150 cells/field of view, p = 0.007) compared with MCF-7 vector control (average of two cells/field of view), whereas MCF-7 CXCR4ΔCTD cells were also invasive compared with vector control (six cells/field of view, p = 0.004; Supplemental Figure S2a). Treatment with AMD3100 (20 μM for 24 h) significantly impaired invasion of MCF-7 CXCR4WT cells (31 cells/field of view, p = 0.0009) and MDA-MB-231 cells (21 cells/field of view), but did not inhibit invasiveness of MCF-7 CXCR4 ΔCTD cells (110 cells/field of view, p = 0.004; Supplemental Figure S2b). This result was expected due to the constitutive activity of CXCR4 in MCF-7 CXCR4 ΔCTD cells, which renders it ligand independent. Furthermore, AMD3100 treatment in presence of CXCL12 significantly decreased invasiveness of MCF-7 CXCR4 WT cells (27.6 cells/field of view, p = 0.0004) and MDA-MB-231 cells (49.4 cells/field of view) to CXCL12 but did not inhibit invasiveness of MCF-7 CXCR4ΔCTD cells to CXCL12 (100 cells/field of view, p = 0.001; Supplemental Figure S2c). AMD3100 treatment decreased invasiveness of MCF-7 CXCR4WT cells and MDA-MB-231 cells in presence of ligand stimulation, suggesting that CXCL12/CXCR4 signaling pathways are involved in invasion. However, due to constitutive activity of CXCR4ΔCTD, MCF-7 CXCR4ΔCTD cells were largely unresponsive to AMD3100 and exhibited high motility and invasion regardless of CXCR4 inhibition.
Targeting MAPK and PI3K pathways alters the mesenchymal properties of MCF-7 CXCR4-expressing cells and MDA-MB-231 cells in three-dimensional reconstituted basement membrane cultures
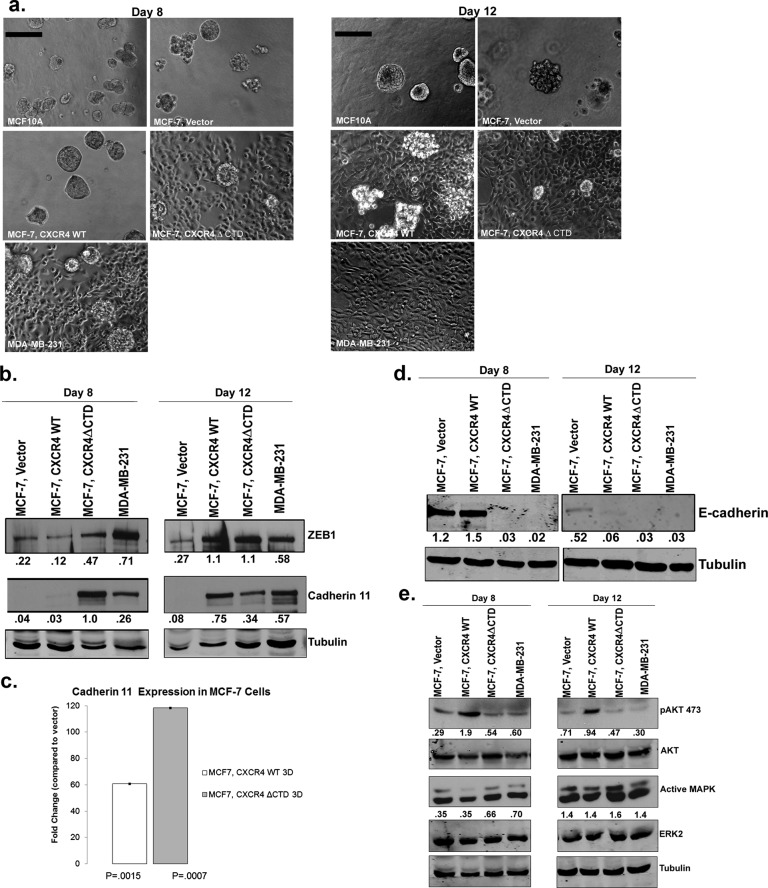
To understand how CXCR4 signaling may contribute to invasion by tumor cells, we cultured MCF-7 vector, MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells in a three-dimensional reconstituted basement membrane matrix (3D rBM; Barcellos-Hoff et al., 1989). After 8 d in culture, MCF-7 vector cells formed rounded, regular colonies similar to what was seen using the immortalized untransformed human mammary epithelial cell line MCF10A (Figure 2a). In contrast, by day 12, overexpression of CXCR4 in MCF-7 CXCR4WT cells resulted in formation of irregular projections, whereas MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells formed stellate projections, with an abundance of single-cell scattering throughout the matrix. As observed in two-dimensional (2D) culture, Western blot analysis from 3D rBM culture of MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells revealed elevated expression of cadherin 11 and ZEB-1 and p120 isoform switching compared with vector (Figure 2, b and c, and Supplemental Figure S3a). Surprisingly, by day 8 in 3D rBM culture, MCF-7 CXCR4WT cells exhibited activation of pAKT473 (Figure 2e), and by day 12, loss of E-cadherin (Figure 2d), gain of cadherin 11 and ZEB-1, and p120 isoform switching (Figure 2b and Supplemental Figure S3a). qRT-PCR analysis demonstrated that cadherin 11 was significantly up-regulated in both MCF-7 CXCR4WT cells (60-fold) and MCF-7 CXCR4ΔCTD cells (120-fold) at day 12 compared with vector control cells (Figure 2c).
FIGURE 2:
MCF-7 CXCR4ΔCTD cells exhibit a stellate phenotype and MCF-7 CXCR4WT cells form predominantly stellate structures after day 8 in 3D rBM culture. (a) Colony formation in 3D rBM culture. MCF-10A, MDA-MB-231, and MCF-7 cell lines were incubated for 8 or 12 d in 3D rBM overlay cultures. Phase contrast images. Bars, 150 μm. (b) Western blot of ZEB-1, cadherin 11, and tubulin from cells grown in 3D rBM cultures at days 8 and 12. Densitometric scans from duplicate assays were quantitated and normalized to the loading control (tubulin). (c) qRT-PCR of cadherin 11 in cells from 2D or 3D rBM cultures at day 12. qRT-PCR analysis of mRNA for cadherin 11 in MCF-7 vector control, MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells from 3D rBM cultures. Data are shown as the fold change in expression of MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells compared with vector control cells, with each gene normalized to β-actin. The difference in expression of cadherin 11 between MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells is statistically significant, based upon a 95% confidence interval. (d) Western blot of E-cadherin and tubulin from cells grown in 3D rBM cultures at days 8 and 12. Densitometric scans from duplicate assays were quantitated and normalized to the loading control (tubulin). (e) Western blot of pAKT473, total AKT, active MAPK, ERK2, and tubulin from cells grown in 3D rBM cultures at days 8 and 12. Densitometric scans from duplicate assays were quantitated, and pAKT473 was normalized to AKT, whereas active MAPK was normalized to ERK2.
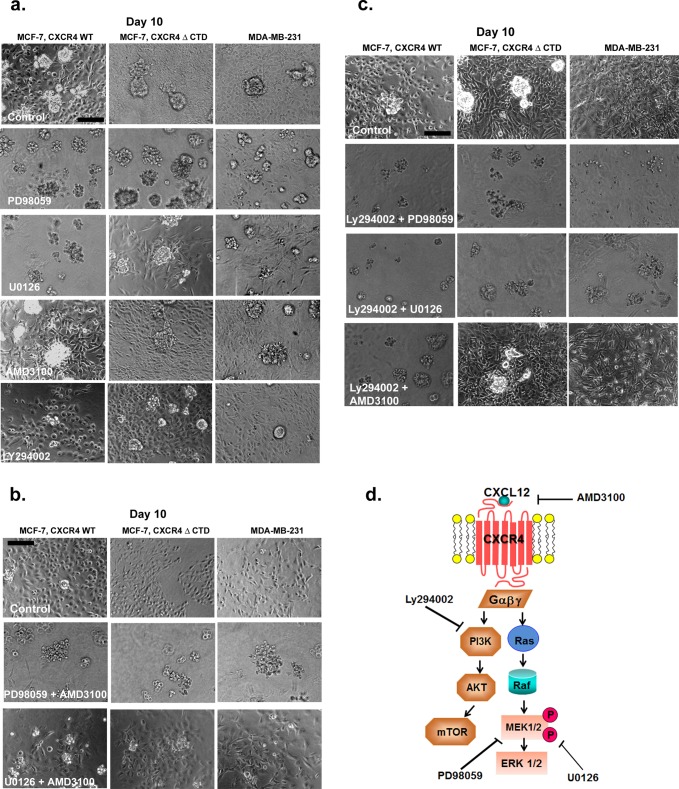
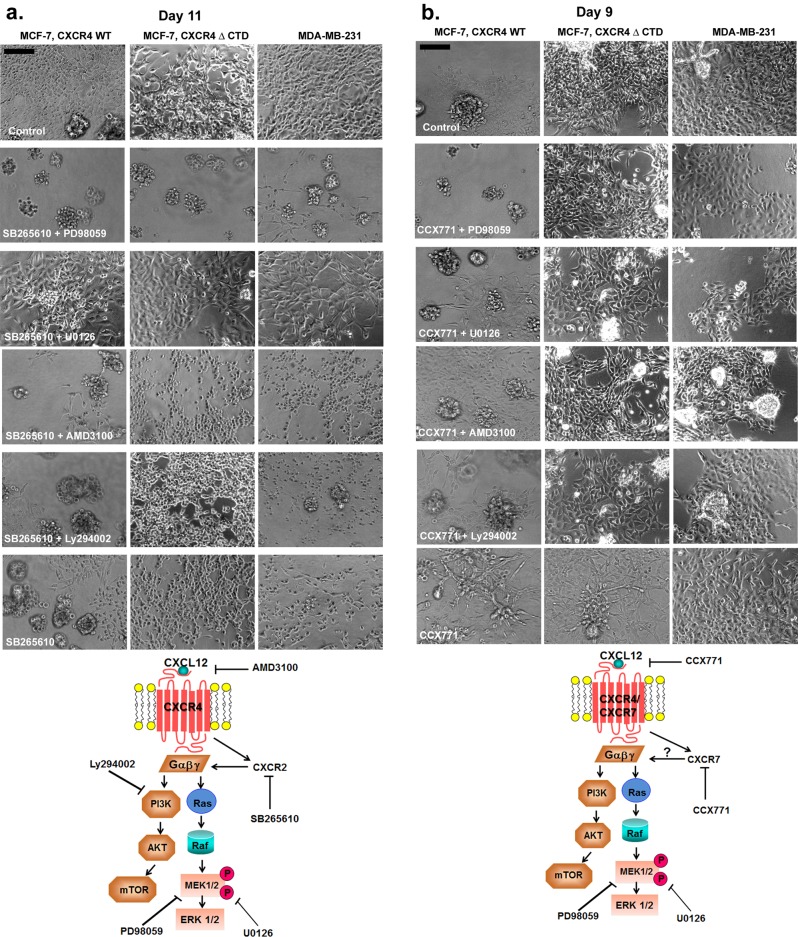
Previous studies demonstrated that MAPK signaling through the effector extracellular signal-regulated kinase 1/2 (ERK1/2) was robustly activated in response to CXCR4 signaling in breast cancer cells (Ueda et al., 2006), which we confirmed in cells grown in a monolayer in 2D culture (Supplemental Figure S3b). Treatment of MCF-7 vector and MCF-7 CXCR4WT cells with the CXCR4 ligand CXCL12 (100 ng/ml) resulted in a time-dependent increase in ERK1/2 phosphorylation (Supplemental Figure S3b), more robustly in MCF-7 CXCR4WT cells than in vector control. However, ERK1/2 phosphorylation was detected in untreated MCF-7 CXCR4ΔCTD cells at the 0-min time point (Supplemental Figure S3b). To further evaluate the biological significance of CXCR4-mediated signaling, we used the 3D rBM assay. To determine whether CXCR4-expressing cells could be induced to revert to a normal acinar phenotype in 3D rBM cultures by inhibition of CXCR4 or its signaling through the MAPK or PI3K pathways, we investigated the effects of inhibitors of CXCR4 (AMD31000), mitogen-activated protein kinase 1 (MEK1; PD98059), MEK1/2 (U0126), and PI3K (LY294002) on the morphology of cells grown in 3D rBM. To examine how treatment influenced the global composition of colonies in the population, we counted the cells of each morphology (stellate, round clusters with or without branching, grape-like clusters, or round, single cells) after treatment. Inhibition of MAPK and PI3K signaling using PD98059 (MEK1), U0126 (MEK1/2), or Ly294002 (PI3K) reduced the number of stellate cells and resulted in round, single cells or grape-like clusters by 10 d in MCF-7 CXCR4 and MDA-MB-231 cells (Figure 3a and Supplemental Figure S4, a–c, p < 0.005). These data suggest that MAPK and PI3K pathways, invoked in response to CXCR4 signaling, are required for morphological changes in response to CXCR4 signaling. However, inhibition with AMD3100 was not sufficient to normalize MCF-7 CXCR4 or MDA-MB-231 cells into a cohesive round colony structure, as cells formed predominately a mixture of round, single cells and stellate cells (Figure 3a and Supplemental Figure S4, a–c, p > 0.005).
FIGURE 3:
Effects of small-molecule inhibitors on the growth of MCF-7 and MDA-MB-231 cells in 3D rBM cultures. (a) MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells were seeded for 2 d and then incubated for 8 d in 3D rBM cultures in the presence of control (DMSO), the MEK1 inhibitor PD98059 (20 μM), the MEK1/2 inhibitor U0126 (10 μM), the CXCR4 inhibitor AMD3100 (40 μM), or the PI3K inhibitor Ly294002 (4 μM). Bars, 150 μm. (b) MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells were incubated for 8 d in 3D rBM cultures in the presence of control (DMSO), PD98059 (10 μM) and AMD3100 (20 μM), or U0126 (10 μM) and AMD3100 (20 μM). Cell lines were treated with inhibitors on day 2, and inhibitors were then added to the medium on alternate days. Phase contrast images. Bars, 150 μm. (c) MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells were incubated for 8 d in 3D rBM cultures in the presence of control (DMSO), Ly294002 (2 μM) and PD98059 (10 μM), Ly294002 (2 μM) and U0126 (10 μM), or Ly294002 (2 μM) and AMD3100 (20 μM). Cell lines were treated with inhibitors on day 2, and inhibitors were then added to the medium on alternate days. Phase contrast images. Bars, 150 μm. (d) Schematic overview of pathway inhibition.
In conclusion, inhibition of CXCR4 was not sufficient to revert the CXCR4-expressing cell lines to a less aggressive phenotype in 3D rBM cultures. However, treatment with inhibitors against MEK1/2, MEK1, or PI3K did significantly reduce the stellate phenotype to rounded, single cells or grape-like structures in MCF-7 CXCR4WT cells and round clusters with branching or grape-like structures in MCF-7 CXCR4ΔCTD cells. In addition, treatment of MDA-MB-231 cells with inhibitors MEK1 and MEK1/2, but not CXCR4 or PI3K, significantly reduced the stellate phenotype to round clusters with branching or grape-like structures. We found that treatment with U0126 (MEK1/2) and PD98059 (MEK1) inhibited MAPK activation, whereas AMD3100 (CXCR4) had no effect on MAPK activation (Supplemental Figure S3c). We infer from this result that in addition to CXCR4 signaling, MEK and PI3K pathways are engaged in the aggressive phenotype of the tumor cells. To test this theory, we treated cells with dual inhibitors of CXCR4 and MEK1 (Figure 3b); CXCR4 and MEK1/2 (Figure 3b); combination of inhibitors against PI3K and MEK1 (Figure 3c); PI3K and MEK1/2 (Figure 3c); and PI3K and CXCR4 (Figure 3c). In MCF-7 CXCR4WT cells, combination of PD98059 (MEK1) and AMD3100 (CXCR4; p = 0.0001), and U0126 (MEK1/2) with AMD3100 (CXCR4; p = 0.0001), induced reversion of the stellate phenotype to rounded, single cells and grape-like clusters (Figure 3b and Supplemental Figure S5a). In both MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells, combination of PD98059 (MEK1) with AMD3100 (CXCR4; MCF-7 CXCR4ΔCTD, p = 0.0009; MDA-MB-231, p = 0.000009) induced reversion of the stellate phenotype to rounded, single cells and grape-like clusters, whereas combination of U0126 (MEK1/2) with AMD3100 (CXCR4; MCF-7 CXCR4ΔCTD, p = 0.031; MDA-MB-231, p = 0.018) resulted in significantly fewer stellate cells compared with dimethyl sulfoxide (DMSO) control (Figure 3b and Supplemental Figure S5, b and c).
Culturing MCF-7 CXCR4 WT cells with dual inhibition of Ly294002 (PI3K) with PD98059 (MEK1; p = 0.0001), LY294002 (PI3K) with U0126 (MEK1/2; p = 0.0001), and LY294002 (PI3K) with AMD3100 (CXCR4; p = 0.0001) resulted in a loss of stellate cells (compared to DMSO control), as cells formed predominately grape-like clusters (Figure 3c and Supplemental Figure S6a). Treatment of MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells with Ly294002 (PI3K) and PD98059 (MEK1; MCF-7 CXCR4ΔCTD, p = 0.0009; MDA-MB-231, p = 0.001), and LY294002 (PI3K) with U0126 (MEK1/2; MCF-7 CXCR4ΔCTD, p = 0.0001; MDA-MB-231, p = 0.0001), resulted in a loss of stellate cells (compared to DMSO control), as cells formed predominately grape-like clusters. In contrast, treatment with LY294002 (PI3K) with AMD3100 (CXCR4; MCF-7 CXCR4ΔCTD, p = 0.258; MDA-MB-231, p = 0.017) did not revert the stellate phenotype in MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells (Figure 3c and Supplemental Figure S6, b and c).
In summary, the stellate phenotype was normalized to a less aggressive phenotype in MCF-7 CXCR4WT cells with the following combinations of inhibitors: MEK1 + CXCR4, MEK1/2 + CXCR4, MEK1 + PI3K, MEK1/2 + PI3K, and PI3K + CXCR4. In contrast, the stellate phenotype was normalized to a less aggressive phenotype in MCF-7 CXCR4ΔCTD cells and MDA-MB-231 cells with the combinations of MEK1 + CXCR4, MEK1 + PI3K, and MEK1/2 + PI3K, whereas inhibitors to MEK1/2 + CXCR4 and PI3K + CXCR4 were less successful in reverting the stellate phenotype. We suspect that signaling through the MAPK pathway (i.e., MEK1/2 to ERK1/2) in response to CXCR4 signaling drives the invasive phenotype of breast tumor cells and can be sustained in a manner that, once initiated, is independent of CXCR4 activation.
CXCR4 signaling in MCF-7 CXCR4-expressing cells results in activation of CXCR2, and targeting CXCR2 in combination with CXCR4 or MEK1 results in less aggressive structures in 3D rBM
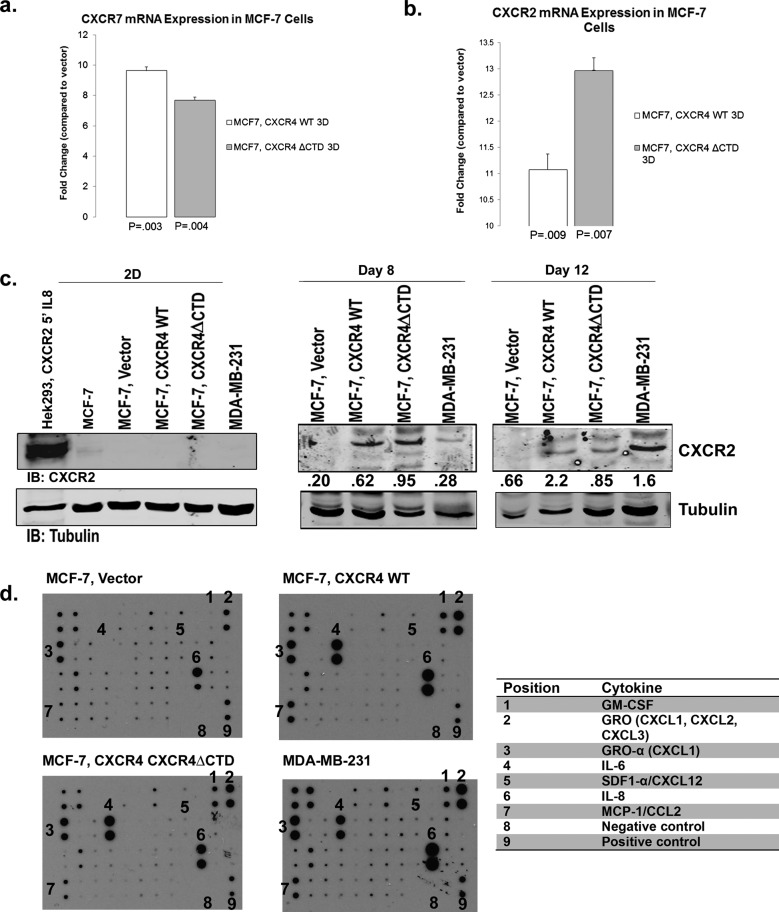
We examined the expression of other CXC receptors based on the cross-talk between ligands and receptors in this family. Using qRT-PCR analysis of RNA harvested from cells cultured in 3D rBM, we found that CXCR7 mRNA was up-regulated 8- to 10-fold in each cell type in 3D rBM compared with vector control (Figure 4a) but was not detected in 2D cultures. In addition, CXCR2 mRNA was up-regulated in MCF-7 CXCR4WT (11-fold) and MCF-7 CXCR4ΔCTD cells (13-fold) compared with vector controls (Figure 4b), whereas the mRNA was not detected in 2D culture. In addition, CXCR2 protein (Figure 4c) was not detected in MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells in 2D cultures. In 3D rBM cultures, CXCR2 protein was detected at day 8 in MCF-7 CXCR4WT and MCF-7 CXCR4 ΔCTD cells and day 12 in MDA-MB-231 cells. Using a cytokine protein array (Supplemental Table S1) with cells cultured in 3D rBM revealed that MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells produced greater amounts of CXCR2 ligands GRO (CXCL1/CXCL2/CXCL3), GROα (CXCL1), and interleukin-8 (CXCL8; Figure 4d). Although CXCR7 ligands CXCL11/ITAQ and CXCL12 (SDF-1α) were not detected, these results do not rule out the possibility that CXCR7 heterodimerized with CXCR4 to modulate its activity (Levoye et al., 2009; Luker et al., 2010; Decaillot et al., 2011). CXCR7 controls CXCL12 gradients required for migration (Dambly-Chaudiere et al., 2007; Valentin et al., 2007; Boldajipour et al., 2008), and it is possible that CXCR7 is playing a role in sequestration of CXCL12 and CXCL11 in 3D rBM cultures. The cytokine array also detected up-regulation of cytokines important in recruitment of myeloid cells to the tumor microenvironment. Interleukin-6 (IL-6), CC chemokine CCL2 (monocyte chemoattractant protein-1), and GM-CSF were detected at high levels in MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells as compared with MCF-7 vector (Figure 4d).
FIGURE 4:
CXCR7 and CXCR2 are up-regulated in MCF-7 cells expressing CXCR4WT or CXCR4∆CTD in 3D rBM cultures. qRT-PCR of CXCR7 (a) and CXCR2 (b) from cells grown in 2D or 3D rBM cultures at day 12. qRT-PCR analysis of mRNA for CXCR7 or CXCR2 in MCF-7 vector control, MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells grown in 2D cultures compared with 3D rBM cultures at day 12. Data are shown as the fold change in expression of MCF-7 CXCR4WT cells and MCF-7 CXCR4ΔCTD cells compared with vector control cells for each gene normalized to the endogenous control β-actin. The difference in expression of CXCR7 and CXCR2 between MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells is statistically significant, based upon a 95% confidence interval. (c) Western blot of CXCR2 and tubulin in cells grown in 2D cultures compared with 3D rBM cultures at days 8 and 12. Cells in 2D culture were stimulated with interleukin-8 (IL8; 100 ng/ml) for 5 min. Densitometric scans from triplicate assays in 2D culture were quantitated and normalized to the loading control (tubulin). Cells in 3D rBM culture were not stimulated with IL8. Densitometric scans from duplicate assays in 3D rBM culture were quantitated and normalized to the loading control (tubulin). (d) Cytokine array analysis of factors identified in conditioned media from 3D rBM cultures at day 8. Antibodies are immobilized on the array in duplicate, and the table lists antibody locations, including positive and negative controls.
Given the up-regulation of CXCR2 and its ligands and CXCR7 in CXCR4-expressing cells in 3D rBM cultures, we investigated the role of CXCR2 and CXCR7 in the aggressive phenotype associated with these cells. We assessed the response of MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells to CXCR2 inhibitor (SB265610) alone and in combination with CXCR4, PI3K, or MEK inhibitors in 3D rBM cultures. Because CXCR2 is up-regulated by day 8 in MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells and by day 12 in MDA-MB-231 cells, CXCR2 inhibition with SB265610 was extended to day 11 in 3D rBM culture. Treatment of MCF-7 CXCR4WT cells with CXCR2 inhibitor (SB265610, p = 0.0005) alone or in combination with inhibitors of CXCR4 (AMD3100, p = 0.0004), PI3K (Ly294002, p = 0.002), or MEK1 (PD98059, p = 0.001) significantly reduced the number of stellate cells and resulted in grape-like clusters, whereas treatment with SB265610 combined with U1026 (MEK1/2, p = 0.001) reduced the number of stellate cells but did not revert the stellate phenotype by 11 d (Figure 5a and Supplemental Figure S7a). Treatment of MCF-7 CXCR4ΔCTD cells with the CXCR2 inhibitor (SB265610, p = 0.0000003) alone or in combination with inhibitor of PI3K (Ly294002, p = 0.0000003) resulted in round, single cells, whereas CXCR2 inhibitor (SB265610) in combination with MEK1 inhibitor (PD98059, p = 0.0000003) resulted in predominately grape-like clusters. Combination of SB265610 with MEK1/2 inhibitor (U0126, p = 0.0004) reduced the number of stellate cells but did revert the stellate phenotype by 11 d (Figure 5a and Supplemental Figure S7b). Treatment of MDA-MB-231 cells with CXCR2 inhibitor (SB265610, p = 0.000005) in combination with inhibitors of CXCR4 (AMD3100, p = 0.00000002), PI3K (Ly294002, p = 0.000000002), and MEK1 (PD98059, p = 0.00003) or MEK1/2 (U0126, p = 0.0003)) significantly reduced the number of stellate cells. Moreover, CXCR2 inhibition (SB265610) in combination with MEK1 (PD98059, p = 0.00003) resulted in predominately grape-like clusters (Figure 5a and Supplemental Figure S7c). We infer from these results that CXCR4 signaling drives activation of CXCR2, and these receptors cooperate to drive EMT and invasion of breast tumor cells.
FIGURE 5:
Effects of CXCR2 or CXCR7 inhibition with the combined inhibition of CXCR4, PI3K, or MAPK on the growth of MCF-7 and MDA-MB-231 cells in 3D rBM cultures. (a) MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells were incubated for 11 d in 3D rBM cultures in the presence of control (DMSO), CXCR2 inhibitor SB265610 (1 μM) and MEK1 inhibitor PD98059 (10 μM), SB265610 (1 μM) and MEK1/2 inhibitor U0126 (10 μM), SB265610 (1 μM) and CXCR4 inhibitor AMD3100 (20 μM), SB265610 (1 μM) and PI3K inhibitor Ly294002 (2 μM), or SB265610 (1 μM) alone. Cell lines were treated with inhibitors on day 2, and inhibitors were then added to the medium on alternate days. Phase contrast images. Bars, 150 μm. (b) MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, and MDA-MB-231 cells were incubated for 9 d in 3D rBM cultures in the presence of control (DMSO), CXCR7 inhibitor CCX771 (1 μM) and PD98059 (10 μM), CCX771 (1 μM) and U0126 (10 μM), CCX771 (1 μM) and AMD3100 (20 μM), or CCX771 (1 μM) and Ly294002 (2 μM). Cell lines were treated with inhibitors on day 2, and inhibitors were then added to the medium on alternate days. Phase contrast images. Bars, 150 μm. Schematic overviews of the pathways inhibited are shown below a and b.
Treatment of MCF-7 CXCR4WT cells with CXCR7 inhibitor (CCX771, p = 0.048) alone or in combination with inhibitors of MEK1/2 (U0126, p = 0.060), CXCR4 (AMD3100, p = 636), or PI3K (Ly294002, p = 0.027) did not significantly reduce the number of stellate cells compared with DMSO treatment for 9 d (Figure 5b and Supplemental Figure S8a). However, CCX771 in combination with MEK1 (PD98059, p = 0.001) significantly reduced the number of stellate cells and resulted in round clusters with branches or grape-like clusters by 9 d (Figure 5b and Supplemental Figure S8a). Treatment of MCF-7 CXCR4ΔCTD cells with CXCR7 inhibitor (CCX771, p = 0.230) or combination of CCX771 and MEK1 (PD98059, p = 0.009), CCX771 and CXCR4 (AMD3100, p = 0.102), or CCX771 and PI3K (Ly294002, p = 0.230) did not significantly reduce the number of stellate cells compared with DMSO treatment (Figure 5b and Supplemental Figure S8b). However, CXCR7 inhibition (CCX771) in combination with MEK1/2 inhibitor (U0126, p = 0.0002) resulted in significantly fewer stellate cells and round clusters with branching (Figure 5a and Supplemental Figure S8b). Treatment of MDA-MB-231 cells with CXCR7 inhibitor (CCX771, p = 0.230) alone did not significantly reduce the number of stellate cells compared with DMSO treatment (Figure 5b and Supplemental Figure S8c). However, the combination of CCX771 with MEK1/2 (U0126, p = 0.00005), MEK1 (PD98059, p = 0.003), CXCR4 (AMD3100, p = 0.001), or PI3K (Ly294002, p = 0.00005) reduced the number of stellate cells compared with DMSO treatment but did not result in less aggressive phenotypic structures by 9 d (Figure 5b and Supplemental Figure S8c).
MCF-7 CXCR4ΔCTD cells express matrix metalloproteinase-2 and metastasize to the lymph nodes
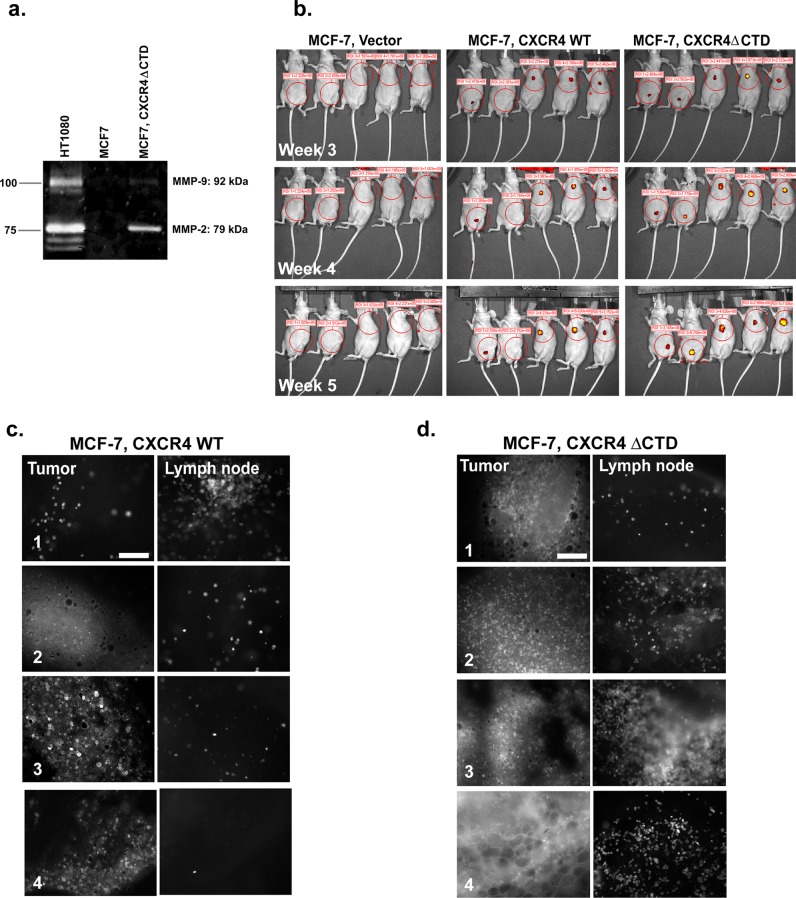
We previously demonstrated that CXCR4ΔCTD promoted metastasis of MCF-7 breast cancer cells to the lungs of tumor-bearing mice (Rhodes et al., 2011b). It has been shown that the CXCL12–CXCR4 axis is an important mediator in activation of matrix metalloproteinases (MMPs), which can facilitate metastasis by proteolyzing the extracellular matrix, thus promoting the migration of tumor cells through the basement membrane and into the lymphovascular system. CXCL12–CXCR4 signaling has been shown to induce MMP-2, MMP-9, and MMP-13 gene expression and secretion to promote the invasion of cancer cells (Fernandis et al., 2004; Samara et al., 2004; Brand et al., 2005; Tang et al., 2008; Yu et al., 2011). Using zymography to test for active MMP-2 and MMP-9, we observed that MMP-2 is up-regulated in 2D cultures of MCF-7 CXCR4ΔCTD cells compared with MCF-7 parental cells (HT1080 is a control cell line; Figure 6a).
FIGURE 6:
MCF-7 CXCR4∆CTD cells express MMP-2 and both MCF-7 CXCR4∆CTD cells and MCF-7 CXCR4WT cells exhibit lymphatic metastasis. (a) Zymographic analysis of MMP-2 and MMP-9. MCF-7 and MCF-7 CXCR4ΔCTD cells were analyzed for MMP-2 and MMP-9 expression using polyacrylamide gels embedded with 0.1 mg/ml gelatin. HT1080 cells are a positive control for MMP-2 and MMP-9 activity. (b) Optical in vivo imaging of nude mice with GFP-expressing MCF-7 vector control, GFP-MCF-7 CXCR4 WT, or GFP-MCF-7 CXCR4ΔCTD tumors at 3, 4, and 5 wk after orthotopic implantation of cells into either the third or the fourth mammary gland. Encircled areas represent regions of interest for assessment of tumor growth. (c) Representative images of primary tumors and lymph node metastases of GFP-MCF-7 CXCR4WT tumors. Fluorescence microscopy of metastases from a tumor bearing GFP-MCF-7 CXCR4WT cells at the inguinal lymph node near the tumor from mouse 1 (xenograft in the fourth mammary gland) and the axillary lymph nodes near the tumor from mice 3–5 (xenograft in the third mammary gland). Tissues from tumor and lymph nodes were dissected and examined using fluorescence microscopy. Bars, 150 μm. (d) Representative images of primary tumors and lymph node metastases of GFP-MCF-7 CXCR4ΔCTD tumors. Fluorescence microscopy of metastases from a tumor bearing GFP-MCF-7 CXCR4ΔCTD cells at the inguinal lymph node near the tumor from mice 1 and 2 (xenograft in the fourth mammary gland) and the axillary lymph nodes near the tumor from mice 3 and 4 (xenograft in the third mammary gland).
To assess the role of CXCR4 signaling in tumor metastasis in greater detail, we injected green fluorescent protein (GFP)–expressing MCF-7 vector, MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells orthotopically in athymic mice in the absence of estrogen supplementation. Tumor development was not detected in MCF-7 vector cells, whereas MCF-7 CXCR4WT and MCF-7 CXCR4ΔCTD cells formed tumors (Figure 6b), as expected from our prior data showing that expression of CXCR4WT or CXCR4ΔCTD in MCF-7 cells enables them to grow in an estrogen-independent manner (Rhodes et al., 2011b). GFP+ tumor cells were detected as early as 3 wk after inoculation (Figure 6b). At the end of week 5, the inguinal lymph node (fourth mammary gland), axillary lymph node (third mammary gland), and lungs were examined for GFP+ cells. Lungs and inguinal draining lymph nodes were examined for the presence of GFP-labeled tumor cells by epifluorescence microscopy. Although GFP+ cells were not detected in the lungs and livers of tumor-bearing mice 5 wk after tumor cell inoculation, GFP+ tumor cells were detected as single cells in draining lymph nodes of MCF-7 CXCRWT and MCF-7 CXCR4ΔCTD tumors (Figure 6, c and d). These data are consistent with the idea that CXCR4 signaling promotes metastasis of breast tumor cells.
MCF-7 CXCR4WT cells migrate in single-cell streams toward the vasculature, whereas MCF-7 CXCR4ΔCTD cells migrate as single-cell entities toward the vasculature in vivo
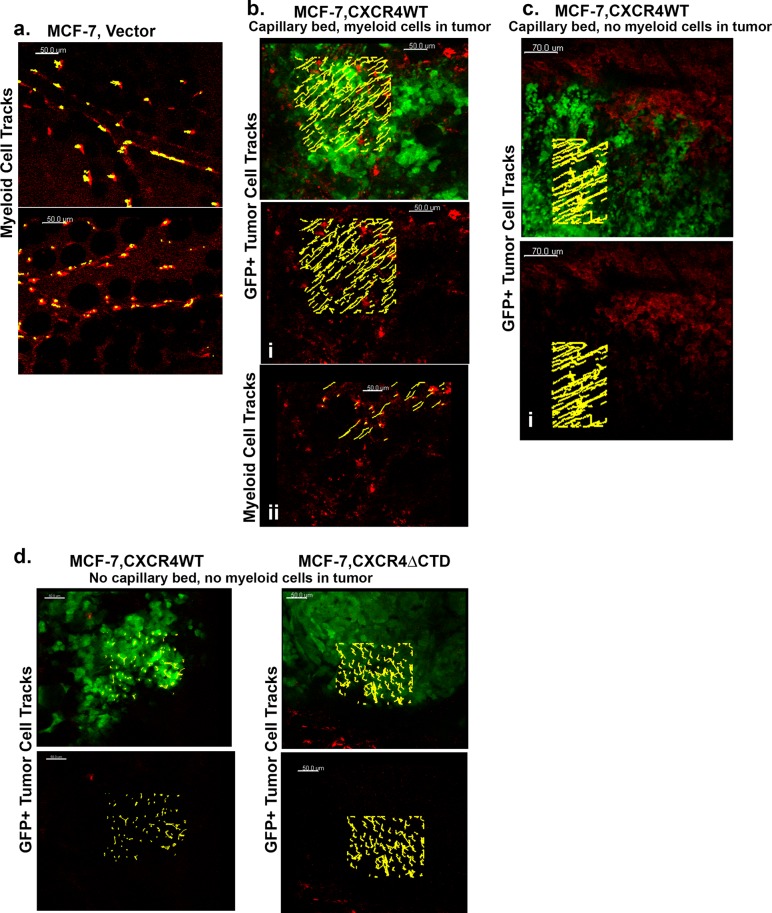
The tumor microenvironment in murine and human cancers is composed of variable numbers of tumor-infiltrating leukocytes that contribute to the metastatic properties of tumors (de Visser et al., 2006). Mammary tumor cells secrete chemokines and other factors that facilitate recruitment of leukocytes to the tumor to modify tumor growth and invasiveness (Bierie et al., 2009; Lazennec and Richmond, 2010). To examine tumor cell migration from the primary tumor to the lymph nodes in vivo with intravital imaging, we orthotopically implanted GFP-expressing MCF-7 vector, MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells in the absence of exogenous estrogen in the fourth mammary fat pad of athymic nude mice 2 wk before imaging. GFP-MCF-7 vector tumors were not detected with intravital imaging in the GFP channel in absence of exogenous estrogen (GFP and Texas red channels are shown; Figure 7a). To analyze the behavior of myeloid cells, rhodamine dextran was intravenously injected to mark the myeloid population. Intravenous injection of rhodamine dextran leaks out of blood vessels over time and is taken up by myeloid cells (Wyckoff et al., 2007). The images in Figure 7a are from two mice and demonstrate that myeloid cells (red) that ingested the rhodamine dextran were nonmigratory. We computed trajectories of individual myeloid cells (myeloid cell tracks presented in yellow) to determine average displacement over time. The trajectories (yellow) of myeloid cells (red) tracked for 20 min with Bitplane Imaris are shown and are nonmigratory (Supplemental Movies S1 and S2). GFP-MCF-7 CXCR4WT tumors were detected in the GFP channel, allowing us to image and analyze migration of tumor cells (green) and myeloid cells (red) over time. Analysis was done in two different mice in several different areas of the tumor microenvironment. Analysis in Figure 7b demonstrates GFP-MCF-7 CXCR4WT cell tracks (yellow, i, GFP channel is off) and myeloid cell tracks (yellow, ii, GFP channel is off) in a tumor with a well-vascularized capillary bed and tumor-infiltrating myeloid cells (red). Figure 7c demonstrates GFP-MCF-7 CXCR4WT cell tracks (yellow, i, GFP channel is off) in a tumor with a well-vascularized capillary bed but lacking tumor-infiltrating myeloid cells. We observed the GFP-MCF-7 CXCR4WT cells migrated in single cell streams toward blood vessels with an average speed of 1.98 μm/s in the tumor in the presence of tumor-infiltrating myeloid cells (Figure 7b), compared with the average speed of 0.98 μm/s in the absence of tumor-infiltrating myeloid cells (Figure 7c). In addition, the dextran-ingesting myeloid cells were migratory in the tumor periphery, with an average speed of 0.5 μm/s (Figure 7b). The most-displaced trajectories (yellow) of single GFP-MCF-7 CXCR4WT cells (green; Figure 7, b and c) and myeloid cells (red; Figure 7b) in the tumor tracked for 20 min are shown (Supplemental Movies S3 and S4). In contrast, GFP- MCF-7 CXCR4WT cells were nonmigratory in tumors in areas remote from the vasculature (Figure 7d), whereas GFP-MCF-7 CXCR4ΔCTD cells displayed random migration with a computed displacement of 0.78 μm/s. The most-displaced trajectories (yellow) of single GFP-MCF-7 CXCR4WT cells (green) and GFP-MCF-7 CXCR4ΔCTD cells (green) in the tumor tracked for 20 min are shown (Figure 7d and Supplemental Movies S5 and S6).
FIGURE 7:
Behavior of GFP- MCF-7 CXCR4WT and GFP- MCF-7 CXCR4ΔCTD cells in vivo. (a–d) Intravital images from a time series of GFP-MCF-7 vector, GFP-MCF-7CXCR4WT, and GFP-MCF-7 CXCR4ΔCTD cells orthotopically implanted in the absence of exogenous estrogen in the fourth mammary fat pad of athymic nude mice 2 wk before imaging. Host vasculature was labeled with 30 μl of 20 mg/ml rhodamine dextran (70 kDa), a skin flap was made to expose the mammary fat pad, and images were acquired with an LSM 510 META inverted confocal microscope with a 20×/0.75 Plan Apochromat objective. (a) GFP-MCF-7 vector tumors were not detected in mice in absence of exogenous estrogen. The GFP and Texas red channels are shown. The trajectories (yellow) of myeloid cells (red) tracked for 20 min with Bitplane Imaris are shown (Supplemental Movies S1 and S2). (b, c) MCF-7 CXCR4WT cells migrate toward blood vessels in single-cell streams. The most-displaced trajectories (yellow) of single GFP-MCF-7 CXCR4WT cells (green) and myeloid cells (red) in the tumor tracked for 20 min are shown (Supplemental Movies S3 and S4). (d) GFP-MCF-7 CXCR4WT are nonmigratory, and GFP-MCF-7 CXCR4ΔCTD cells display random migration in tumors without a vasculature. The most-displaced trajectories (yellow) of single GFP-MCF-7 CXCR4WT cells (green) and GFP-MCF-7 CXCR4ΔCTD cells (green) in the tumor tracked for 20 min are shown (Supplemental Movies S5 and S6).
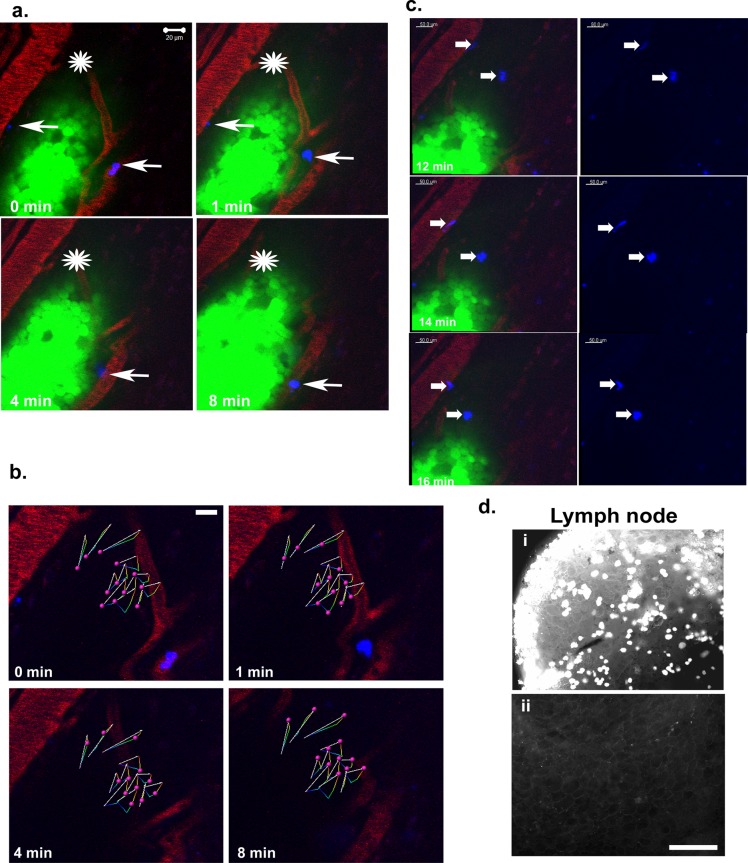
Because the production of CXCL1 and CXCL8 by CXCR4-expressing MCF-7 cells (as shown in 3D rBM; Figure 4d) can attract myeloid cells, especially neutrophils, into the tumor, we used intravital imaging to examine the interaction between leukocytes and GFP-expressing MCF-7 CXCR4ΔCTD cells. Promyelocytic HL60 cells differentiated along the neutrophil lineage (dHL60; Newburger et al., 1979) were labeled with DiIC18(5)-DS Cy5 (pseudocolored blue) and injected in the femoral vein before imaging. Time-lapse imaging revealed migration of GFP-MCF-7 CXCR4ΔCTD cells toward rhodamine-labeled vessels, with an active leading edge (Figure 8, a and b; an asterisk marks the area of reference, to act as a landmark to identify the direction of cell movement in Figure 8a; arrows point to dHL60 cells) and the presence of GFP-labeled tumor cells in draining lymph nodes (but not contralateral lymph nodes; Figure 8d). GFP-MCF-7 CXCR4ΔCTD cells migrated with an average speed of 6.63 μm/s (Figure 8b). The dHL60 cells (pseudocolored blue, average of two cells in the vasculature adjacent to the tumor) trafficked through the vasculature (red) and intravasated into the tumor microenvironment (Figure 8a). After time the dHL60 cells (average of two cells migrated in the tissue toward the tumor cells) were identified in front of the leading edge of migrating tumor cells (Figure 8c and Supplemental Movie S7). These results demonstrate that CXCR4 signaling drives invasion and motility of breast cancer cells, which may lead to heightened metastasis.
FIGURE 8:
MCF-7 CXCR4ΔCTD cells migrate toward blood vessels and metastasize to the lymph nodes. (a) Intravital images from a time series of GFP-MCF-7 CXCR4ΔCTD cells orthotopically implanted in the fourth mammary fat pad of athymic nude mice 2 wk before imaging. Differentiated HL60 cells were labeled with DiI Cy5 (blue) and injected into the vasculature via a catheter in the femoral vein. Host vasculature was labeled with 30 μl of 20 mg/ml rhodamine dextran (70 kDa), a skin flap was made to expose the mammary fat pad, and images were acquired 2 h after injection of labeled HL60 cells with an LSM 510 META inverted confocal microscope with a 40×/1.3 Plan Apochromat objective. An asterisk is placed over the area of reference to act as a landmark to identify the direction of cell movement. Differentiated HL60 cells (average of two cells in the vasculature adjacent to the tumor) labeled with DiI Cy5 (blue) are indicated by arrows. (b) The 12 most-displaced trajectories of single GFP-MCF-7 CXCR4ΔCTD cells (pink spheres) in the migrating leading edge were tracked over time with Bitplane Imaris. The arrows represent displacement of tracked cells, and colored lines are dragon tails that represent displaced trajectory of the cells tracked over time (Supplemental Movie S7). (c) Differentiated HL60 cells (average of two cells migrated in the tissue toward the tumor cells) labeled with DiI Cy5 (blue) are indicated by arrows. The dHL60 cells migrated toward the leading edge of the migrating GFP-MCF-7 CXCR4ΔCTD tumor cells over the indicated time periods. (d) GFP-MCF-7 CXCR4ΔCTD cell localization in lymph node metastasis. GFP+ tumor cells were detected in draining lymph nodes (i) but not contralateral lymph nodes (ii) of the tumor-bearing mouse. Bars, 150 μm.
DISCUSSION
The up-regulation of CXCR4 in the early stages of breast cancer supports a role for CXCR4 in tumor progression to metastasis. We demonstrate here that CXCR4 signaling within luminal mammary carcinoma cells in 3D rBM culture results in up-regulation of cytokines, chemokines, and chemokine receptors important for tumor migration and metastasis. Therefore it is imperative to determine whether therapies that inhibit chemokines will be useful for inhibiting breast cancer metastasis. MCF-7 cells that express CXCR4WT have epithelial morphology in 2D culture characteristic of luminal MCF-7 cells but in 3D rBM culture undergo transition to a mesenchymal morphology. In contrast, MCF-7 cells that express a constitutively active CXCR4 have a mesenchymal morphology characteristic of a basal-like phenotype in both 2D and 3D rBM cultures. Basal-like tumors preferentially metastasize to distant organs, such as the lung and brain (Rodriguez-Pinilla et al., 2006), so it is not surprising that MCF-7 CXCR4ΔCTD cells metastasize to the lungs (Rhodes et al., 2011b).
In this study, continuous signaling through CXCR4ΔCTD switches MCF-7 cells from an epithelial to a mesenchymal phenotype. Similarly, MCF-7 CXCR4WT cells cultured in 3D rBM exhibit an EMT phenotype characterized by up-regulation of ZEB-1, an E-cadherin repressor in breast carcinoma cells (Eger et al., 2005), loss of E-cadherin, and gain of cadherin 11. This phenotype correlates with a p120 isoform switch from isoform 2 to the invasive isoform 1, which is analogous to features in metastatic breast cancer cells. In agreement with prior reports, forced expression of E-cadherin in MCF-7 CXCR4ΔCTD cells in 2D or 3D rBM cultures did not convert the cells to an epithelial phenotype (Supplementary Figures S9 and S10; Wang et al., 2002). The observed differences between MCF-7 CXCR4WT cells in 2D and 3D rBM cultures demonstrate the importance of the 3D matrix in the integration of signals mediated by both cell-to-cell and cell-to-matrix interactions. Using 3D rBM cultures, Wang et al. (2002) demonstrated that reversion of metastatic human breast cancer cell lines to a nonmalignant phenotype requires specific pairs of inhibitors applied together, as single inhibitors induce a partial phenotypic reversion, indicating that signaling pathways require intervention at multiple sites to elicit reversion. In a 3D rBM environment, we show that combined inhibition of CXCR4 and MEK1, PI3K and MEK1, or PI3K and MEK1/2, but not combined inhibition of CXCR4 and PI3K, induced reversion of the aggressive phenotype associated with MCF-7 CXCR4ΔCTD and MDA-MB-231 cells. These data imply that CXCR4-independent activation of MEK1 or MEK1/2 works in concert with CXCR4 to drive EMT in MCF-7 cells.
The demonstration that CXCR7, a receptor for CXCL12 and CXCL11, may be involved in breast cancer (Burns et al., 2006) is intriguing, since this receptor has been postulated to be a decoy receptor (Balabanian et al., 2005a; Burns et al., 2006; Boldajipour et al., 2008). CXCR7 has a scavenging function in breast cancer cells (Luker et al., 2010) and increases their survival and adhesion but does not increase their growth (Burns et al., 2006). However, the CXCR7 antagonist CCX754 reduces tumor growth (Burns et al., 2006), and CXCR7 knockdown in breast cancer cells reduces both tumor growth and lung metastasis (Miao et al., 2007). In contrast, Hernandez et al. (2011) demonstrated that CXCR7 overexpression decreased in vivo invasion, intravasation, and metastasis of mammary adenocarcinoma cells, though it enhanced primary tumor growth and angiogenesis. Our 3D rBM studies demonstrate that, although CXCR4 expression in breast cancer cells induces expression of CXCR7, inhibition of CXCR7 with CCX771 in combination with inhibition of PI3K, MAPK, or CXCR4 did not result in less aggressive phenotype structures. The ligands for CXCR7 were not available in the 3D rBM cultures but would be in vivo, so expression of CXCR7 may affect breast cancer growth and metastasis.
Here we demonstrate that CXCR4 expression induced expression of CXCR2 and ligands for CXCR2 in 3D rBM cultures. Inhibition of CXCR2 in combination with inhibition of CXCR4 induced reversion of the stellate phenotype to a less aggressive phenotype of CXCR4-expressing cells in 3D rBM cultures. Combined inhibition of CXCR2 with MEK1/2, MEK1, or PI3K reduced the number of stellate cells, whereas CXCR2 + MEK1 resulted in grape-like structures. It is possible that CXCR2 continues to activate MEK when PI3K or CXCR4 is inhibited. Thus inhibition of MEK and CXCR4, CXCR2 and CXCR4, CXCR2 and MEK, or PI3K and MEK reversed the aggressive phenotype. Our studies show that inhibition of PI3K in combination with CXCR2 or PI3K in combination with MEK1/2 reversed the stellate phenotype of CXCR4-expressing MCF-7 cells in 3D rBM cultures.
In addition, we demonstrate that CXCR4 expression induced IL-6, CCL2, and GM-CSF expression in 3D rBM cultures. These cytokines are involved in recruitment of myeloid cells to the tumor microenvironment, tumor growth and metastasis, and disease outcome (Tamm et al., 1989; Yamashita et al., 1994; Asgeirsson et al., 1998; Arihiro et al., 2000; Qian et al., 2011; Dethlefsen et al., 2013). Stable expression of IL-6 in MCF-7 cells bestows the ability to form tumors that display loss of epithelial markers (Asgeirsson et al., 1998) and have advanced tumor grade (Asgeirsson et al., 1998; Sullivan et al., 2009). Thus, by up-regulating other cytokines and chemokines and/or their receptors, CXCR4 is able to amplify its ability to enhance invasion and metastasis of breast tumor cells.
The cause of death in cancer patients is associated with metastatic dissemination rather than growth of the primary tumor. Therefore better understanding of the chemokines and chemokine receptors involved, not only in overall metastasis, but also in site-specific metastasis, is essential. In mammary cancer, metastasizing cancer cells enter lymphatic vessels that drain the mammary gland and disseminate to other organs. In human breast cancer, CXCR4 is associated with axillary lymph node metastasis (Kato et al., 2003; Kang et al., 2005b; Su et al., 2006; Klevesath et al., 2013). Our experiments revealed that GFP+ CXCR4-expressing cells migrated to the lymph nodes as single-cell entities. Intravital imaging demonstrated that GFP-MCF-7 CXCR4WT cells migrate in single cell streams toward the vasculature and are nonmigratory in areas of the tumor that lack a blood supply. Whereas GFP-MCF-7 CXCR4ΔCTD cells migrate as single cells toward the vasculature in the tumor, they display random migration in areas of the tumor that lack a blood supply. Furthermore, CXCR4-expressing cells were highly invasive in Matrigel invasion assays. Inhibition of CXCR4 with AMD3100 significantly decreased invasion of MCF-7 CXCR4WT cells but not MCF-7 CXCR4ΔCTD cells, suggesting that signaling through constitutively active CXCR4 is ligand independent. In 3D rBM culture, CXCR4-expressing cells formed irregular stellate projections, with single-cell scattering throughout the matrix. Thus continuous activation of CXCR4 maintains MCF-7 cells in a mesenchymal state for single-cell migration and metastasis to lymph nodes and lungs. The EMT phenotype is amplified by up-regulation of CXCR2, which works in concert with CXCR4, PI3K, and MAPK activation in 3D rBM cultures to facilitate EMT.
On the basis of observations here, we give a schematic diagram to illustrate the mechanistic pathways associated with the EMT phenotype in CXCR4-expressing cell lines (Supplemental Figure S11). Our data indicate that inhibition of CXCR2 and CXCR4 together or inhibition of either receptor combined with inhibitors of MEK and/or PI3K may be useful in inhibiting the metastasis of CXCR4- and CXCR2-expressing breast cancers. PI3K inhibitors are in clinical development, with early trials ongoing for breast cancer (Miller et al., 2011), and a phase I trial with the CXCR4 inhibitor POL6326 in combination with eribulin in patients with metastatic breast cancer was to be completed in December 2013 (www.clinicaltrialsfeeds.org). Other CXCR4 antagonists are in clinical trials for cancer (Debnath et al., 2013; Domanska et al., 2013). Our data predict that combining CXCR4 antagonists with MEK inhibitors, PI3K inhibitors, or CXCR2 inhibitors may offer therapeutic advantages for CXCR4-positive breast cancer.
MATERIALS AND METHODS
Cell culture
MCF-7, HT1080, Hs578T, MDA-MB-231, HEK293, MCF10A, HL60, and Phoenix cells were obtained from the American Type Culture Collection (Manassas, VA) Collection and maintained according to the supplier's protocols. Generation of stable MCF-7 cell lines with pBMN-IRES-EGFP (vector), pBMN-CXCR4WT-IRES-EGFP, and pBMN-CXCR4ΔCTD-IRES-EGFP was previously described (Ueda et al., 2006). PCR products encoding CXCR4WT (1071 base pairs) and CXCR4ΔCTD (967 base pairs) were subcloned into the pBMN-IRES-EGFP vector, which contains a long terminal repeat (LTR) and the Psi (Ψ) consensus sequence for viral packaging. E-cadherin–expressing MCF-7 CXCR4ΔCTD cells were infected with retrovirus using conditioned media from Phoenix E-cadherin-pBMN-IRES-EGFP cells as described previously (Ireton et al., 2002). The mesenchymal phenotype of MCF-7 CXCR4ΔCTD cells was confirmed by phase contrast microscopy (Supplemental Figure S1, a and b). The triple-negative, CXCR4+ metastatic breast cancer cell line MDA-MB-231 served as control for monitoring the EMT phenotype. CXCR2-expressing HEK293 cells were generated as described previously (Yang et al., 1997).
Quantitative real-time PCR
RNA was isolated using TRIzol (Invitrogen, Grand Island, NY). cDNAs were synthesized using the iScript cDNA Synthesis RT-PCR kit (1708890; Bio-Rad, Hercules, CA), according to the user manual. cDNA was amplified by qRT-PCR using IQ Real Time Sybr Green PCR supermix (Bio-Rad). All experiments were performed in triplicate. For β-actin, E-cadherin, cadherin 11, ZEB-1, ZEB-2, TWIST-1, TWIST-2, slug, snail, vimentin, CXCR4, CXCR2, and CXCR7, primers were purchased from SA Biosciences (Frederick, MD). Data are shown as fold changes (−ΔΔCt) with 95% confidence limits compared with vector control. Each gene was normalized to β-actin control. Data were analyzed using analysis of variance (ANOVA) on −ΔCt, with individual mean comparisons of vector control, and active intervention groups were estimated using linear contrasts from the ANOVA. We consider an individual comparison statistically significant if p < 0.017 to control the experimentwise error rate at 5%.
3D morphogenesis assays
Cultures were dissociated with Accutase (Innovative Cell Technologies, San Diego, CA) and seeded on top of a thin layer of Matrigel (BD Biosciences, Bedford, MA). MCF10A cells were seeded on growth factor–reduced Matrigel, and MCF-7 vector, MCF-7 CXCR4WT, MCF-7 CXCR4ΔCTD, MDA-MB-231, and MCF-7 CXCR4ΔCTD E-cadherin cells were seeded on Matrigel. All cells were seeded at a density of 2000 cells/cm2 and overlaid with the appropriate 2% Matrigel diluted in culture medium, as described previously (Debnath et al., 2003). Cultures were monitored over time with phase contrast microscopy. Tumor cells that formed clusters from single cells are referred to as colonies. Morphology of colonies was assessed as cohesive round colonies with no branching; round colonies with chains or branching extensions of cells; single, round cells; grape-like; or stellate. The presence of five or more cellular extensions per colony was considered positive. Each cell line was plated in triplicate, and each experiment was repeated three times. Statistical significance among groups was compared using the two-sample t test. p < 0.017 was considered statistically significant following the Bonferroni correction to control the experimentwise error rate <5%.
Cytokine array from 3D rBM cultures
Cells were seeded on top of a thin layer of Matrigel as described and then serum starved overnight on day 7. Conditioned media were collected on day 8, and cell debris was removed by centrifugation. Cytokine arrays were performed according to the manufacturer's instructions (RayBiotech, Norcross, GA).
Animal studies
Balb/C nu/Foxn1 athymic nude mice at 3 wk of age were purchased from Harlan Sprague Dawley (Indianapolis, IN). The studies were approved by the Institutional Animal Care and Use Committee (IACUC) at Vanderbilt University Medical Center, and all protocols were IACUC approved. To establish orthotopic implants of mammary tumor cells in female athymic nude mice, GFP-expressing MCF-7 vector, MCF-7 CXCR4WT, or MCF-7 CXCR4ΔCTD cells were dissociated with cell-dissociation buffer (Invitrogen) and resuspended in 30 µl of sterile saline solution, and 5 × 105 cells were injected subcutaneously in the fourth mammary gland of mice. For optical in vivo imaging of mammary tumors, mice were anesthetized with 2.5% isoflurane for fluorescence imaging in the IVIS 200 imaging system (Xenogen Corp., Alameda, CA) with GFP filter at 1.5-cm depth and 1-s exposure.
For confocal intravital imaging of nude mice with mammary implants 2 wk after implantation of GFP-MCF-7 vector, GFP-MCF-7 CXCR4WT, and MCF-7 CXCR4ΔCTD cells, mice were anesthetized with 2.5% isofluorane, and a skin flap was made to expose the mammary fat pad. Host vasculature was labeled with 30 µl of 20 mg/ml rhodamine dextran (70 kDa; Molecular Probes, Eugene, OR) via tail vein injection. Images were acquired with an LSM 510 META inverted confocal microscope with a 10×/0.30 Plan Neofluar objective or 20×/0.75 Plan Apochromat objective. Time-lapse images were taken every 10 s for 20 min. In another experiment, 2 wk after MCF-7 CXCR4ΔCTD cells were implanted in nude mice, HL60 cells differentiated along a neutrophil lineage (dHL60) were labeled with 2 µM DiIC18(5)-DS Cy5 (Molecular Probes), and 1 × 106 cells were injected via a catheter in the femoral vein. Host vasculature was labeled with 30 µl of 20 mg/ml rhodamine dextran (70 kDa) via tail vein injection. After injection of labeled dHL60 cells, images were acquired with an LSM 510 META inverted confocal microscope with a 40×/1.3 Plan Apochromat objective. Time-lapse images were taken every 10 s for 20 min. The number of disseminating GFP-expressing CXCR4ΔCTD MCF-7 cells was determined after the mice were killed. Inguinal lymph nodes from the tumor-bearing and contralateral sides of the mouse were dissected and examined whole with an epifluorescence microscope to detect GFP-positive cells.
Image analysis and cell tracking of GFP-expressing MCF-7 CXCR4ΔCTD and MCF-7 CXCR4 WT cells in vivo
The data set was analyzed with Bitplane Imaris and corrected for respiratory motion artifacts with drift correction and spot detection at a minimum diameter of 8 µm and tracking algorithm (autoregressive motion) with a maximum radius of 50 µm. The image set was acquired using a region of interest for which cells from the migrating leading edge were tracked. This more accurately covers one microenvironment than the entire image field. Cell displacement was measured for comparing single-cell migration, since cells rarely migrate in straight lines.
The Supplemental Methods describes reagents and established procedures for Western blot, invasion assays, immunofluorescence, and zymography.
Supplementary Material
Acknowledgments
We are grateful to Yukiko Ueda, Jim Wahl, Albert Reynolds, Andries Zijlstra, and Sarah Kurley for cells, reagents, clones, and technical assistance with 3D rBM cultures. We thank ChemoCentryx (Mountain View, CA) for the CXCR7 inhibitor, CCX771. We thank Linda Horton and Krystle Fordyce for technical assistance, lab colleagues for helpful discussions of this project, and members of the Vanderbilt Cell Imaging Shared Resource for assistance with confocal microscopy and Bitplane Imaris software. We are grateful to Charles Manning and the Vanderbilt University Institute of Imaging Science and the Center for Small Animal Imaging for assistance in optical in vivo imaging. We also thank Barbara Fingleton for critical editorial comments regarding the manuscript. This work was funded by a VA Career Scientist award to A.R., National Cancer Institute Grant CA34590 to A.R., Vanderbilt-Ingram Cancer Center Support Grant CA68485, the Ingram Professorship to A.R., Vascular Biology Training Grant T32-HL0775 (P. Bock, PI) to T.S., and American Cancer Society Postdoctoral Award PF-11-092-01-CSM to T.S.
Abbreviations used:
- 2D
two-dimensional
- 3D rBM
three-dimensional reconstituted basement membrane
- EMT
epithelial–mesenchymal transition
- ERK1/2
extracellular signal-regulated kinase 1/2
- GFP
green fluorescent protein
- GM-CSF
granulocyte–macrophage colony stimulating factor
- IL-6
interleukin-6
- MEK1
mitogen-activated protein kinase 1
- MMP
matrix metalloproteinase
- PI3K
phosphatidylinositol 3-kinase
- WT
wild type
- ZEB-1
zinc finger E box–binding homeobox 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0360) on January 8, 2014.
T.S., R.C., and A.R. designed the experiments and wrote the manuscript: S.W. designed and consulted for imaging studies; T.S. and Y.S. performed experiments; T.S. analyzed data; and G.A. performed statistical analysis.
The authors declare no conflicts of interest.
REFERENCES
- Ali S, O'Boyle G, Mellor P, Kirby JA. An apparent paradox: chemokine receptor agonists can be used for anti-inflammatory therapy. Mol Immunol. 2007;44:1477–1482. doi: 10.1016/j.molimm.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Arihiro K, Oda H, Kaneko M, Inai K. Cytokines facilitate chemotactic motility of breast carcinoma cells. Breast Cancer. 2000;7:221–230. doi: 10.1007/BF02967464. [DOI] [PubMed] [Google Scholar]
- Asgeirsson KS, Olafsdottir K, Jonasson JG, Ogmundsdottir HM. The effects of IL-6 on cell adhesion and e-cadherin expression in breast cancer. Cytokine. 1998;10:720–728. doi: 10.1006/cyto.1998.0349. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005a;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Balabanian K, et al. WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood. 2005b;105:2449–2457. doi: 10.1182/blood-2004-06-2289. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Chavey C, Andrieu C, Busson M, Vacher S, Le Corre L, Guinebretiere JM, Burlinchon S, Lidereau R, Lazennec G. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr Relat Cancer. 2007;14:1039–1052. doi: 10.1677/erc.1.01301. [DOI] [PubMed] [Google Scholar]
- Bierie B, Chung CH, Parker JS, Stover DG, Cheng N, Chytil A, Aakre M, Shyr Y, Moses HL. Abrogation of TGF-beta signaling enhances chemokine production and correlates with prognosis in human breast cancer. J Clin Invest. 2009;119:1571–1582. doi: 10.1172/JCI37480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Brand S, Dambacher J, Beigel F, Olszak T, Diebold J, Otte JM, Goke B, Eichhorst ST. CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp Cell Res. 2005;310:117–130. doi: 10.1016/j.yexcr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Burns JM, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath B, Xu S, Grande F, Garofalo A, Neamati N. Small molecule inhibitors of CXCR4. Theranostics. 2013;3:47–75. doi: 10.7150/thno.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Decaillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem. 2011;286:32188–32197. doi: 10.1074/jbc.M111.277038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen C, Hojfeldt G, Hojman P. The role of intratumoral and systemic IL-6 in breast cancer. Breast Cancer Res Treat. 2013;138:657–664. doi: 10.1007/s10549-013-2488-z. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- Diaz GA. CXCR4 mutations in WHIM syndrome: a misguided immune system. Immunol Rev. 2005;203:235–243. doi: 10.1111/j.0105-2896.2005.00226.x. [DOI] [PubMed] [Google Scholar]
- Domanska UM, Kruizinga RC, Nagengast WB, Timmer-Bosscha H, Huls G, de Vries EG, Walenkamp AM. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur J Cancer. 2013;49:219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- Fernandis AZ, Prasad A, Band H, Klosel R, Ganju RK. Regulation of CXCR4-mediated chemotaxis and chemoinvasion of breast cancer cells. Oncogene. 2004;23:157–167. doi: 10.1038/sj.onc.1206910. [DOI] [PubMed] [Google Scholar]
- Gulino AV. WHIM syndrome: a genetic disorder of leukocyte trafficking. Curr Opin Allergy Clin Immunol. 2003;3:443–450. doi: 10.1097/00130832-200312000-00005. [DOI] [PubMed] [Google Scholar]
- Gulino AV, et al. Altered leukocyte response to CXCL12 in patients with warts hypogammaglobulinemia, infections, myelokathexis (WHIM) syndrome. Blood. 2004;104:444–452. doi: 10.1182/blood-2003-10-3532. [DOI] [PubMed] [Google Scholar]
- Hernandez L, Magalhaes MA, Coniglio SJ, Condeelis JS, Segall JE. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011;13:R128. doi: 10.1186/bcr3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez PA, Gorlin RJ, Lukens JN, Taniuchi S, Bohinjec J, Francois F, Klotman ME, Diaz GA. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- Ierano C, et al. A point mutation (G574A) in the chemokine receptor CXCR4 detected in human cancer cells enhances migration. Cell Cycle. 2009;8:1228–1237. doi: 10.4161/cc.8.8.8250. [DOI] [PubMed] [Google Scholar]
- Ireton RC, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Mansel RE, Jiang WG. Genetic manipulation of stromal cell-derived factor-1 attests the pivotal role of the autocrine SDF-1-CXCR4 pathway in the aggressiveness of breast cancer cells. Int J Oncol. 2005a;26:1429–1434. [PubMed] [Google Scholar]
- Kang H, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. The elevated level of CXCR4 is correlated with nodal metastasis of human breast cancer. Breast. 2005b;14:360–367. doi: 10.1016/j.breast.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Kato M, Kitayama J, Kazama S, Nagawa H. Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res. 2003;5:R144–150. doi: 10.1186/bcr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Choi U, Whiting-Theobald NL, Linton GF, Brenner S, Sechler JM, Murphy PM, Malech HL. Enhanced function with decreased internalization of carboxy-terminus truncated CXCR4 responsible for WHIM syndrome. Exp Hematol. 2005;33:460–468. doi: 10.1016/j.exphem.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Klevesath MB, Pantel K, Agbaje O, Provenzano E, Wishart GC, Gough P, Pinder SE, Duffy S, Purushotham AD. Patterns of metastatic spread in early breast cancer. Breast. 2013;22:449–454. doi: 10.1016/j.breast.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- Liang Z, Wu T, Lou H, Yu X, Taichman RS, Lau SK, Nie S, Umbreit J, Shim H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64:4302–4308. doi: 10.1158/0008-5472.CAN-03-3958. [DOI] [PubMed] [Google Scholar]
- Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, et al. WHIM syndrome caused by a single amino acid substitution in the carboxy-tail of chemokine receptor CXCR4. Blood 120, 181–189. 2012. [DOI] [PMC free article] [PubMed]
- Luker KE, Steele JM, Mihalko LA, Ray P, Luker GD. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci USA. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13:224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Newburger PE, Chovaniec ME, Greenberger JS, Cohen HJ. Functional changes in human leukemic cell line HL-60. A model for myeloid differentiation. J Cell Biol. 1979;82:315–322. doi: 10.1083/jcb.82.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Cancer Lett. 2007;256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes LV, et al. Effects of SDF-1-CXCR4 signaling on microRNA expression and tumorigenesis in estrogen receptor-alpha (ER-alpha)-positive breast cancer cells. Exp Cell Res. 2011a;317:2573–2581. doi: 10.1016/j.yexcr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes LV, et al. Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011b;71:603–613. doi: 10.1158/0008-5472.CAN-10-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pinilla SM, Sarrio D, Honrado E, Hardisson D, Calero F, Benitez J, Palacios J. Prognostic significance of basal-like phenotype and fascin expression in node-negative invasive breast carcinomas. Clin Cancer Res. 2006;12:1533–1539. doi: 10.1158/1078-0432.CCR-05-2281. [DOI] [PubMed] [Google Scholar]
- Samara GJ, Lawrence DM, Chiarelli CJ, Valentino MD, Lyubsky S, Zucker S, Vaday GG. CXCR4-mediated adhesion and MMP-9 secretion in head and neck squamous cell carcinoma. Cancer Lett. 2004;214:231–241. doi: 10.1016/j.canlet.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Schmid BC, Rudas M, Rezniczek GA, Leodolter S, Zeillinger R. CXCR4 is expressed in ductal carcinoma in situ of the breast and in atypical ductal hyperplasia. Breast Cancer Res Treat. 2004;84:247–250. doi: 10.1023/B:BREA.0000019962.18922.87. [DOI] [PubMed] [Google Scholar]
- Schuller U, Koch A, Hartmann W, Garre ML, Goodyer CG, Cama A, Sorensen N, Wiestler OD, Pietsch T. Subtype-specific expression and genetic alterations of the chemokinereceptor gene CXCR4 in medulloblastomas. Int J Cancer. 2005;117:82–89. doi: 10.1002/ijc.21116. [DOI] [PubMed] [Google Scholar]
- Su YC, Wu MT, Huang CJ, Hou MF, Yang SF, Chai CY. Expression of CXCR4 is associated with axillary lymph node status in patients with early breast cancer. Breast. 2006;15:533–539. doi: 10.1016/j.breast.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28:2940–2947. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Cardinale I, Krueger J, Murphy JS, May LT, Sehgal PB. Interleukin 6 decreases cell-cell association and increases motility of ductal breast carcinoma cells. J Exp Med. 1989;170:1649–1669. doi: 10.1084/jem.170.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CH, Tan TW, Fu WM, Yang RS. Involvement of matrix metalloproteinase-9 in stromal cell-derived factor-1/CXCR4 pathway of lung cancer metastasis. Carcinogenesis. 2008;29:35–43. doi: 10.1093/carcin/bgm220. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Neel NF, Schutyser E, Raman D, Richmond A. Deletion of the COOH-terminal domain of CXC chemokine receptor 4 leads to the down-regulation of cell-to-cell contact, enhanced motility and proliferation in breast carcinoma cells. Cancer Res. 2006;66:5665–5675. doi: 10.1158/0008-5472.CAN-05-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Wang F, Hansen RK, Radisky D, Yoneda T, Barcellos-Hoff MH, Petersen OW, Turley EA, Bissell MJ. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- Yamashita J, Hideshima T, Shirakusa T, Ogawa M. Primary tumor levels of interleukin-6 in relation to tumor burden in human breast-cancer. Oncol Rep. 1994;1:1185–1187. doi: 10.3892/or.1.6.1185. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Anastasiadis PZ. p120 catenin is essential for mesenchymal cadherin-mediated regulation of cell motility and invasiveness. J Cell Biol. 2006;174:1087–1096. doi: 10.1083/jcb.200605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Schraw WP, Mueller SG, Richmond A. Interruption of G protein-coupling in CXCR2 does not alter ligand binding, but eliminates ligand-activation of GTPgamma35S binding, calcium mobilization, and chemotaxis. Biochemistry. 1997;36:15193–15200. doi: 10.1021/bi971594u. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yu T, Wu Y, Helman JI, Wen Y, Wang C, Li L. CXCR4 promotes oral squamous cell carcinoma migration and invasion through inducing expression of MMP-9 and MMP-13 via the ERK signaling pathway. Mol Cancer Res. 2011;9:161–172. doi: 10.1158/1541-7786.MCR-10-0386. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Mueller BM, DiScipio RG, Schraufstatter IU. Akt plays an important role in breast cancer cell chemotaxis to CXCL12. Breast Cancer Res Treat. 2008;110:211–222. doi: 10.1007/s10549-007-9712-7. [DOI] [PubMed] [Google Scholar]
- Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.