The Sec14-like phosphatidylinositol transfer protein Sfh3 associates with bulk LDs in vegetative cells but targets to a neutral lipid hydrolase-rich LD pool during sporulation. Sfh3 inhibits LD utilization by a PtdIns-4-phosphate–dependent mechanism, and this inhibition prevents prospore membrane biogenesis in sporulating cells.
Abstract
Lipid droplet (LD) utilization is an important cellular activity that regulates energy balance and release of lipid second messengers. Because fatty acids exhibit both beneficial and toxic properties, their release from LDs must be controlled. Here we demonstrate that yeast Sfh3, an unusual Sec14-like phosphatidylinositol transfer protein, is an LD-associated protein that inhibits lipid mobilization from these particles. We further document a complex biochemical diversification of LDs during sporulation in which Sfh3 and select other LD proteins redistribute into discrete LD subpopulations. The data show that Sfh3 modulates the efficiency with which a neutral lipid hydrolase-rich LD subclass is consumed during biogenesis of specialized membrane envelopes that package replicated haploid meiotic genomes. These results present novel insights into the interface between phosphoinositide signaling and developmental regulation of LD metabolism and unveil meiosis-specific aspects of Sfh3 (and phosphoinositide) biology that are invisible to contemporary haploid-centric cell biological, proteomic, and functional genomics approaches.
INTRODUCTION
Lipid droplets (LDs) are important energy-storage organelles in eukaryotic cells. These particles are composed of a neutral lipid core consisting primarily of triacylglycerides (TAGs) and sterol esters (SEs) surrounded by a phospholipid monolayer and a coat of associated proteins (Murphy and Vance, 1999). The unilocular LD, a hallmark feature of human white adipocytes, occupies up to 90% of cell volume (Pilch et al., 2007), and regulation of LD metabolism is an important factor in obesity and its attendant health problems, such as type II diabetes and cardiovascular disease. LDs also exhibit remarkable functional diversity. For example, LDs contribute to cell cycle control (Kurat et al., 2009), act as temporary storage depots for unfolded membrane proteins (Welte, 2007), and act as sites of ecosanoid production in macrophages (Silva et al., 2009; Weibel et al., 2009). LD utilization is also key to productive infection by pathogenic agents such as Chlamydia and hepatitis C virus (Kumar et al., 2006; Ogawa et al., 2009). An understanding of the complex regulation of how lipid-based chemical energy is stored and used in cells requires detailed understanding of how LD metabolism is controlled.
Current models suggest that LDs form in discrete regions of the endoplasmic reticulum (ER) via aggregation of neutral lipids between the two phospholipid leaflets of the ER bilayer. This progressive coalescence drives budding of mature LDs from ER membranes (Martin and Parton, 2006; Wolins et al., 2006). Evidence in support of this mechanism includes the ER residence of enzymes that catalyze the final steps of TAG and SE synthesis (e.g., diacylglycerol acyltransferases and acyl-CoA:cholesterol acyltransferases; Yen et al., 2008; Chang et al., 2009) and the bidirectional trafficking of proteins between LDs and the ER (Zehmer et al., 2009; Jacquier et al., 2011). Finally, high-resolution imaging experiments reveal that yeast LDs remain tethered to the ER, further arguing for an ER origin of LDs (Szymanski et al., 2007).
Although the enzymology of neutral lipid synthesis is well studied and conserved from yeast to humans (Turkish and Sturley, 2007), rather less is known about the pathway of LD assembly or control of LD homeostasis (Adeyo et al., 2011). The most detailed information regarding control of LD metabolism is culled from the white adipocyte system, in which the major regulatory factor is perilipin (Plin 1), the founding member of the mammalian perilipin, ADRP, TIP47 (PAT) protein family. Plin 1 is a peripheral LD protein that serves dual roles in regulating LD homeostasis. Under conditions of basal lipolysis, Plin 1 restricts lipase accessibility to LD lipids (Londos et al., 1999). In response to hormonal stimulation, Plin 1 is phosphorylated by protein kinase A and activates TAG lipase and hormone-sensitive lipase (Sztalryd et al., 2003; Tansey et al., 2003; Subramanian et al., 2004; Londos et al., 2005; Moore et al., 2005; Lass et al., 2006; Zimmermann et al., 2009). Rather less is known about the other PAT family members, Plin 2 (ADRP), Plin 3 (TIP47), Plin 4 (S3-12), and Plin 5 (OXPAT; Brasaemle, 2007; Bickel et al., 2009; Kimmel et al., 2010), and Plin-independent mechanisms for regulating LD metabolism also exist. Drosophila mutants lacking all Plins exhibit abnormal body fat distribution yet display surprisingly functional body fat regulation (Beller et al., 2010). Moreover, no Plin family members have been described in Caenorhabditis elegans or Saccharomyces cerevisiae. How do these organisms regulate LD production and consumption? Cyclin-dependent kinases play a role in controlling LD metabolism (Kurat et al., 2009), but little is known about how lipid signaling interfaces with this process.
Here we describe a novel link between phosphoinositide signaling and LD metabolism. This discovery was prompted by functional analyses of Sfh3, a nonclassical member of the Sec14-like phosphatidylinositol transfer protein (PITP) superfamily whose members help specify the outcomes of phosphoinositide signaling in cells (Schaaf et al., 2008; Bankaitis et al., 2009). We report that Sfh3 associates with bulk LDs in vegetative cells but targets to a neutral lipid hydrolase-rich LD pool during a developmentally regulated program of meiotic membrane biogenesis. Furthermore, we demonstrate that Sfh3 is an inhibitor of LD utilization, that this LD homeostatic activity of Sfh3 operates via a phosphatidylinositol (PtdIns)-binding– and PtdIns-4-phosphate (PtdIns-4-P)–dependent mechanism, and that Sfh3-mediated regulation of LD metabolism is of developmental consequence. These results provide new insights into how a Sec14-like PITP couples phosphoinositide signaling with LD homeostasis. The data also reveal unexpectedly complex heterogeneities in the LD cohort of yeast diploid cells engaged in a developmental program of nutrient stress–induced membrane biogenesis.
RESULTS
Sfh3 is a nonconventional Sec14-like PITP
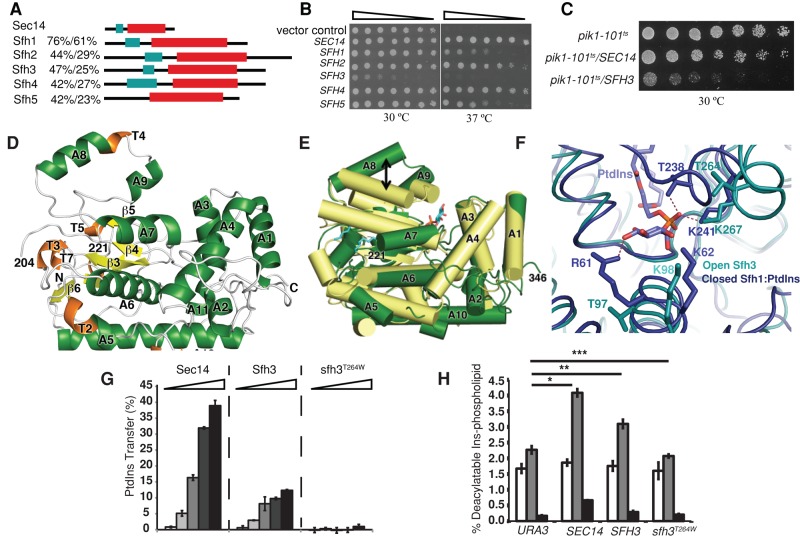
To assess functional similarities between Sec14 and its yeast homologues, we overexpressed Sfh1–-Sfh5 in sec14ts yeast and compared their abilities to rescue sec14ts lethality at nonpermissive temperature. Sfh3 exhibited particularly unusual behavior in this assay, in that its enhanced expression failed to rescue growth of yeast at 37ºC. Indeed, elevated Sfh3 expression (Sfh3OE) was strongly deleterious to proliferation of sec14ts yeast at normally permissive temperatures (30ºC; Figure 1B), even though Sfh3OE exerted only very modest effects on growth of wild-type (WT) yeast at 30 or 37°C (unpublished data). That the deleterious effects were related to phosphoinositide signaling is supported by our observation that pik1ts yeast compromised for activity of the trans-Golgi network (TGN)/endosomal PtdIns 4-OH kinase (Pik1) were similarly sensitive to Sfh3OE (Figure 1C). These in vivo data demonstrate that Sfh3 antagonized Sec14/Pik1 signaling in vivo.
FIGURE 1:
Sfh3 exhibits unique functional properties. (A) Domain organization of Sec14 and Sfh proteins. Primary sequence comparisons (similarity/identity) and domain organizations (N-terminal tripod domain, green box; lipid-binding domain, red box). (B) Sfh3 overexpression fails to rescue the growth of sec14-1ts mutant. Equivalent numbers of yeast sec14-1ts cells expressing the designated genes from episomal vectors were spotted in threefold dilution series onto SD agar and incubated at the permissive and restrictive temperatures of 30 and 37°C, respectively. Images were taken after 48-h incubations. (C) Sfh3 overexpression inhibits the growth of temperature sensitive pik1-101ts at permissive temperature of 30°C. Same amount of pik1-101ts cells containing indicated genes on multicopy plasmids were spotted in twofold dilution series on SD agar and incubated at 30°C for 48 h before images were taken. (D–F) Structural characterization of Sfh3. (D) Ribbon diagram of the Sfh3 crystal structure with α-helices in green, 310 helices in orange, and β-strands in yellow. (E) Superposition of Sfh3 (green) on Sfh1 (gold). Helices are shown as solid rods. Movement of gating helix A8 between open Sfh3 and closed Sfh1 conformers is designated by the arrow. (F) The PtdIns (magenta) binding pocket in Sfh1 (cyan) is superposed onto the corresponding residues in Sfh3 (green). Residues within 4.2 Å of the PtdIns headgroup are shown in stick representation. (G) Sfh3 phospholipid-transfer activities. Purified recombinant Sec14, Sfh3, and Sfh3T264W were assayed for PtdIns-transfer activity in a 0.004-, 0.2-, 1-, 5-, and 25-μg step series of protein. Average values and SD (n = 4). (H) Sfh3 potentiates PtdIns-4-P production in vivo. Strain CTY303 (sec14∆cki1∆) carrying the indicated genes on yeast episomal expression vectors was radiolabeled to steady state with [3H]inositol. Deacylated phosphoinositides were resolved and quantified (PtdIns-3-P, open bars; PtdIns-4-P, gray bars; PtdIns-4,5-P2, black bars). Average values and SD (n = 4). Data derived from PtdIns-4-P levels of URA3 plasmid control and SEC14, URA3 plasmid control, and SFH3, URA3 plasmid control and sfh3T264W were compared by t test: *p = 0.000797; **p = 0.009545; ***p = 0.300888.
To gain insight into the functional differences between Sec14 and Sfh3, we solved a high-resolution Sfh3 crystal structure. Gel filtration and equilibrium sedimentation analyses demonstrated that recombinant Sfh3 (expected Mr = 40 kDa) purified as a dimer (Supplemental Figure S1, A and B). Diffraction-quality crystals of native and selenomethionine-substituted Sfh3 were produced and the structure solved to 1.93-Å resolution with excellent canonical properties (Table 1; Protein Data Bank ID [PDB] 4M8Z). In spite of the relatively low sequence similarity, Sfh3 adopts a Sec14 fold consisting of 11 α-helices, 7 310-helices and 6 β-strands (Figure 1D). Superposition of the Sfh3 structure onto that of holo-Sfh1 bound either to PtdIns or PtdCho displays a root-mean-square deviation of 3.9 Å over 189 equivalent atoms. The Sfh3 gating helix A8 is displaced ∼15.6 Å from the position of the cognate Sfh1 structural unit (Figure 1E), indicating that Sfh3 crystallized in a “open” conformation analogous to that reported for the phospholipid-free apo-Sec14.
TABLE 1:
Data collection and refinement statistics.
| SeMet-Sfh3 | |
|---|---|
| SAD wavelength, Å | 0.97820 |
| Resolution, Å | 1.93 (2.0– 1.93) |
| Space group | P212121 |
| Unit cell dimensions, Å | a = 52.5, b = 114.7, c = 144.7, α = β = γ = 90.0° |
| Number of reflections | 338,750 |
| Rsym (highest shell),a % | 7.3 (28.3) |
| Completeness (highest shell), % | 99.9 (99.9) |
| Average redundancy (highest shell), % | 5.1 (5.0) |
| I/σ | 28.9 (6.0) |
| Monomers per asymmetric unit (AU) | 2 |
| Number of protein atoms/AU | 2605 |
| Number of waters/AU | 194 |
| Rworking (Rfree),b % | 18.9 (22.4) |
| Average B factors, Å2 | |
| Protein | 19.5 |
| Water | 23.9 |
| Root-mean-square deviations | |
| Bond lengths, Å | 0.019 |
| Bond angles, deg | 1.6 |
Parentheses indicate highest shell.
aRsym = Σ |I − 〈I〉|/Σ |I|, where I is the observed intensity and <I> is the average intensity of several symmetry-related observations.
bRworking = Σ ||Fo| − |Fc||/ Σ |Fo| and Rfree = Σ ||Fo| − |Fc||/ Σ |Fo| for 7% of the data not used at any stage of the structural refinement, where Fo and Fc are the observed and calculated structure factors, respectively.
Superposition of Sec14/Sfh1 PtdIns-binding module onto the Sfh3 structure showed that the PtdIns-binding strategy is conserved in Sfh3. Accordingly, we generated the sfh3T264W mutant specifically defective in PtdIns binding due to a steric incompatibility between the bulky Trp side chain and the PtdIns headgroup phosphate (Figure 1F). Sfh3 is an active PtdIns-transfer protein in vitro, with a specific PtdIns-transfer activity ∼30% that of Sec14, and, as expected, sfh3T264W was strongly defective in PtdIns-transfer activity (Figure 1G). The mutant sfh3T264W is otherwise well behaved, as it 1) was stably expressed both as a bacterial recombinant protein and in yeast; 2) like Sfh3, purifies cleanly as a dimer (Supplemental Figure S1C); and 3) is shown by circular dichroism analyses to be well structured, with no indication of being prone to malfolding (Supplemental Figure S1, D and E).
Like Sec14, Sfh3 potentiated PtdIns-4-P production in vivo. This property was assessed in a sec14Δ cki1Δ ‘bypass Sec14’ strain, which maintains basal phosphoinositide mass as a result of absence of Sec14. The major PtdIns and phosphoinositide species were measured upon reconstitution of Sec14, Sfh3, or sfh3T264W expression in the sec14Δ cki1Δ strain, and PtdIns-4-P levels were elevated approximately twofold relative to basal control by Sec14 expression (Figure 1H). By comparison, reconstitution of the system with Sfh3OE evoked an ∼1.5-fold increase in bulk PtdIns-4-P relative to basal controls. Basal PtdIns-4-P levels were indifferent to sfh3T264WOE (Figure 1H), and sfh3T264WOE had no effect on growth of sec14-1ts yeast (unpublished data). We thereby consider sfh3T264W OE to be a functional null.
Novel features of the Sfh3 fold
Whereas the core fold is conserved between Sec14 and Sfh3, the “open” structures differed in several major respects (Supplemental Figures S2 and S3). These differences are detailed in the Supplemental Text. Four highlights are summarized here. First, the string motif lies behind the β-sheet floor of the lipid-binding pockets of Sec14-like proteins, and this substructure both reinforces the floor of the phospholipid-binding pocket and harbors critical components of the conformational switch elements that gate access to the binding pocket. The Sfh3 string motif was extended relative to Sec14/Sfh1 by a β-strand (B6) and two helices (Supplemental Figure S2A). Second, interpretable electron density for the N-terminal half of helix A7 (210VPGNSKIP217) was missing from the Sfh3 electron density map (Supplemental Figure S2B). In apo-Sec14 and holo-Sfh1 structures, the corresponding motif is well ordered and produces a pronounced bend in the cognate helix (Sha et al., 1998; Schaaf et al., 2008). The presumed conformational flexibility of the 210VPGNSKIP217 motif rendered the Sfh3 lipid-binding pocket wider and shallower than those of the PtdIns/PtdCho-binding proteins Sec14 and Sfh1 (Supplemental Figure S2B). Third, electrostatic charge distribution across the Sfh3 lipid-binding pocket exhibited a polarity reverse to those of the Sec14 and Sfh1 lipid-binding pockets (Supplemental Figure S2C). Thus the Sfh3 lipid-binding pocket defines a chemical environment radically different from the pocket chemical environments of the classic Sec14 and Sfh1 PITPs. Finally, alignment of 66 fungal Sfh3 orthologues demonstrated that PITPs of this class unanimously diverge from Sec14 orthologues in the PtdCho headgroup coordinating region, accounting for why Sfh3 is not a PtdCho-transfer protein (Supplemental Figure S3).
Sfh3 associates with LDs
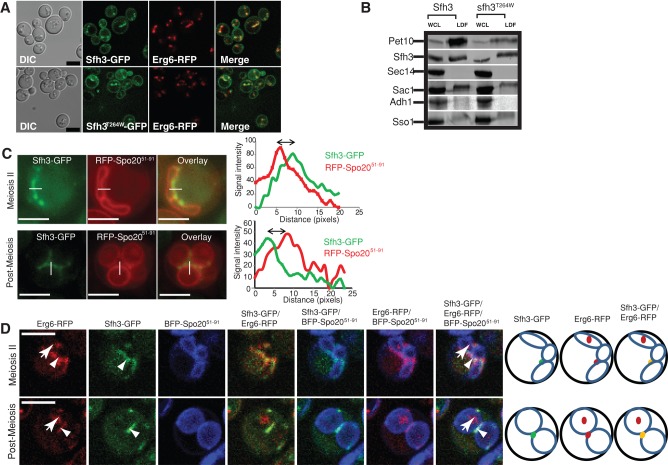
The striking functional differences between Sfh3 and Sec14 indicate that the functional coupling of these PITPs is very different, even though these proteins share similar structural folds and both stimulate PtdIns-4-P synthesis in vivo. In that regard, Sfh3 was previously annotated as an LD-associated protein based on imaging experiments using Sfh3-EGFP and Nile red fluorescence pairs (Schnabl et al., 2003). Confident assignment of those punctate structures as LDs using Nile red as LD marker is problematic since Nile red and enhanced green fluorescent protein (EGFP) both excite at 488 nm and exhibit emission overlap (Greenspan et al., 1985; Wolinski and Kohlwein, 2008). However, we found that Sfh3-GFP and the Erg6-RFP LD marker protein colocalized in living yeast, with an Sfh3-GFP pool also detected on plasma membrane and/or peripheral ER (Figure 2A). The sfh3T264W-GFP PtdIns-binding mutant similarly homed to LDs. Biochemical experiments supported the imaging data, as both Sfh3 and sfh3T264W fractionated with highly purified LDs (Figure 2B). We therefore conclude that Sfh3 is a bona fide LD protein. Using Pet10, an LD marker protein (Wang and Lee, 2012), to normalize the data, we found that sfh3T264W showed a reproducible 1.5-fold enrichment in LD fractions relative to Sfh3. That cytosolic pools were observed for both Sfh3 and sfh3T264W suggests that both proteins engage in dynamic associations with LDs and that PtdIns-binding does not play a determining role in controlling Sfh3 association with, or dissociation from, LD surfaces.
FIGURE 2:
Sfh3 is an LD-associated protein. (A) Yeast expressing Sfh3-GFP or sfh3T264W-GFP and Erg6-RFP from their endogenous promoters were cultured to logarithmic growth phase in minimum medium. Fluorescence images are shown along with their corresponding DIC images. Scale bar, 5 μm. (B) Purified LD fractions were prepared from sfh3Δ strains carrying SFH3 or sfh3T264W CEN expression plasmids, as indicated. Pet10, Sfh3, Sec14, Adh1, Sso1, and Sac1 were visualized by immunoblotting using specific polyclonal antibodies against each protein. Equal cell equivalents of whole-cell lysate (WCL) and purified LD fraction were loaded for each individual query protein blot. Purified LD fractions loads had ∼10 cell equivalents per 1 equivalent in the WCL fraction in each protein query blot. In all cases, Sec14 served as dual TGN protein and PITP control. Adh1 served as a cytosolic control. Sso1 and Sac1 served as plasma membrane and ER controls, respectively. (C, D) Sfh3 associates with a specific LD population during meiosis. (C) Diploid cells expressing Sfh3-GFP and the red fluorescence PSM marker RFP-Spo2051-91 were examined during and after completion of meiosis II. Right, line scans for both Sfh3-GFP and RFP-Spo2051-91 signal intensities across the PSM. Sfh3-GFP labeled LDs align exclusively along the ascal side of the expanding PSM, and the distance offset between Sfh3-GFP and RFP-Spo2051-91 peaks (indicated by the double-headed arrow) is 280 ± 61 nm. (D) Cells expressing Sfh3-GFP, Erg6-RFP, and the PSM marker mTagBFP-Spo2051-91 were examined during meiosis II (top) and after its completion (bottom). Also shown are overlays between the various signals. Arrowheads identify LDs labeled with both Sfh3-GFP and Erg6-RFP. Those LDs associate with the ascal side of PSM. Arrows identify LDs labeled with Erg6-RFP but not Sfh3-GFP. These droplets are inside the lumen of the PSM-limited compartment. Right, cartoon representations of signals identified by the arrows or arrowheads. Green dots represent Sfh3-GFP signal; red dots represent Erg6-RFP signals; yellow dots represent the colocalization between Sfh3-GFP and Erg6-RFP. PSMs are shown as blue oblongs (during meiosis II) and blue circles (completion of meiosis II).
Developmental regulation of Sfh3 targeting to LDs
Although Sfh3 associated with all LDs in haploid (see earlier discussion) and vegetative diploid (unpublished data) cells, different results were obtained when Sfh3 distribution was examined in sporulating diploids. Sporulation is a natural developmental response of yeast to nutrient starvation. During sporulation, four haploid nuclei are produced within the mother cell cytoplasm by meiosis. Each nucleus is packaged within a novel envelope: the prospore membrane (PSM). Formation of the PSM requires execution of a high-capacity program for internal membrane synthesis, one that demands reconfiguration of membrane trafficking through the secretory pathway and is essential for production of viable spores (Neiman, 1998). PSM biogenesis further requires developmentally regulated hydrolysis of PtdCho to PtdOH catalyzed by phospholipase D (Rose et al., 1995; Rudge et al., 1998).
In sporulating yeast, Sfh3 localized to a series of tightly clustered LDs distributed along the ascal side of PSMs labeled with RFP-Spo2051-91 (Nakanishi et al., 2007; Figure 2C). This distribution was reminiscent of electron microscope studies that documented a close apposition of LDs along the forming PSM in sporulating cells (Lynn and Magee, 1970). Triple-label imaging experiments demonstrated that, in contrast to vegetative cells (Figure 2A), Sfh3 targeted to a specific subset of Erg6-red fluorescent protein (RFP)–marked LDs that were closely associated with the PSM (Figure 2D). As was the case in vegetative cells, the LD-resident Erg6-RFP associated with all LDs in sporulating yeast (Figure 3A).
FIGURE 3:
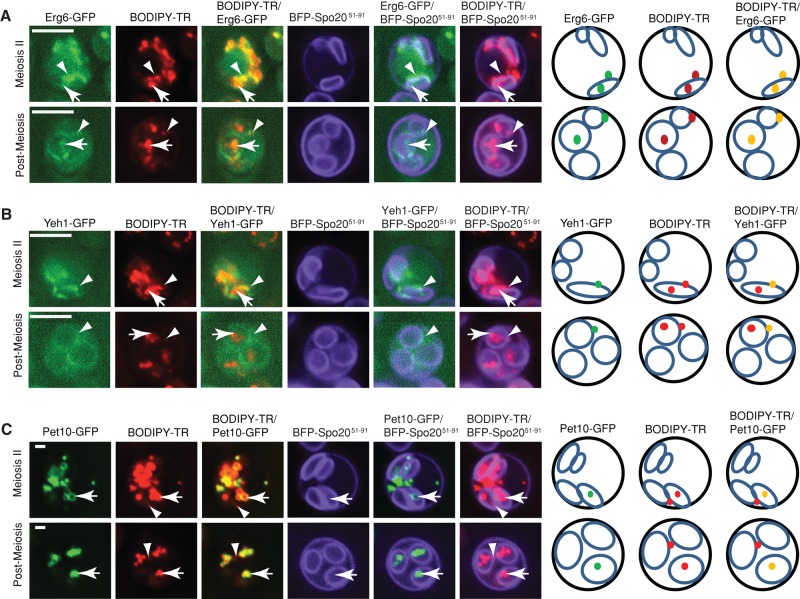
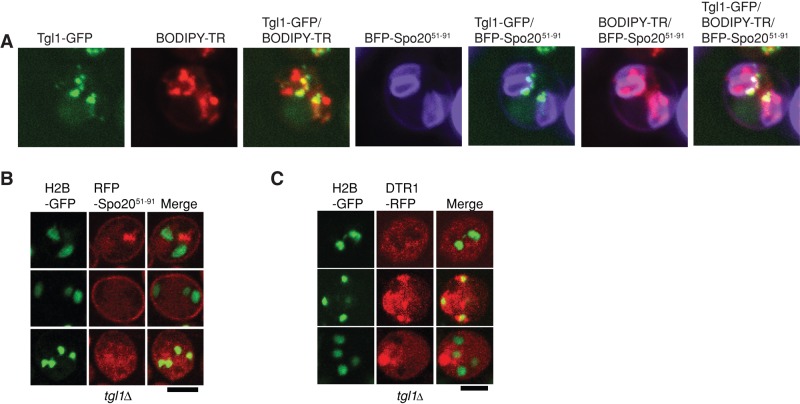
LD heterogeneity during sporulation. (A) LDs were stained with BODIPY-TR. PSM was labeled with mTagBFP-Spo2051-91. LD proteins were tagged with GFP. Arrows highlight GFP protein localized within the PSM, and arrowheads highlight GFP protein associated with the ascal side of the PSM. Right, cartoon representations of signals identified by the arrows or arrowheads. Green dots represent GFP signal; red dots represent LDs stained with BODUPY-TR; yellow dots represent the merged signal. PSMs are shown as blue oblongs (during meiosis II) and blue circles (completion of meiosis II). (A) Erg6-GFP associates with all the LDs in sporulating yeast. (B) Yeh1-GFP–labeled LDs associate with the ascal side of PSM. (C) Pet10-GFP LDs distribute to the space enclosed by the PSM.
To further characterize the unexpected LD heterogeneity in meiotic cells, we examined the localization profiles of the other major LD resident proteins during meiosis II and in postmeiotic cells. Again, the PSMs were labeled with a fluorescent marker, whereas LDs were specifically stained with BODIPY-TR methyl ester. Three distinct localization patterns were discerned in these analyses. Class I LD proteins (Erg6, Tgl3, Bsc2, Fat1, Tgl5) showed extensive overlap with the general LD stain during meiosis II, and class 1 LD proteins partitioned both to spore and ascal cytoplasms in postmeiotic cells (Figure 3A). Class II LD proteins (Sfh3, Yeh1, Tgl4, Rrt8, Tgl1) were selectively concentrated on a PSM-associated LD pool during meiosis II and were excluded from the spore cytoplasm in postmeiotic cells (Figure 3B). Class III LD proteins (Pet10, Erg7) displayed a profile inverse to that of Sfh3—that is, localization to an LD pool not consistently associated with the PSM during meiosis II and entirely contained within the spore cytoplasm in postmeiotic cells (Figure 3C). Thus, our results identified at least two subpools of LDs and three classes of LD proteins in yeast undergoing meiosis. One subpool, marked by Sfh3, associated specifically with the ascal face of the forming PSM. The second, marked by Pet10, was inherited by the spores. Every LD protein analyzed here decorated all LDs in vegetative cells (unpublished data).
Sfh3 inhibits spore formation
To determine whether Sfh3 activity modulates the sporulation program, we constructed isogenic homozygous diploid strains expressing variable levels of Sfh3. These diploids were first evaluated for sporulation competence after 5 d of nutrient deprivation. Both WT and sfh3Δ diploids exhibited excellent sporulation frequencies, and sfh3Δ diploids were modestly, but reproducibly, more efficient in this regard (Supplemental Table S1). Sfh3OE diploids were severely sporulation deficient, however, with only a minor fraction of cells presenting morphologically recognizable spores. Sfh3OE-dependent inhibition of sporulation required Sfh3 PtdIns-binding activity, as sfh3T264WOE diploids exhibited wild-type sporulation frequencies.
To identify the stage at which the sporulation program fails in Sfh3OE diploids, we compared the efficiencies for completing meiotic divisions with sporulation efficiencies in end-point imaging assays after 24 and 48 h of nutrient deprivation. Completion of meiosis II in sporulating was diagnosed by formation of four haploid nuclei. WT diploids exhibited a consistent lag between completion of meiosis II and formation of visible spores (10.7 ± 1.2 vs. 7.0 ± 0.5% at 24 h and 21.2 ± 2.8 vs. 14.0 ± 1.8% at 48 h; Supplemental Table S2). sfh3Δ diploids demonstrated no such time lag. This difference between completion of meiosis II and efficiency of spore formation was strongly increased in Sfh3OE diploids, with 4.9 ± 0.8% versus 0.9 ± 0.1% at 24 h and 7.5 ± 1.8% versus 2.0 ± 0.4% at 48 h.
Sfh3 inhibits PSM biogenesis
The inability of Sfh3OE diploids to form morphologically recognizable spores upon completion of meiosis II suggested failure in construction of the PSMs that package each haploid nucleus, a failure made more interesting by the specific targeting of Sfh3 to PSM-associated LDs. PSM biogenesis requires developmentally regulated hydrolysis of PtdCho to PtdOH catalyzed by phospholipase D (Rose et al., 1995; Rudge et al., 1998). Thus, formation of this structure was evaluated both by Nomarski (differential interface contrast [DIC]) optics and by monitoring intracellular distribution of the RFP-Spo2051-91 PtdOH biosensor. In vegetative cells, RFP-Spo2051-91 localizes to the plasma membrane. During sporulation, RFP-Spo2051-91 assembles into a small cytosolic horseshoe structure located near the spindle poles, and the marked structure subsequently progresses through a series of discrete morphological transformations until it resolves into an envelope that surrounds the nucleus (Nakanishi et al., 2006; Diamond et al., 2009).
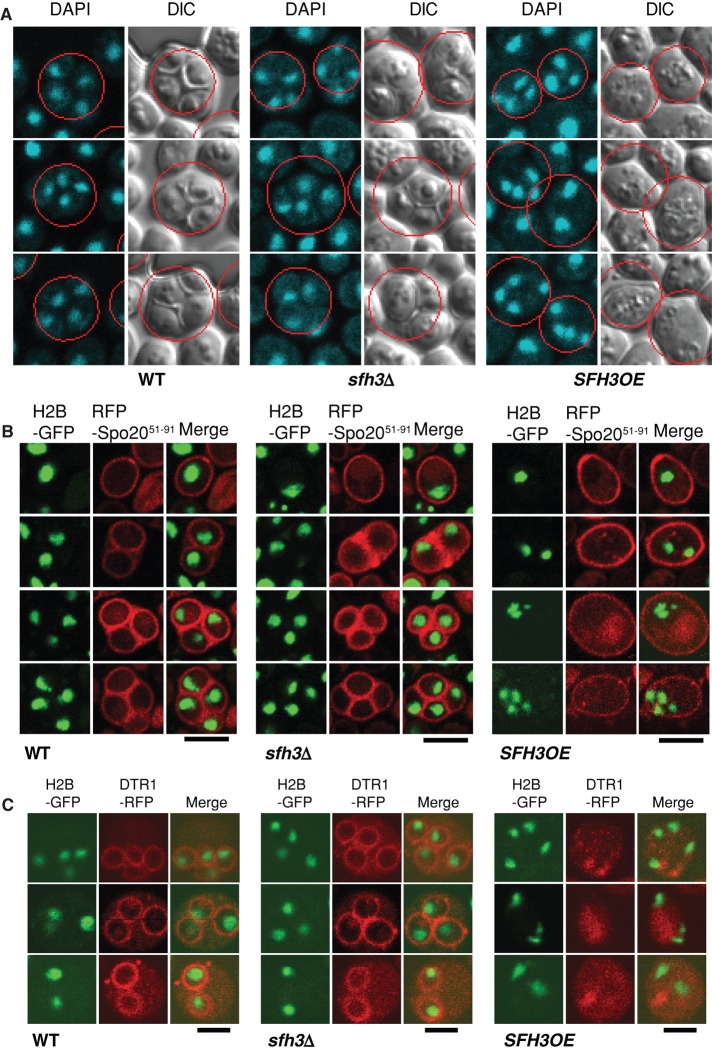
The four haploid nuclei of sporulated WT and sfh3Δ diploids were efficiently encapsulated by morphologically correct spore walls, and H2B-GFP/RFP-Spo2051-91 double-label imaging confirmed the DIC results (Figure 4, A and B). In both WT and sfh3Δ diploids, RFP-Spo2051-91 specifically labeled the plasma membrane of uninucleate cells. By contrast, the reporter lined the spore plasma membranes of multinucleate cells with mature spores. Multinucleate cells lacking spore walls were prevalent in meiotic Sfh3OE diploids (∼75% of sporulating cells; Figure 4B). In those cells, RFP-Spo2051-91 was not associated with replicated genomes. Even during early stages of sporulation, organized relocalization of RFP-Spo2051-91 reporter to juxtanuclear positions (diagnostic of active PSM biogenesis) was only rarely observed (∼10% of sporulating cells).
FIGURE 4:
SFH3OE compromises PSM formation. (A) Diploid yeast (S288C) were transferred to starvation medium for 4 d and imaged. DNA was labeled with 4′,6-diamidino-2-phenylindole. Cells with four nuclei are identified by red circles. Cells that had produced mature spores were readily apparent by DIC microscopy for WT and sfh3∆ strains but not for the isogenic SFH3OE derivative. (B) WT, sfh3∆, and SFH3OE cells expressing RFP-Spo2051-91 were imaged. Yeast genomes were marked with histone H2B-GFP. For all strains, RFP-Spo2051-91 localized to the plasma membrane (PM) in vegetative cells (single nuclei). For sporulating WT and sfh3∆ cells (tetrads, triads, and dyads), RFP-Spo2051-91 labeled PSMs but remained targeted to the PMs of SFH3OE cells that had completed meiosis II (multiple nuclei). Bar, 5 μm. (C) WT, sfh3∆, and SFH3OE diploid cells expressing Dtr1-RFP were imaged after transfer to starvation medium for 12 h. Dtr1-RFP labeled PSMs of WT and sfh3∆ cells (tetrads, triads, and dyads) but remained cytosolic in SFH3OE cells that had completed Meiosis II (multiple nuclei). Bar, 5μm.
The RFP-Spo2051-91 data diagnosed failures in PSM biogenesis under Sfh3OE conditions. Given that RFP-Spo2051-91 peripherally associates with PSMs, we confirmed the biogenic defects by following a PSM integral membrane protein marker during the developmental switch (Dtr1-RFP; Felder et al., 2002; Morishita and Engebrecht, 2008). Whereas forming Dtr1-marked PSMs were on ready display during early stages of sporulation in WT cells, such productive events were rarely observed in isogenic Sfh3OE contexts (∼5% of sporulating cells; Figure 4C).
Sfh3OE supports robust phospholipase D activity
PSM biogenesis requires developmentally regulated hydrolysis of PtdCho to PtdOH catalyzed by phospholipase D (Rose et al., 1995; Rudge et al., 1998). One trivial mechanism for why Sfh3OE might evoke PSM biogenic defects is that Sfh3OE inhibits PLD activation. Indeed, the vegetative phenotypes of Sfh3OE yeast (e.g., enhancement of sec14ts-associated growth defects; see Figure 1B) and the impaired ability of Sfh3OE diploids to complete sporulation were superficially consistent with PLD insufficiencies (Honigberg et al., 1992; Rose et al., 1995; Rudge et al., 1998; Xie et al., 1998). Moreover, PLD deficiencies nullify all mechanisms for “bypass Sec14” (Xie et al., 1998), and a cardinal phenotype of Sfh3OE vegetative cells was ablation of the “bypass Sec14” phenotypes associated with functional inactivation of 1) the sterol-binding protein Kes1 and 2) the CDP-choline pathway for PtdCho biosynthesis (kes1Δ and cki1Δ; Supplemental Figure S4A). Both qualitative and quantitative measurements of PLD activity by choline release assay indicated that PLD activity was not compromised by Sfh3OE (Supplemental Figure S4, B and C). Instead, Sfh3OE potentiated PLD activity (compare choline release in sec14ts cki1 vs. sec14ts cki1 SFH3OE strains; Supplemental Figure S4C). This potentiation was consistent with the functional antagonism between Sec14 and Sfh3 (Figure 1B); given Sec14 deficiencies activate PLD by unknown mechanisms (Sreenivas et al., 1998; Xie et al., 1998; Routt et al., 2005).
Ergosterol ester hydrolase defects partially phenocopy Sfh3OE
Of the four class II LD proteins that colocalized with Sfh3 to PSM-associated LDs, two are ergosterol ester hydrolases (Yeh1; Tgl1) and one is a TAG hydrolase (Tgl4). This striking partitioning (documented for Tgl1 in Figure 5A) suggested that PSM-associated LDs are uniquely poised for fueling PSM expansion. This idea was consistent with other evidence that LDs associated with the ascal face of the PSM are consumed during spore morphogenesis (Lin et al., 2013). We analyzed the effects of Tgl1 deficiency on PSM formation as an independent test of whether PSM-associated LD utilization was required for expansion of these membranes. Indeed, of the tgl1Δ diploid yeast cells that completed meiosis II, a major fraction failed to package the haploid nuclei within a spore wall (50%). When earlier stages of the sporulation program were monitored, ∼70% of the sporulating tgl1Δ diploid cells exhibited defects in the packaging of haploid nuclei within well-formed PSMs (marked with RFP-Spo2051-91 or Dtr1-RFP; Figure 5, B and C). Thus Tgl1-deficient diploids recapitulated Sfh3OE-like defects in PSM biogenesis, albeit in a less penetrant manner. The similarities evident in the PSM biogenic and sporulation defects of tgl1Δ/tgl1Δ and Sfh3OE diploids indicated that Sfh3 and Tgl1 play opposing roles in regulating lipid incorporation from LDs into developing PSMs. When coupled with the coincident enrichment of Sfh3 and neutral lipid hydrolases to a PSM-associated LD pool, these data suggest that Sfh3 antagonized neutral lipid mobilization from this specific LD subpopulation.
FIGURE 5:
Defects in the LD-resident Tgl1 lipase compromise PSM formation. (A) Tgl1 is associated with LDs that line the ascal face of the PSM. LDs were stained with BODIPY-TR. PSM was labeled with mTagBFP-Spo2051-91. Tgl1 was tagged with GFP. (B, C) Homozygous tgl1∆ diploid cells are defective in PSM formation. Homozygous tgl1Δ diploids coexpressing H2B-GFP and either (A) RFP-Spo2051-91 or (B) Dtr1-RFP were imaged. Cells with multiple nuclei that failed to enclose the meiotic products with PSMs were a dominant phenotype for these cells.
Sfh3 activity and LD homeostasis
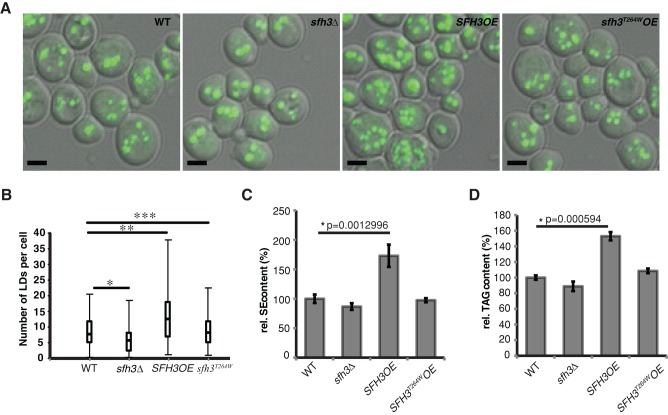
The selective localization of Sfh3 to an LD subpool in sporulating cells precluded direct functional characterization of how Sfh3 modulates LD metabolism during the meiotic program. Thus the relationships between Sfh3 activity and LD homeostasis were surveyed with BODIPY staining in vegetative haploid cells grown with glucose as carbon source and Sfh3 dosage as experimental variable (Figure 6A). The distributions of cellular LD load for isogenic WT, sfh3Δ, and Sfh3OE cells are shown in Figure 6B. Whereas LD number/cell was highly variable under these culture conditions, enhanced Sfh3 production clearly increased the LD load of cells relative to WT controls. This phenotype was particularly evident in the significantly increased incidence of cells loaded with >20 LDs in the Sfh3OE yeast population. These effects were not recapitulated in sfh3T264WOE yeast, which exhibited WT distributions of LD load (these sfh3T264WOE cells still express endogenous Sfh3), or in sfh3Δ cells, which exhibited reduced LD loads. The imaging data were buttressed by biochemical measurements that recorded ∼2- and ∼1.5-fold increases in SE and TAG mass, respectively, in Sfh3OE cells (Figure 6, C and D). Enhanced expression of Sec14, Sfh1, Sfh2, Sfh4, or Sfh5 failed to exert discernible effects on LD load (Supplemental Figure S5A), highlighting Sfh3 specificity in LD homeostatic control. Given that PITP specificity correlated with a unique targeting to LDs, we conclude that the LD phenotype reported a direct functional interface between Sfh3 and LD metabolism.
FIGURE 6:
LDs accumulate in SFH3OE cells. (A) Wild-type, sfh3Δ, SFH3OE, and sfh3T264W OE yeast were cultured in minimum medium to stationary phase before staining with BODYPY493/503. Representative images from each strain. Bar, 2 μm. (B) Whole-cell LD numbers were determined for ∼200 cells for each strain shown in A. Per-cell LD load distributions are shown as box plots composed of low, high, median, quartile I, and quartile III thresholds for cellular LD number in each group. Data from WT vs. sfh3Δ, WT vs. SFH3OE, and WT vs. sfh3T264WOE were compared by t test: *p = 6.92E-08; **p = 9.14E-07; ***p = 0.361, respectively. (C, D) Yeast of indicated genotypes were cultured in SD minimal medium with 2% glucose for 40 h. Whole-cell lipids were extracted and resolved by TLC. Total SE and TAG were quantified. SE (C) and TAG (D) contents of indicated yeast strains relative to WT. Values represent averages from three independent experiments plotted as mean average ± SD.
LD lipidomics
No gross derangements in LD structure/morphology were observed in sfh3Δ and Sfh3OE yeast relative to WT cells (Supplemental Figure S5B). To probe for biochemical changes, we profiled the glycerophospholipid and neutral lipid compositions of LDs purified from WT, sfh3Δ, and Sfh3OE yeast (Supplemental Tables S3–S5). No significant differences were recorded in the lipidomic profiles. In all cases, PtdCho was the most abundant phospholipid species, followed by PtdEtn, PtdSer, PtdIns, PtdOH, and PtdGro in rank order (Supplemental Figure S6A). PtdIns was underrepresented in LDs relative to bulk membrane (20 mol%), whereas PtdEtn and PtdSer were enriched in LDs relative to bulk membranes (15 and 6 mol%, respectively; unpublished data). SE and TAG molecular species profiles were also similar in LDs purified from WT, Sfh3OE, and sfh3Δ yeast (Supplemental Figure S6, B and C), and the TAG:ergosterol ester (ES) and neutral lipid:phospholipid ratios were also essentially the same (TAG:ES and neutral lipid:phospholipid stoichiometries of ∼4:1 and ∼23:1, respectively, in LDs purified from all strains). We conclude that LD expansion in Sfh3OE cells was metabolically coherent, in that it reflected a balanced increase in mass for all major LD lipid species.
Sfh3 inhibits LD utilization
We sought to distinguish whether Sfh3 enhanced neutral lipid allocation into the LD pool or retarded neutral lipid mobilization from LD stores. A [3H]oleate pulse-radiolabeling regimen assessed whether LD expansion in Sfh3OE cells resulted from elevated neutral lipid synthesis. Those experiments demonstrated that WT, sfh3Δ, and Sfh3OE cells exhibit comparable rates of [3H]oleate incorporation into TAG and SE (Figure 7A). To analyze LD consumption, we measured lipolysis-mediated glycerol release for WT, sfh3Δ, and Sfh3OE yeast challenged with glucose and nitrogen deprivation. Whereas WT cells showed rapid and sustained release of free glycerol upon starvation, this response was strongly diminished in starved Sfh3OE cells. No reductions in glycerol release were recorded for sfh3T264WOE cells relative to WT (these cells express endogenous Sfh3), whereas sfh3Δ cells produced the most vigorous release of free glycerol upon nutrient deprivation (Figure 7B).
FIGURE 7:
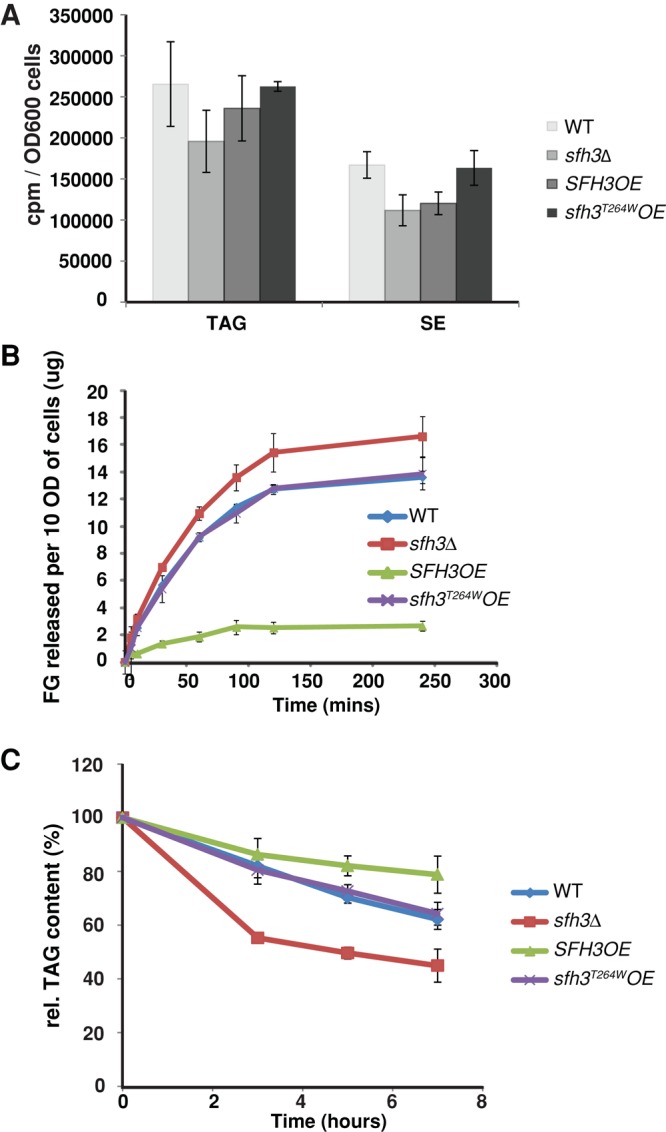
Sfh3 inhibits LD utilization. (A) Incorporation of [3H]oleate into neutral lipid synthesis in indicated strains. Logarithmic-growth-phase cells were pulsed with [3H]oleate, and incorporation of [3H]oleate into the indicated neutral lipid classes was quantified. Averages from three independent experiments. Error bars represent SDS. (B) Yeast of indicated genotype were cultured in SD minimal media for 40 h before transfer to starvation medium. Free glycerol release to the medium was measured at the indicated time points (10 OD600 = 1.5 × 108 cells). (C) Yeast of indicated genotypes were cultured in SD minimal media for 40 h before transfer into fresh 2% glucose medium supplemented with 10 μg/ml cerulenin. Indicated lipids were quantified at the indicated time points.
In vivo TAG catabolism was independently measured as reporter of LD utilization. Again, Sfh3OE cells showed delayed TAG degradation relative to WT cells when shifted from stationary phase into fresh medium dosed with cerulenin, a potent inhibitor of fatty acid synthesis (Figure 7C). Reciprocally, sfh3Δ cells exhibited substantially increased rates of TAG degradation. Taken together, these data established that Sfh3 inhibits neutral lipid mobilization from LD stores.
Elevated PtdIns-4-P inhibits LD utilization and spore formation
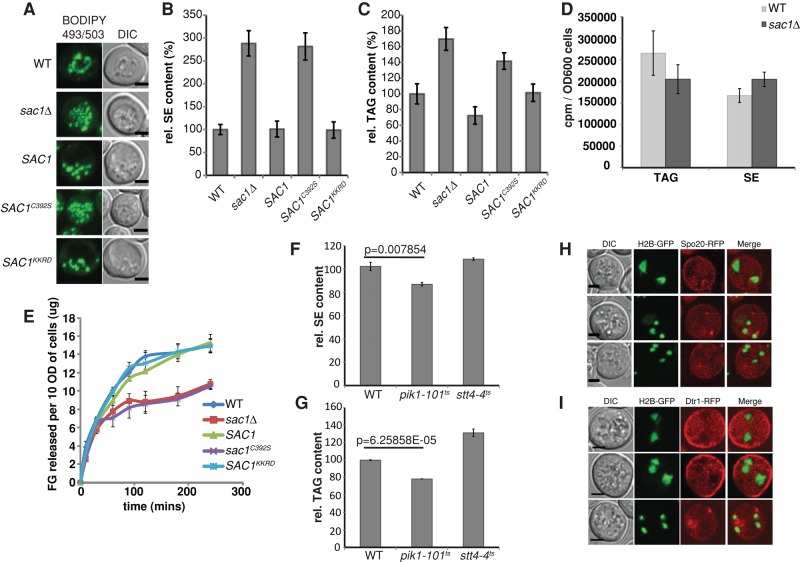
The PtdIns-binding requirement for Sfh3 to inhibit PSM formation in sporulating diploid cells and bulk LD utilization in vegetative cells suggested that a pool of PtdIns-4-P is channeled toward inhibition of LD utilization. If so, Sfh3-independent elevation of PtdIns-4-P is predicted to phenocopy Sfh3OE with regard to LD homeostasis. Sac1, the major yeast PtdIns-4-P phosphatase, is an integral membrane protein of the ER and Golgi systems, and sac1Δ mutants accumulate high levels of PtdIns-4-P (Guo et al., 1999; Rivas et al., 1999). Indeed, BODIPY 493/503 staining reported a threefold to fourfold increase in LD load in sac1Δ cells compared with WT (Figure 8A). Expression of the catalytic-dead sac1C392S failed to reverse LD accumulation in sac1Δ cells, establishing a Sac1 phosphoinositide phosphatase activity requirement for LD homeostasis. Our finding that Sac1KKRD (a catalytically active enzyme that fails to exit the ER; Kurat et al., 2006) restored normal LD homeostasis suggested that LD accumulation in sac1Δ cells was provoked via an ER-localized PtdIns-4-P pool (Figure 8A).
FIGURE 8:
Elevated PtdIns-4-P inhibits LD utilization. (A) Cells of indicated genotype were reconstituted for Sac1, sac1C392S, or Sac1KKRD expression (with appropriate CEN plasmid vectors, as indicated). LDs were stained with BODIPY493/503, and whole-cell projections of confocal Z-stacks are shown. (B, C) Yeast of indicated genotype were cultured in minimum medium with 2% glucose for 40 h. Whole-cell lipids were extracted and resolved by TLC. Total SE and TAG were quantified. Comparisons of the relative SE (B) and TAG (C) contents of the indicated yeast strains. The averages from three independent experiments are plotted as mean ± SD. (D) Incorporation of [3H]oleate into neutral lipid synthesis was measured in the indicated strains. WT and sac1Δ cells exhibited similar neutral lipid synthesis rates. (E) Cells of indicated genotype were cultured in minimal SD media before shift to starvation medium. Free glycerol release was measured at 0, 10, 30, 60, 90, 120, and 240 min after shift. (F, G) Pik1, but not Stt4, deficiencies result in decreased neutral lipid content. WT, pik1-101ts, and stt4-4ts strains were cultured in YPD to stationary phase. Each culture was then divided into two equal aliquots. One aliquot was cultured at 30ºC for another 3 h and the other was shifted to 37ºC for 3 h. Neutral lipids were extracted, resolved, and quantified. SE (F) and TAG (G) mass for indicated strains cultured at 37ºC relative to those cultured at 30ºC. (H, I) sac1Δ homozygous diploid cells are incompetent for PSM biogenesis. Diploid sac1Δ cells coexpressing H2B-GFP and (H) RFP-Spo2051-91 or (I) Dtr1-RFP and were imaged. Instead of enclosing the newly formed haploid genome, the indicated PSM markers stay on the PM or in cytosol.
Biochemical measurements were congruent with the imaging data. Neutral lipid analyses reported ∼3- and 1.5-fold increase in the SE and TAG mass, respectively, in sac1Δ cells relative to isogenic WT controls (Figure 8, B and C). Expression of Sac1KKRD, but not of the catalytic-dead sac1C392S, corrected neutral lipid accumulation defects associated with sac1Δ, confirming that Sac1 phosphoinositide phosphatase activity was essential for maintenance of proper neutral lipid homeostasis. [3H]oleate pulse-radiolabeling experiments reported that sac1Δ mutants did not exhibit increased rates of neutral lipid synthesis (Figure 8D). Instead, glycerol release measurements confirmed that LD homeostatic derangements reflected diminished LD utilization in sac1Δ and sac1C392S cells and that expression of either Sac1 or Sac1KKRD corrected these LD consumption defects (Figure 8E). To identify which of the two major yeast PtdIns 4-OH kinases (Pik1 and Stt4) might generate the PtdIns-4-P pool that inhibits LD utilization, we measured neutral lipid contents in pik1-101ts and stt4-4ts strains at both permissive and nonpermissive temperatures. Inactivation of the Pik1 PtdIns 4-OH kinase, but not the Stt4 PtdIns 4-OH kinase, consistently evoked ∼20% reduction in both SE and TAG mass (Figure 8, F and G).
The concepts that an expanded PtdIns-4-P pool inhibits LD utilization and that LD catabolism is essential for PSM biogenesis predicted that Sac1 deficiencies will evoke sporulation defects in the face of an actively engaged meiotic program. Indeed, homozygous sac1Δ diploids recapitulated the Spo− phenotypes of Sfh3OE diploids. Of the sporulating sac1Δ diploid cells that completed meiosis II within 96 h, as reported by the presence of four H2B-GFP-labeled nuclei, only ∼1% presented PSM-encapsulated haploid genomes (Figure 8, H and I).
DISCUSSION
Here we link PITP-regulated PtdIns-4-P signaling with control of LD metabolism by demonstrating that the nonclassical PITP Sfh3 associates with LDs in yeast and inhibits neutral lipid mobilization from those particles in a PtdIns-4-P–dependent mechanism. We further document an unexpectedly complex heterogeneity of LD pools during diploid yeast sporulation and that Sfh3-mediated regulation of LD utilization influences the efficiency of this developmental stress response. Given that key aspects of Sfh3 biological function are uniquely manifested in diploid yeast cells, these activities are invisible to the types of haploid-centric studies that dominate contemporary cell biological, functional genomics, and proteomics efforts in the yeast model. The collective data offer novel insights into how Sec14-like PITPs integrate phosphoinositide signaling with broader territories of the lipid metabolome.
Sfh3 and LD metabolism
Several lines of genetic and biochemical evidence identify Sfh3 as an inhibitor of neutral lipid mobilization from LDs. Because Sfh3 is required for neither LD production nor degradation, we interpret Sfh3 as one component of a tuning (or timing) mechanism for modulating rates of LD consumption. We propose that this Sfh3/LD relationship is the principle that underlies the superficial association of Sfh3 activity with pleiotropic drug resistance phenotypes, an association that depends on how the experiments are performed. It is from this perspective that we comment on annotation of Sfh3 as Pdr16 on the basis of pleiotropic drug resistance phenotypes associated with sfh3Δ mutants (van den Hazel et al., 1999). We concur with van den Hazel et al. (1999) that sfh3Δ mutants show a specific hypersensitivity to chronic challenge with sublethal doses of azole (e.g., miconazole, ketoconazole, fluconazole) and polyene (amphotericin B) inhibitors of ergosterol biosynthesis (unpublished observations). These results also agree with competitive fitness studies that report selective extinction of sfh3Δ cells from a genetically diverse population challenged with azole drugs (Anderson et al., 2009). However, we find that sfh3Δ cells are more resistant than are WT cells to acute high-dose pulses of azoles. The converse behaviors are on display upon chronic and acute azole challenges of Sfh3OE cells, respectively (unpublished observations). We interpret these paradoxical drug resistance phenotypes as manifestations of accelerated depletion of LD ergosterol ester reserves when sfh3Δ cells are confronted by sterol stress. That is, accelerated LD consumption in shf3Δ cells is protective under acute conditions of incapacitated sterol synthesis but is deleterious in physiological contexts of chronic sterol stress.
Developmental programs for LD utilization
Sfh3 tuning/timing mechanisms would be advantageous under conditions in which vigorous fatty acid flux must be delicately balanced with the need to shelter precious cellular resources from oxidative damage. Such a situation is confronted by diploid yeast dually challenged with carbon and nitrogen deprivation. Under those conditions, diploid cells induce a robust program of PSM biogenesis in which a massive flux of fatty acid and lipid is channeled toward the ordered sequestration of the haploid genomes produced by meiosis. Sfh3 localization to a specific subpopulation of LDs associated with the ascal side of the PSM suggests that LD metabolism is closely coordinated with PSM expansion. Supporting evidence comes from our demonstration that PSM biogenesis is compromised in Sfh3OE and tgl1Δ diploid cells.
Finally, our demonstration that LD components differ between individual LD pools in the same cell during sporulation (i.e., a developmental stress response unique to diploid yeast cells) indicates that LD subpools are functionally diversified during meiosis. The striking enrichment of neutral lipid hydrolases to the PSM-associated LDs suggests that this LD subpool is uniquely engineered for consumption into PSM biogenesis and that Sfh3 tunes the rate and/or timing of neutral lipid mobilization from this specific LD pool. This novel concept is invisible to haploid yeast studies that dominate contemporary cell biological and functional genomics efforts and to LD proteomics studies that can only report bulk-averaged profiles. Whereas this Sfh3 tuning/timing role is dispensable for formation of viable spores, it is interesting to ask whether spore hardiness or meiotic genome quality is affected when spores are produced in the absence of Sfh3.
PtdIns-4-P and LD metabolism
Sfh3 biological activity as negative regulator of neutral lipid mobilization from LD stores requires its ability to bind PtdIns and to stimulate PtdIns-4-P production. These data identify Sfh3-dependent PtdIns-4-P signaling as physiological antagonist of LD utilization. Our demonstration that catalytic inactivation of Sac1, the major yeast PtdIns-4-P phosphatase, recapitulates the biological and metabolic effects of enhanced Sfh3 activity buttresses this conclusion. In accord with concepts for how Sec14-like PITPs regulate PtdIns-4-P signaling (Schaaf et al., 2008; Bankaitis et al., 2009), we posit that Sfh3 stimulates production of a specific PtdIns-4-P pool dedicated to control of LD utilization. Although Sac1 deficiencies closely phenocopy Sfh3OE, one difference is that rates of TAG lipolysis are more strongly inhibited by Sfh3OE than by Sac1 deficiencies. Given that Sac1 catalytic activity is relevant to LD utilization, we interpret this difference to reflect an inefficient “spilling” of excess PtdIns-4-P in sac1Δ mutants into the LD-relevant PtdIns-4-P pool. In LD utilization experiments that measure initial rates of lipolysis, such an inefficient mechanism would present attenuated effects on LD consumption rates. However, the effects would still be apparent in steady-state measurements (i.e., bulk TAG and SE measurements).
What is the nature of the proposed Sfh3-regulated PtdIns-4-P pool whose signaling is channeled to LD metabolism? One possibility is the operant PtdIns-4-P pool resides on the LD surface, although attempts to detect such a pool using PtdIns-4-P biosensors (e.g., FAPP1 PH domain) failed. Alternatively, PtdIns-4-P regulation of LD metabolism might be ectopically transmitted from another organelle, as LDs are closely apposed to ER membranes (Binns et al., 2006; Zehmer et al., 2009). Rescue of sac1Δ LD defects by an ER-trapped Sac1 PtdIns-4-P phosphatase supports such a possibility. The TGN/endosomal system, which harbors significant Pik1-dependent PtdIns-4-P pools, is another candidate source for transmitting PtdIns-4-P signaling to LDs. Either way, as we find no obvious lipid specificity in Sfh3-mediated inhibition of LD utilization (there is a balanced accumulation of all LD constituents in Sfh3OE cells), Sfh3-mediated regulation is not directed at one particular class of lipase. Instead, lipolysis is generally inhibited.
A role for PtdIns-4-P in control of LD metabolism is consistent with other observations. For example, a potential link between PtdIns-4-P and LD metabolism is suggested in cells infected with hepatitis C virus. During infection, elevation in cellular PtdIns-4-P coincides with LD accumulation, and increased PtdIns-4-P and LD accumulations are both required for production of infectious virus (Miyanari et al., 2007; Fukasawa, 2010; Zhang et al., 2012). Moreover, proteomic analyses identify ARF GTPase, subunits of the COPI complex, and Rab GTPases as constituents of the LD surface (Binns et al., 2006; Bartz et al., 2007), and genome-wide screens in yeast and Drosophila link the activities of these same membrane-trafficking regulators to LD homeostasis (Szymanski et al., 2007; Beller et al., 2008; Fei et al., 2008; Guo et al., 2008). Although the underlying mechanisms remain to be established, it is suggested that recruitment of an active vesicle biogenic machinery to LD surfaces promotes lipolysis by stimulating fission of small LDs from mother LDs. Such fission events are proposed to facilitate lipase access to LD lipids (Walther and Farese, 2009; Brasaemle and Wolins, 2012). Our data demonstrating an antagonistic involvement of PtdIns-4-P in LD lipolysis suggest that activity of these components on the LD surface might be inhibited, either directly or indirectly, by an Sfh3-dependent PtdIns-4-P pool.
PITP specificity in LD homeostasis
Sec14-like PITPs function not as transfer proteins, but as PtdIns-presentation scaffolds that potentiate the biologically insufficient activities of PtdIns 4-OH kinases (Schaaf et al., 2008; Bankaitis et al., 2009). Cell biological and structural data indicate that classic Sec14-like PITPs (Sec14, Sfh1) present PtdIns to PtdIns 4-OH kinases in a manner that requires priming by a second ligand (PtdCho) and that the chemical nature of that priming ligand instructs channeling of the expanded phosphoinositide pool to a specific physiological outcome (Schaaf et al., 2008; Bankaitis et al., 2009; Nile et al., 2010; Kono et al., 2013). That is, individual PITP action specifies functionally distinct PtdIns-4-P pools.
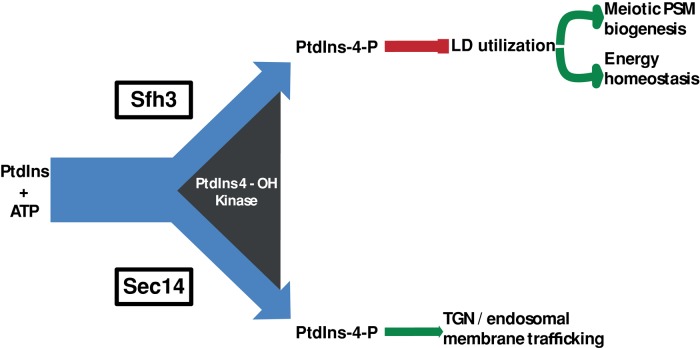
In this regard, the PITP specificities in LD homeostatic circuits are striking. No other yeast Sec14-like PITP, including the related Sfh4, can execute Sfh3-like functions in control of LD utilization. Although this functional specificity likely involves the LD-targeting properties of Sfh3, another contributing factor may be the mechanism by which Sfh3 stimulates PtdIns-4-P synthesis. The lipid-binding pockets of Sfh3 and its orthologues present structural and chemical environments that accommodate PtdIns binding but otherwise radically differ from those of Sec14 orthologues (e.g., in the PtdCho-binding barcode). These structural distinctions forecast that Sfh3-like proteins prime PtdIns-4-P synthesis in response to distinct sets of lipophiles. In that regard, the antagonistic effects of Sfh3 activity on essential Sec14- and Pik1-dependent vegetative functions 1) highlight that Sfh3 and Sec14 channel PtdIns-4-P synthesis/signaling toward distinct biological outcomes and 2) indicate that this differential channeling is competitive. We interpret these data as indicating that bulk PtdIns-4-P synthesis/signaling is functionally partitioned by PITPs, in this case according to the fractional influences of Sfh3 versus Sec14 on total cellular Pik1 PtdIns 4-OH kinase activity (Figure 9).
FIGURE 9:
Differential partitioning of PtdIns-4-P signaling outcomes by Sfh3 and Sec14. The PtdIns-4-P pools generated by action of a PtdIns 4-OH kinase (we presently favor Pik1) are channeled toward different biological outcomes. The pool generated by collaboration of Sec14 with the PtdIns 4-OH kinase channels to regulation of TGN/endosomal membrane trafficking, whereas the pool whose production is potentiated by Sfh3 is channeled toward control of LD utilization. We propose this to be a competitive design, as the fractional balance between Sfh3/kinase interactions and Sec14/kinase interactions will determine allocation of PtdIns-4-P signaling power toward specific cellular outcomes. This concept is on display in cells with reduced Sec14 or Pik1 and increased Sfh3 activities (Figure 1, B and C). A tunable PITP/lipid kinase balance affords considerable flexibility to the cellular PtdIns-4-P signaling landscape.
MATERIALS AND METHODS
Reagents
Standard reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA). All lipids were purchased from Avanti (Alabaster, AL). LD540 was provided by Christoph Thiele (Max Planck Institute of Molecular and Cellular Biology, Dresden, Germany).
Yeast strains and media
Yeast strains CTY182 (MATa ura3-52 lys2-801 Δhis3-200), CTY1-1A (MATa ura3-52 lys2-801 Δhis3-200 sec14-1ts), and CTY303 (MATa ura3-52 cki1 Δhis3-200 sec14ΔP::HISG) were previously described (Bankaitis et al., 1989, 1990; Cleves et al., 1991). Yeast strains BY4741 (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0) and sfh3Δ (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 sfh3Δ::KanMX) were purchased from Thermo Scientific Open Biosystems (Huntsville, AL). Yeast strains AN117-4B (MATα arg4-NspI his3SK ho::LYS2 leu2 lys2 rme1::LEU2 trp1::hisG ura3) and AN117-16D (MATa his3/SK ho::LYS2 leu2 lys2 trp1::hisG ura3) were previously reported (Neiman et al., 2000). Sfh3OE strains (MATα his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 PMA1-SFH3::URA3) and (MATα arg4-NspI his3SK ho::LYS2 leu2 lys2 rme1::LEU2 trp1::hisG ura3 PMA1-SFH3::URA3) were constructed by integrating the SFH3 open reading frame under control of the PMA1 promoter into the ura3Δ0 locus of strains BY4741 and AN117-4B. Strains SFH3::GFP (HIS3) and ERG6::RFP (HIS3) were constructed by genomic integration of GFP or RFP at the C-terminus of Sfh3 or Erg6 by homologous recombination. Strains containing SFH3::GFP (HIS3) ERG6::RFP (HIS3) were constructed by standard crosses and tetrad dissection. All the diploid yeast were generated by standard crosses. Plasmid pRS426-R20 is a PTEF2-RFP-Spo2051-91 construct.
Media included YPD (1% yeast extract, 2% bactopeptone, and 2% glucose), SD (0.67% nitrogen base, 2% glucose with amino acids supplemented), YPA (2% peptone, 1% yeast extract, 2% potassium acetate), and starvation medium (2% potassium acetate with amino acid supplements).
Protein expression and purification
His8-Sfh3 was purified from Escherichia coli cultured in media supplemented with selenomethionine instead of methionine. Cell lysates were incubated with TALON metal affinity resin (Clontech, Mountain View, CA), followed by elution with imidazole (Ren et al., 2011).
Gel filtration
Purified recombinant proteins were subjected to gel filtration chromatography on a S200 (10/300) column (Bio-Sciences, Piscataway, NJ) preequilibrated with buffer (25 mM Na2HPO4, 300 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM NaN3, pH 7.5) run at 0.4 ml/min. Fractions (0.5 ml) containing peak A280 nm absorbance eluted between 12 and 15 ml were pooled, and protein content was quantified using SDS–PAGE gel electrophoresis with internal bovine serum albumin standards.
Phospholipid transfer
Transfer assays measured [3H]PtdIns transport from rat liver microsomes to PtdCho liposomes or [14C]PtdCho from liposomes (98 mol% PtdCho, 2mol% PtdIns) to bovine heart mitochondria (Aitken et al., 1990; Bankaitis et al., 1990; Schaaf et al., 2008).
Confocal microscopy
Yeast cells were cultured in SD medium for 20 h and mounted onto Slab medium (SD medium with 20% gelatin) for microscopic analysis (63×/oil immersion objective; numerical aperture 1.3). Z-stack images were collected using a Zeiss LSM510 (Zeiss, Jena, Germany) and compiled to generate three-dimensional projections for visualizing all LDs.
Imaging of sporulating cells
For localization of different LD proteins, strains carrying C-terminal GFP fusions to the protein examined were mated to strain AN117-4B carrying pRS426-mTagBFP-Spo2051-91 as a prospore membrane marker (Neiman et al., 2000; Huh et al., 2003; Lin et al., 2013). The resulting diploids were sporulated and stained with 5 mM BODIPY-TR methyl ester (Life Technologies, Carlsbad, CA) to visualize LDs. Images were collected on a Zeiss AxioObserver Z.1 microscope. All images were processed using AxioVision 4.7 software.
Glycerol release assay
Yeast were cultured in SD medium for 40 h. Cells were then collected, washed three times with water, and resuspended at ∼2.5 OD/ml in 2% potassium acetate supplemented with the required amino acids. Supernatants were collected at different time points, and free glycerol concentrations were determined using Free Glycerol Reagent (F6428; Sigma-Aldrich).
Lipid droplet purification
LDs were purified as described (Leber et al., 1994). Briefly, yeast lysates were prepared in 12% Ficoll 400 solution, followed by centrifugation for 60 min at 28,000 rpm in an SW28 swing bucket rotor (Beckman Coulter, Brea, CA). The floating layer was collected from the top of the gradient and subjected to a second round of flotation through a discontinuous gradient (12% Ficoll 400 overlaid with 8% Ficoll 400). The floating layer was collected and refloated through a final discontinuous gradient (8% Ficoll 400 overlaid with 0.25 M sorbitol). The LD fraction was collected from the top of the gradient.
Transmission electron microscopy
Cells were cultured in SD minimal media, and 10 OD600 of cells were pelleted, fixed with 3% glutaraldehyde, converted to spheroplasts by digestion with Zymolyase, and stained with 2% OsO4 (Adamo et al., 2001). Samples were dehydrated and embedded in Spurr's resin, and 60-nm ultrathin sections were cut with a diamond knife. The sections were stained with 2% uranyl acetate and lead citrate and examined with a Tecnai 12 (FEI, Hillsboro, OR) electron microscope.
Neutral and phospho lipidomics
Quantitative profiling of neutral lipids (Hutchins et al., 2008) and phospholipids was performed as described (Ivanova et al., 2007; Myers et al., 2011). Experimental details are provided in the Supplemental Methods.
Phosphoinositide analyses
Cells were cultured in SD medium, radiolabeled with [3H]inositol to steady state (>20 h), and collected by precipitation with trichloroacetic acid. Lipids were extracted and deacylated by incubating cell pellet with methylamine reagent (10% methylamine, 45% methanol, and 11% n-butanol) at 53°C for 1 h. Deacylated glycerophosphoinositols were dissolved in water, extracted twice with n-butanol/petroleum ether/ethyl formate (20:4:1 vol/vol/vol), and resolved and quantified by high-performance liquid chromatography (Stolz et al., 1998).
Phospholipase D measurements
PLD activities were assayed by quantifying release of free choline into growth medium using a coupled choline oxidase assay (Li et al., 2000). Details are provided in the Supplemental Methods.
Sporulation assay
Diploid yeast cells were cultured in YPD overnight to stationary phase. Cells were washed with water, transferred to YPA medium for another 24 h, and washed with water one final time before being resuspended in starvation medium for at least 2 d.
Crystallization and structure determination
Sfh3 crystals were grown by sitting drop vapor diffusion at 22°C from solutions containing 1 μl of selenomethionine-labeled protein at 5 mg/ml and 1 μl of crystallant: 21% (wt/vol) polyethylene glycol, 5% (wt/vol) glycerol, 100 mM ammonium sulfate, and 100 mM ammonium acetate (pH 5.6). Crystals were grown in cryoprotectant and flash-cooled in liquid N2. Data to 1.9-Å resolution were collected at the South East Regional Collaborative Access Team (SER-CAT, Argonne National Laboratory, Lemont, IL) and processed and scaled with HKL2000 (Otwinowski and Minor, 1997). Experimental phases were determined by single-wavelength anomalous dispersion (SAD) in SGXpro (Fu et al., 2005). Model building and refinement used phase-combine maps in COOT (Emsley and Cowtan, 2004) and REFMAC 5 (Collaborative Computational Project N, 1994), respectively. The final model contains two molecules of Sfh3 (residues 17–345). Residues 205–220 were not modeled due to lack of interpretable electron density.
Phylogenetic and structural bioinformatics
Fungal Sec14-like homologues were identified using BLAST pairwise similarity searches (Altschul et al., 1997). Protein sequences were aligned using MUSCLE (Edgar, 2004), and multiple sequence alignments (MSAs) were edited according to structural alignment of Sec14 and Sfh3 crystal structures. Sequences were separated into Sfh3 and Sec14 orthologues (or monophyletic groups) using neighbor-joining phylogenetic analysis (Saitou and Nei, 1987; Larkin et al., 2007) calculated from the MSA. Highly conserved residues were determined independently for each set of orthologues. Highly conserved residues for Sec14 and Sfh3 orthologues were mapped onto the Sec14 structure (PDB ID 1AUA) and Sfh3 structure, respectively. Lipids were docked into lipid-binding cavities of the open forms of Sec14 and Sfh3 based on a superposition with the PtdIns- and PtdCho-bound structures of Sfh1 (PDB IDs 3B7N and 3B7Q, respectively).
Circular dichroism
Purified proteins (0.2 mg/ml) were dialyzed against 4 l of 10 mM potassium phosphate buffer, pH 7.5. Spectra were collected between 180 and 250 nm on an AVIV 62DS Circular Dichroism spectrometer (Aviv Instruments, Lakewood, NJ) using a 1-mm–path length cuvette.
Lipid analysis
Yeast strains were grown in synthetic medium (with 2% glucose) to stationary phase. Lipids were isolated from ∼100 OD of cells by the method of Folch et al. (1957) with modifications. Briefly, lipids were extracted with chloroform: methanol (2:1) and washed with 1 M KCl. Lipid extracts were resolved by TLC on silica gel 60 plates (Merck, Darmstadt, Germany) in petroleum ether/diethyl ether/acetic acid (80:20:1) and visualized with methanolic MnCl2, followed by plate charring. Band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD) after digital scanning. Standards included trioleylglycerol, dioleylglycerol, and cholesterol stearate (Sigma-Aldrich).
Analyses of TAG degradation
In vivo analyses of TAG turnover were performed as reported (Kurat et al., 2006), with modifications. Cells were cultured in minimum medium with 2% glucose for 30 h before transfer to fresh minimum medium to a cell density of A600 nm ≈ 2 in the presence of 10 μg/ml cerulenin. Aliquots (20 ml) were collected for lipid analyses at the appropriate time points.
[3H]oleate pulse radiolabeling
Incorporation of [3H]oleate into TAG and SE species was quantified as previously described (Oelkers et al., 2000). Log-phase cultures cultured in YPD were pulsed with [3H]oleate (1 μCi/ml) for 30 min at 30ºC. Total lipids were extracted, resolved by TLC, and stained with iodine vapor. TAG and SE species were identified using trioleylglycerol and cholesterol stearate (Sigma-Aldrich) as standards and individually harvested, and radiolabel in each fraction was determined by liquid scintillation counting.
Supplementary Material
Acknowledgments
This work was supported by Grant GM44530 from the National Institutes of Health (V.A.B.) and the Robert A. Welch Foundation (V.A.B.). J.R. was supported by a postdoctoral fellowship from the American Heart Association (11POST6880007). A.M.N. was supported by National Institutes of Health Grant GM72540, D.M.E. and the University of Utah Protein Interactions Core Facility were supported by National Institutes of Health Grant GM82545. T.J.L. and R.C.M. were supported by National Institutes of Health Grant GM069338, and P.T.I., D.S.M., and H.A.B. were supported by National Institutes of Health U54 GM069338 and PO1 ES013125. We thank Chao-wen Wang (Institute of Plant and Microbial Biology, Taiwan) for Pet10 antibody and Henrik Dohlman (University of North Carolina at Chapel Hill) for kindly facilitating the pulse-labeling experiments. We are also grateful to Michael Kay (University of Utah, Salt Lake City, UT) and Joel Goodman (UT–Southwestern Medical Center, Dallas, TX) for helpful discussions and comments and Lora Yanagisawa (Texas A&M Health Science Center, College Station, TX) for critical comments on the manuscript. We acknowledge the University of North Carolina Lineberger Comprehensive Cancer Center Genome Analysis and Nucleic Acids Core facilities.
Abbreviations used:
- BFP
blue fluorescent protein
- DAG
diacylglycerol
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- ES
ergosterol ester
- LD
lipid droplet
- PITP
phosphatidylinositol transfer protein
- PLD
phospholipase D
- PSM
prospore membrane
- PtdCho
phosphatidylcholine
- PtdEtn
phosphatidylethanolamine
- PtdGro
phosphatidylglycerol
- PtdIns
phosphatidylinositol
- PtdIns-4-P
phosphatidylinositol-4-phosphate
- PtdOH
phosphatidic acid
- PtdSer
phosphatidylserine
- RFP
red fluorescent protein
- SE
sterol ester
- Sfh3OE
Sfh3 overexpression
- TAG
triacylglycerol
- TGN
trans-Golgi network
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-11-0634) on January 8, 2014.
The authors declare no financial conflicts.
REFERENCES
- Adamo JE, Moskow JJ, Gladfelter AS, Viterbo D, Lew DJ, Brennwald PJ. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken JF, van Heusden GP, Temkin M, Dowhan W. The gene encoding the phosphatidylinositol transfer protein is essential for cell growth. J Biol Chem. 1990;265:4711–4717. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JB, Sirjusingh C, Syed N, Lafayette S. Gene expression and evolution of antifungal drug resistance. Antimicrob Agents Chemother. 2009;53:1931–1936. doi: 10.1128/AAC.01315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Malehorn DE, Emr SD, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci 35, 150–160. 2009. [DOI] [PMC free article] [PubMed]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 2010;12:521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. 2012;287:2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab. 2009;297:E1–E9. doi: 10.1152/ajpendo.90926.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Diamond AE, Park JS, Inoue I, Tachikawa H, Neiman AM. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:134–145. doi: 10.1091/mbc.E08-06-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder T, Bogengruber E, Tenreiro S, Ellinger A, Sa-Correia I, Briza P. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:799–810. doi: 10.1128/EC.1.5.799-810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Koliwad S, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fu ZQ, Rose J, Wang BC. SGXPro: a parallel workflow engine enabling optimization of program performance and automation of structure determination. Acta Crystallogr D Biol Crystallogr. 2005;61:951–959. doi: 10.1107/S0907444905010541. [DOI] [PubMed] [Google Scholar]
- Fukasawa M. Cellular lipid droplets and hepatitis C virus life cycle. Biol Pharm Bull. 2010;33:355–359. doi: 10.1248/bpb.33.355. [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Conicella C, Espositio RE. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics. 1992;130:703–716. doi: 10.1093/genetics/130.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res. 2008;49:804–813. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Ohto U, Hiramatsu T, Urabe M, Uchida Y, Satow Y, Arai H. Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 2013;340:1106–1110. doi: 10.1126/science.1233508. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Cocchiaro J, Valdivia RH. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 2006;16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- Li X, Routt S, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch D, Bankaitis VA. Identification of a novel family of nonclassical yeast PITPs whose function modulates activation of phospholipase D and Sec14p-independent cell growth. Mol Biol Cell. 2000;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Kim C, Smith SO, Neiman AM. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 2013;9:e1003700. doi: 10.1371/journal.pgen.1003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- Londos C, Sztalryd C, Tansey JT, Kimmel AR. Role of PAT proteins in lipid metabolism. Biochimie. 2005;87:45–49. doi: 10.1016/j.biochi.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Lynn RR, Magee PT. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J Cell Biol. 1970;44:688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Moore HP, Silver RB, Mottillo EP, Bernlohr DA, Granneman JG. Perilipin targets a novel pool of lipid droplets for lipolytic attack by hormone-sensitive lipase. J Biol Chem. 2005;280:43109–43120. doi: 10.1074/jbc.M506336200. [DOI] [PubMed] [Google Scholar]
- Morishita M, Engebrecht J. Sorting signals within the Saccharomyces cerevisiae sporulation-specific dityrosine transporter, Dtr1p, C terminus promote Golgi-to-prospore membrane transport. Eukaryot Cell. 2008;7:1674–1684. doi: 10.1128/EC.00151-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- Myers DS, Ivanova PT, Milne SB, Brown HA. Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim Biophys Acta. 2011;1811:748–757. doi: 10.1016/j.bbalip.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Morishita M, Schwartz CL, Coluccio A, Engebrecht J, Neiman AM. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J Cell Sci. 2006;119:1406–1415. doi: 10.1242/jcs.02841. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Suda Y, Neiman AM. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J Cell Sci. 2007;120:908–916. doi: 10.1242/jcs.03405. [DOI] [PubMed] [Google Scholar]
- Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM, Katz L, Brennwald PJ. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics. 2000;155:1643–1655. doi: 10.1093/genetics/155.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile AH, Bankaitis VA, Grabon A. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clin Lipidol. 2010;5:867–897. doi: 10.2217/clp.10.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Hishiki T, Shimizu Y, Funami K, Sugiyama K, Miyanari Y, Shimotohno K. Hepatitis C virus utilizes lipid droplet for production of infectious virus; Proc Jpn Acad B Phys Biol Sci; 2009. pp. 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pilch PF, Souto RP, Liu L, Jedrychowski MP, Berg EA, Costello CE, Gygi SP. Cellular spelunking: exploring adipocyte caveolae. J Lipid Res. 2007;48:2103–2111. doi: 10.1194/jlr.R700009-JLR200. [DOI] [PubMed] [Google Scholar]
- Ren J, Schaaf G, Bankaitis VA, Ortlund EA, Pathak MC. Crystallization and preliminary X-ray diffraction analysis of Sfh3, a member of the Sec14 protein superfamily. Acta Crystallogr F Struct Biol Cryst Commun. 2011;67:1239–1243. doi: 10.1107/S1744309111027096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routt SM, Ryan MM, Tyeryar K, Rizzieri K, Roumanie O, Brennwald PJ, Bankaitis VA. Nonclassical PITPs activate phospholipase D via an Stt4p-dependent pathway and modulate function of late stages of the secretory pathway in vegetative yeast cells. Traffic. 2005;6:1157–1172. doi: 10.1111/j.1600-0854.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Morris AJ, Engebrecht J. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J Cell Biol. 1998;140:81–90. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schaaf G, et al. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl M, Oskolkova OV, Holic R, Brezna B, Pichler H, Zagorsek M, Kohlwein SD, Paltauf F, Daum G, Griac P. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur J Biochem. 2003;270:3133–3145. doi: 10.1046/j.1432-1033.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14p. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- Silva AR, Pacheco P, Vieira-de-Abreu A, Maya-Monteiro CM, D'Alegria B, Magalhaes KG, de Assis EF, Bandeira-Melo C, Castro-Faria-Neto HC, Bozza PT. Lipid bodies in oxidized LDL-induced foam cells are leukotriene-synthesizing organelles: a MCP-1/CCL2 regulated phenomenon. Biochim Biophys Acta. 2009;1791:1066–1075. doi: 10.1016/j.bbalip.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Subramanian V, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey JT, Huml AM, Vogt R, Davis KE, Jones JM, Fraser KA, Brasaemle DL, Kimmel AR, Londos C. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J Biol Chem. 2003;278:8401–8406. doi: 10.1074/jbc.M211005200. [DOI] [PubMed] [Google Scholar]
- Turkish A, Sturley SL. Regulation of triglyceride metabolism. I. Eukaryotic neutral lipid synthesis: “many ways to skin ACAT or a DGAT”. Am J Physiol Gastrointest Liver Physiol. 2007;292:G953–G957. doi: 10.1152/ajpgi.00509.2006. [DOI] [PubMed] [Google Scholar]
- van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Lee SC. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J Cell Sci. 2012;125:2930–2939. doi: 10.1242/jcs.100230. [DOI] [PubMed] [Google Scholar]
- Weibel GL, Joshi MR, Wei C, Bates SR, Blair IA, Rothblat GH. 15(S)-lipoxygenase-1 associates with neutral lipid droplets in macrophage foam cells: evidence of lipid droplet metabolism. J Lipid Res. 2009;50:2371–2376. doi: 10.1194/jlr.M900081-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Wolinski H, Kohlwein SD. Microscopic analysis of lipid droplet metabolism and dynamics in yeast. Methods Mol Biol. 2008;457:151–163. doi: 10.1007/978-1-59745-261-8_11. [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht JA, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci USA. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hong Z, Lin W, Shao RX, Goto K, Hsu VW, Chung RT. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PloS One. 2012;7:e32135. doi: 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Lass A, Haemmerle G, Zechner R. Fate of fat: the role of adipose triglyceride lipase in lipolysis. Biochim Biophys Acta. 2009;1791:494–500. doi: 10.1016/j.bbalip.2008.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.