Abstract
HIV-1 (Human immunodeficiency virus type 1)׳s infection is considered as one of most harmful disease known by human, the survivability rate of the host reduced significantly when it developed into AIDS. HIV drug resistance is one of the main problems of its treatment and several drug designs have been done to find new leads compound as the cure. In this study, in silico virtual screening approach was used to find lead molecules from the library or database of natural compounds as HIV-1 protease inhibitor. Virtual screening against Indonesian Herbal Database with AutoDock was performed on HIV-1 protease. From the virtual screening, top ten compounds obtained were 8-Hydroxyapigenin 8-(2",4"-disulfatoglucuronide), Isoscutellarein 4'-methyl ether, Amaranthin, Torvanol A, Ursonic acid, 5-Carboxypyranocyanidin 3-O-(6"-O-malonyl-beta-glucopyranoside), Oleoside, Jacoumaric acid, Platanic acid and 5-Carboxypyranocyanidin 3-O-beta-glucopyranoside.
Keywords: Indonesian herbal database, HIV-1 protease, molecular docking, virtual screening
Background
HIV (Human Immunodeficiency Virus) is still becoming global health major concern. More than 34 million of people were estimated living with HIV in 2011. Its infection could develop into AIDS (acquired immunodeficiency syndrome), marked as one of the most destructive disease in modern era. UNAIDS had estimated 1.7 million adult and child deaths due to AIDS in 2011 alone. HIV has high resistance characteristic due to its high rate of replication (109-1010 virions per person per day) and error-prone polymerase, and 5-10 mutations estimated to occur in each HIV life cycle [1, 2]. Unless some steps in the cycle are interrupted by specific treatment, the infection will spread throughout the body leading to deterioration of the host׳s immune system [3].
Maturation is an essential step in HIV life cycle, triggered by proteolytic cleavage of polyproteins by viral protease [4]. The critical role of HIV protease in the virus cycle was demonstrated as early as 1988 and within a decade, the first inhibitor of the enzyme, saquinavir, was already licensed for use in the treatment of HIV infection [5, 6]. As for 2012, not less than 9 protease inhibitors were approved by FDA to be used as medication to HIV [7]. The development of those protease inhibitors were a notable example of structure aided drug design and suggest there are more to be attained from it. New lead of protease inhibitor could be developed using this approach, including lead from natural product. Indonesia is recognized for its biodiversity, ranked second after Brazil. It consist more than 30,000 plant species out of 40,000 species known all over the world. At least 9,600 species have pharmacological activity [8]. Previous research has gathered some of the three dimensional data of Indonesian herbal compounds and published it as Indonesian Herbal Database (HerbalDB) (http://herbaldb.farmasi.ui.ac.id) [9].
In this research, we applied virtual screening as one of in silico structure aided drug design method to HerbalDB. This research employed structure-based screening (molecular docking) with AutoDock [10] and PyRx [11]. Top ten compounds obtained from the virtual screening of HerbalDB will be considered as hits and will be represented in ranks. The aim of this research is to find feasible AutoDock virtual screening parameter as HIV-1 protease inhibitor and to search the potential candidates from HerbalDB using the parameters obtained.
Methodology
This research was done using literature study and virtual screening by molecular docking (structure-based virtual screening). The experimental design for our experiment was as follow:
Preparing the HIV-1 PR Protein Structure:
The macromolecule (HIV-1 PR) three dimensional structure were obtained from the Protein Data Bank website www.pdb.org [12]. The inclusion criteria for our target macromolecule were wild type / non-mutant protein and in a complex with ligan. The exclusion criteria were having resolution >2.5 Å and incomplete chain. Macromolecule was downloaded with the *.pdb format for further processes. The chosen macromolecule then has to be optimized for virtual screening preparation. The macromolecule cleaned from solute, marker and cofactor molecules. Gasteiger partial charges and AutoDock forcefield were applied.
Preparing Ligand Structure:
Ligands used for this research were classified into two groups. The first group consists of ligands that were obtained from DUD [13]. We specifically downloaded controls for HIV-1 protease. It is consisted of 62 positive controls (simply called as DUD ligands) and 2038 negative controls (called as decoys). The data have *.mol2 format. The second ligands groups used in this research was obtained from HerbalDB. It contains known molecules from some plants that were used for herbal medicine in Indonesia. The molecules have *.mol and *.mol2 format. It were converted to *.pdbqt format so it will be available to use by AutoDock. All ligands (controls from DUD and ligands from HerbalDB) were converted from *.mol2 format into *.pdbqt format using Open Babel [14].
Optimizing the Virtual Screening Parameter:
Validation of Virtual screening using DUD database ligands and decoys was done using AutoDock 4.2 in PyRx [10, 11]. The screening result will be tabulated and analyzed from its Receiver Operating Characteristic (ROC) curve and Enrichment Factors (EF) to search for feasible parameters which will gives accurate prediction without compensating too much times and computer costs.
Virtual Screening of Indonesian Herbal Database:
After obtaining the parameter from the optimization, virtual screening was done to all structures of HerbalDB using AutoDock in PyRx.We sorted the screening result based on binding affinities and then ranked it to get top ten compounds that will be considered as hits in this reseach. Visualization was also done using AutoDockTools and PyMol to see the interaction between the ligand and HIV-1 protease.
Result & Discussion
The virtual screening target chosen for this research is wild HIV-1 protease with pdb identity 3OXC [15]. It meets our inclusion criteria as the virtual screening target. After it was downloaded from the protein data bank website, it was all cleaned from water molecules and other ligand chemical components such as saquinavir, formic acid and sulfate ion.
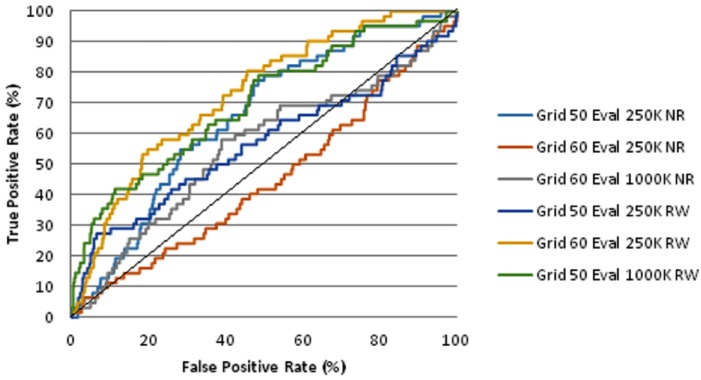
The ligand binding site was determined from the position of 3OXC ligand saquinavir. The XYZ coordinates is 5.192, -4.557, 14.799 for x-axis, y-axis, and z-axis, respectively. We varied gridbox parameter to see which one that gives the best EF1% and ROC AUC. We also compare the result of the screening if it was done completely without water molecule and retaining water molecules in the scope of macromolecule binding site. The enrichment and ROC AUC of DUD database virtual screening using AutoDock was shown in Table 1 (see supplementary material). The best EF1% was obtained with the parameter as follow: retaining water molecules, maximum energy of evaluation 1,000,000 and gridbox 50 × 50 × 50 units (spacing 0.375 Angstrom per unit). The EF score is better than the EF shown in the DUD site, 11.9 vs 4.7, respectively. The best AUC was obtained from the 60×60×60 gridbox and 250,000 maximum energy of evaluation. Parameter that gives best AUC and best EF both have better AUC (0.7275 and 0.6992, respectively) compared to the AUC from previous research (0.69 for AutoDock). The screening using AutoDock also suggest us that retaining water molecules in binding site could give better Enrichment Factor. The ROC curve of AutoDock screening result was shown in Figure 1. We prefer to use the parameter that gives best EF rather than the parameter that gives the best AUC for virtual screening of herbal database. This is given that early EF will be more useful than AUC as the whole compounds of database was rarely synthetized or extracted. The compounds that were highlighted as the top ranks from the screening result will be synthetized and extracted instead.
Figure 1.
Receiver Operating Characteristic Curve of DUD Database Virtual Screening using AutoDock. K = kilo (thousand), NR = Not retaining water, RW = Retains water at binding site.
Virtual screening of HerbalDB using AutoDock results were given on the Table 2 (see supplementary material). The top ten ranked compound were listed as hits from this screening. The binding energy of the top ten ranked compounds were varies from -18.74 kcal/mol to -11.79 kcal/mol. The first and second compounds are flavone glycosides that come from the fruit of Helicteres isora [16, [17]. Otake and colleagues have done research to examine anti HIV-1 activity of 30 plants extract. They concluded that water extract of Helicteres isora fruit have anti HIV-1 activity, but the compound and mechanism of action wasn׳t determined from the research [18]. Amaranthin, the compound that was ranked third, is a lectin group pigment that could be found in Amaranthus caudatus, A. tricolor, Celosia argentea and C. cristata seed [16, [19]. Ursonic acid in the fifth rank is a member of pentacylic triterphene that could be found in Angelica ursica Maxim, Ficus macrocarpa, Cordia multispicata, Myrica rubra, and Lantara camara Linn [16, [20, [21, [22, [23, [24]. From literature search, we haven׳t found any research about the activity of amaranthin and ursonic acid as protease inhibitor or anti HIV-1. 5-Carboxypyranocyanidin 3-O-(6"-Omalonyl- beta-glucopyranoside) and 5-Carboxypyranocyanidin 3-O-beta-glucopyranoside were in rank 6 and 10, respectively. Both are flavonoid glycoside from Allium cepa [16, [25]. Allium cepa has been patented to be used in HIV/AIDS treatment by Goldman et al [26]. Oleoside in the rank 5 is secoiridoid glucoside from Oleaceae familia plants [16, [27]. The leaf extract of this plant has been used to suppress the acute infection and HIV-1 cell to cell infection [28].
In the eighth rank is jacoumaric acid, a triterpene compound. It can be found in Jacaranda caucana and Psidium guajava [16, [29, [30]. There isn׳t any research for anti HIV activity for the compound yet. Platanic acid is found in Syzigium claviflorum, Platanus occidentalis, Melaleuca ericifolia, Melaleuca leucadendron [16, [31, [32, [33]. Fujioka and Kashiwada (1994) have researched the effect of betulinic acid and platanic acid as anti-HIV agent from Syzigium claviflorum plant. Platanic acid shows anti HIV activity with EC50 6.5 µM [34].
From this research, we concluded that the optimum result for HIV virtual screening by AutoDock were given by the parameter gridbox 50 × 50 × 50 units, 0.375 Angstrom per unit, gridbox centre at 5.192, -4.557, 14.799 for x-axis, y-axis, and zaxis, respectively, 1,000,000 numbers of evaluation and retaining water molecule near the binding site. The parameter gives better EF1% than the benchmark EF1%. We also done virtual screening from HerbalDB and we obtained top ten compounds as hits. Some of the compound, such as 8- Hydroxyapigenin 8-(2",4"-disulfatoglucuronide), Isoscutellarein 4'-methyl ether, and oleoside came from the plant that have been studied before and shows anti-HIV activity but there isn׳t any research about the mechanism or the contributing compound. From this research, we could get some insight about the compound and they need to be assayed in vitro to search more about its potential.
Supplementary material
Acknowledgments
We are grateful for funding provided by the Directorate General of Higher Education, Ministry of Education and Culture Republic of Indonesia through the National Strategic Research Project 2013 to Arry Yanuar.
Footnotes
Citation:Yanuar et al, Bioinformation 10(2): 052-055 (2014)
References
- 1. www.fda.gov/downloads/Drugs/Guidances/ucm071173.pdf.
- 2.Basavapathruni A, Anderson KS. FASEB J. 2007;21:3795. doi: 10.1096/fj.07-8697rev. [DOI] [PubMed] [Google Scholar]
- 3.Brik A, Wong CH. Org Biomol Chem. 2003;1:5. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 4.Fun A, et al. Retrovirology. 2011;8:70. doi: 10.1186/1742-4690-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohl NE, et al. Proc Natl Acad Sci USA. 1988;85:4686. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. http://archive.hhs.gov/news/press/1995pres/951207.html.
- 7. http://aidsinfo.nih.gov/contentfiles/ApprovedMedstoTr eatHIV_FS_en.pdf.
- 8. http://perpustakaan.depkes.go.id:8180/bitstream//1234 56789/561/3/KMK381-0307-G.pdf.
- 9.Yanuar A, et al. Int J Comp Sci. 2011;8:180. [Google Scholar]
- 10.Morris GM, et al. J Comput Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf LK, et al. Chemical & Engineering News. 2009;87:31. DOI:10.1021/cen-v087n028.p031. [Google Scholar]
- 12.Berman HM, et al. Nucleic Acid Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang N, et al. J Med Chem. 2006;49:6789. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Boyle NM. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tie Y, et al. Proteins. 2007;67:232. doi: 10.1002/prot.21304. [DOI] [PubMed] [Google Scholar]
- 16.Afendi FM, et al. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- 17.Kamiya K, et al. Phytochemistry. 2001;57:297. doi: 10.1016/s0031-9422(01)00005-x. [DOI] [PubMed] [Google Scholar]
- 18.Otake T, et al. Phytother Res. 1995;9:6. DOI:10.1002/ptr.2650090103. [Google Scholar]
- 19.Schliemann W, et al. Phytochemistry. 2001;58:159. doi: 10.1016/s0031-9422(01)00141-8. [DOI] [PubMed] [Google Scholar]
- 20.Bohlmann F, et al. Tetrahedron Lett. 1968;9:3947. DOI:10.1016/S0040-4039(00)72373-2. [Google Scholar]
- 21.Kuroyanagi M, et al. Chem Pharm Bull (Tokyo) 2001;49:954. doi: 10.1248/cpb.49.954. [DOI] [PubMed] [Google Scholar]
- 22.Lansky EP, Newman RA. JE thnopharmacol. 2007;109:177. doi: 10.1016/j.jep.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa T, et al. J Nat Med. 2007;61:112. DOI: 10.1007/s11418-006-0105-8. [Google Scholar]
- 24.Begum S, et al. Chem Pharm Bull (Tokyo) 2008;56:1317. doi: 10.1248/cpb.56.1317. [DOI] [PubMed] [Google Scholar]
- 25.Fossen T, Andersen ØM. Phytochemistry. 2003;62:1217. doi: 10.1016/s0031-9422(02)00746-x. [DOI] [PubMed] [Google Scholar]
- 26.Goldman SP, et al. US Patent. 2002 [Google Scholar]
- 27.Jensen SR, et al. Phytochemistry. 2002;60:213. doi: 10.1016/s0031-9422(02)00102-4. [DOI] [PubMed] [Google Scholar]
- 28.Lee-Huang S, et al. Biochem Biophys Res Commun. 2003;307:1029. doi: 10.1016/s0006-291x(03)01292-0. [DOI] [PubMed] [Google Scholar]
- 29.Ogura M, et al. Phytochem. 1977;16:286. DOI:10.1016/S0031-9422(00)86809-0. [Google Scholar]
- 30.Begum S, et al. Phytochemistry. 2002;61:399. doi: 10.1016/s0031-9422(02)00190-5. [DOI] [PubMed] [Google Scholar]
- 31.Abdel Bar FM, et al. J Nat Prod. 2008;71:1787. doi: 10.1021/np800360a. [DOI] [PubMed] [Google Scholar]
- 32.Lee CK. J Nat Prod. 1998;61:375. doi: 10.1021/np9606052. [DOI] [PubMed] [Google Scholar]
- 33.Ruzicka L, Lamberton AH. Helv Chim Acta. 1940;23:1338. DOI: 10.1002/hlca.194002301157. [Google Scholar]
- 34.Fujioka T, et al. J Nat Prod. 1994;57:243. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.