Abstract
The epithelium-specific ETS (ESE)-1 transcription factor is induced in chondrocytes by interleukin-1β (IL-1β). We reported previously that early activation of EGR-1 by IL-1β results in suppression of the proximal COL2A1 promoter activity by displacement of Sp1 from GC boxes. Here we report that ESE-1 is a potent transcriptional suppressor of COL2A1 promoter activity in chondrocytes and accounts for the sustained, NF-κB-dependent inhibition by IL-1β. Of the ETS factors tested, this response was specific to ESE-1, since ESE-3, which was also induced by IL-1β, suppressed COL2A1 promoter activity only weakly. In contrast, overexpression of ETS-1 increased COL2A1 promoter activity and blocked the inhibition by IL-1β. These responses to ESE-1 and ETS-1 were confirmed using siRNA-ESE1 and siRNA-ETS1. In transient cotransfections, the inhibitory responses to ESE-1 and IL-1β colocalized in the –577/–132 bp promoter region, ESE-1 bound specifically to tandem ETS sites at 403/ 381 bp, and IL-1-induced binding of ESE-1 to the COL2A1 promoter was confirmed in vivo by ChIP. Our results indicate that ESE-1 serves a potent repressor function by interacting with at least two sites in the COL2A1 promoter. However, the endogenous response may depend upon the balance of other ETS factors such as ETS-1, and other IL-1-induced factors, including EGR-1 at any given time. Intracellular ESE-1 staining in chondrocytes in cartilage from patients with osteoarthritis (OA), but not in normal cartilage, further suggests a fundamental role for ESE-1 in cartilage degeneration and suppression of repair.
The chondrocyte is a specialized mesenchymal cell that synthesizes matrix proteins responsible for the tensile strength and resistance to mechanical loading of the articular cartilage (Poole, 2005). In adult articular cartilage, the chondrocytes maintain a low turnover rate of replacement of cartilage matrix proteins. Collagen turnover has been estimated to occur with a half-life of greater than 100 years, whereas the glycosaminoglycan constituents on the aggrecan core protein are more readily replaced and the half-life of aggrecan subfractions is in the range of 3–24 years. Nevertheless, chondrocytes in vivo respond to structural changes in the surrounding cartilage matrix as occurs during the initial stages of osteoarthritis (OA) when increased cell proliferation and synthesis of matrix proteins, proteinases, and cytokines are observed. The early changes in synthetic activity are viewed as an attempt to regenerate the matrix with cartilage-specific components, including types II, IX, and XI collagens and aggrecan, and the presence of collagens not normally found in adult articular cartilage is evidence of phenotypic modulation (Sandell and Aigner, 2001). Genomic and proteomic analyses of global gene expression cartilage have confirmed the increased levels of type II collagen (COL2A1) mRNA and protein in OA cartilage (Bau et al., 2002; Hermansson et al., 2004), possibly associated with the increased levels of anabolic factors such as bone morphogenetic proteins and inhibin βA/activin, members of the TGF-β superfamily (Fukui et al., 2003; Hermansson et al., 2004), as well as prostaglandins (Tchetina et al., 2006). In later stages when catabolic processes are dominant, the complex composition of the articular cartilage matrix laid down during development is more difficult for the chondrocyte to replicate, particularly once there is severe damage to the collagen network.
The proinflammatory cytokines, interleukin-1β (IL-1β) and tumor necrosis factor (TNF)-α, are involved in the destruction of the articular cartilage in joint diseases such as rheumatoid arthritis and OA (Lubberts and van den Berg, 2003; Kobayashi et al., 2005). The chondrocyte is the cellular target of cytokine action in cartilage, and IL-1β and TNF-α colocalize with matrix metalloproteinase (MMP) production in superficial regions of OA cartilage (Tetlow et al., 2001). IL-1β suppresses the expression of a number of genes associated with the differentiated chondrocyte phenotype, including COL2A1 (Goldring et al., 1988; Okazaki et al., 2002). The upregulation by IL-1β of MMP13, cyclooxygenase (COX) 2, and nitric oxide synthase (NOS) 2 gene expression in chondrocytes and other cell types is mediated by the induction and activation a number of transcription factors, including NF-κB, C/EBPβ, and δ, AP-1 family members, and certain ETS factors. The ETS factors constitute a family of at least 30 members that play central roles in regulating genes involved in development, differentiation, and cell proliferation (Verger and Duterque-Coquillaud, 2002). Several ETS factors, including ETS-1 and PEA3, cooperate with AP-1 in regulating MMP gene expression (Mengshol et al., 2001; Vincenti and Brinckerhoff, 2002; Tower et al., 2003).
ESE-1, also known as ESX, ELF3, ERT, and JEN, is a novel ETS factor that is restricted to epithelial tissues under physiological conditions (Oettgen et al., 1997, 1999a). We reported recently that ESE-1 is expressed in non-epithelial tissues undergoing inflammation such as rheumatoid synovium and in chondrocytes, as well as glioma cells, smooth muscle cells, synovial fibroblasts, osteoblasts, and monocyte-macrophages, after treatment with IL-1β, TNFα, or lipopolysaccharide (LPS). This induction relies on the translocation of NF-κB, p65/p50, to the nucleus and transactivation of the ESE-1 promoter via a high affinity NF-κB binding site (Grall et al., 2003). Furthermore, following induction, ESE-1 can directly activate transcription of NOS2 (Rudders et al., 2001) and COX2 (Grall et al., 2005) by binding to two or more functional ETS sites in the respective promoters. Together these studies indicate that increased expression of these IL-1β-induced genes is mediated indirectly by NF-κB via activation of ESE-1, which then serves as a primary transcription factor involved in upregulating promoter activity.
In a previous study, we found that the early growth response (EGR)-1 zinc finger protein, after induction and activation by IL-1β, inhibits COL2A1 promoter activity by binding to the –131/+125 bp core promoter and preventing the interactions among CBP, Sp1 and TATA-binding proteins (Tan et al., 2003). Since Egr-1 did not account fully for the suppression of COL2A1 activity, we hypothesized that this early transient response might permit further sustained repression by IL-1β-induced factors that bind to upstream promoter sequences. Furthermore, we could find no functional binding site for NF-κB. In the present study, we found that ESE-1 is induced by IL-1β by an NF-κB-dependent mechanism. ESE-1 binds strongly to the COL2A1 promoter, accounting partially for the IL-1β-dependent inhibition. Of the ETS factors tested, this response was specific to ESE-1, since ESE-3, which was also induced by IL-1β, suppressed COL2A1 promoter activity only weakly. In contrast, overexpression of ETS-1 increased COL2A1 promoter activity and blocked the inhibition by IL-1. These results suggest for the first time a mechanism involving a balance among ETS factors in the control of COL2A1 transcription and a loss of balance during inflammation due to the induction of ESE-1, a factor not expressed normally in cartilage.
Materials and Methods
Cell culture
Articular cartilage was obtained as discarded surgical material with approval by the Institutional Review Board and primary chondrocyte cultures were prepared, as described previously (Goldring, 2004). The immortalized human chondrocyte cell lines T/C-28a2 and C-28/I2 (Goldring et al., 1994a) were cultured in Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 (1/1, v/v; Life Technologies) containing 10% fetal calf serum (FCS; Biowhitaker, Walkersville, MD) and passaged using trypsin–EDTA solution (Invitrogen, Carlsbad, CA) at >95% confluency every 5–6 days. For experiments, confluent monolayer cultures were changed to medium containing 1% Nutridoma-SP (Roche Applied Science, Indianapolis, IN) and incubated overnight prior to treatment with IL-1β at 200–500 pg/ml for the times indicated.
Adenoviral infection
Adenoviral infections were performed in subconfluent C-28/I2 cultures 2 or 3 days after passaging. Transduction with adenovirus was performed for 90 min using a multiplicity of infection of 1:125 in a 1-ml volume of serum-free medium per well in six-well plates. After infection the cells were incubated for 4 h in medium containing 10% FCS. The cultures were then changed to medium containing 1% Nutridoma, incubated for 1 h, and treated with IL-1β for 18 h. The IκBα adenovirus was kindly provided by Dr. Fionula Brennan (Foxwell et al., 1998).
RNA extraction and RT-PCR analysis
Total RNA was isolated by a one-step extraction procedure using the RNeasy Mini Kit (Qiagen, Valencia, CA), and 1.0 μg was reversed transcribed in 20 μl containing final concentrations of 2.4 IU/μl of MuLv Reverse Transcriptase (RT), 2.5 μM of oligo d(T)16, and 1 U/μl of RNase Inhibitor, all obtained from PerkinElmer Life Sciences, as described previously (Robbins et al., 2000). The primers for human ESE-1 were 5′-ACCTGGATCCCACTGAT-GGCAAGCTC-3′ (sense) and 5′-CCGACTCTGGAGAACCTC-TTCCTCC-3′ (antisense) derived from the mRNA sequence (GenBankTM accession U73844) (Oettgen et al., 1997). The primers for human ESE-2 were 5′-CTGCCTTCTCTTGCCTT-GAAAGCC-3′ (sense) and 3′-ATTGAAAGTACAGGTAC-CGCCGC-3′ (antisense) (Oettgen et al., 1999b). The primers for human ESE-3 were 5′-CCTGGACACCAACCAGCTGGATGC-3′ (sense) and 5′-CCTGAAGACGCCCTCAGATCGGTC-3′(antisense) (Kas et al., 2000). The primers for COL2A1, COX2, NOS2, and GAPDH were described previously (Robbins et al., 2000). The PCR amplification mixture contained 2 μl of the RT product in a final volume of 50 μl containing 1 mM MgCl2, 200 μM dNTPs, 0.2 μM of each sense and antisense primers, and 2.5 U of Taq DNA polymerase (Promega, Madison, WI). Within an experiment, the same RT mix was used for the different PCR reactions to enable comparisons, and amplification curves were performed between 25 and 35 cycles to assure assay of products in the exponential phase, using the MJR Research PTC-200 Peltier thermal cycler. Following an initial step at 95°C for 2 min, each cycle consisted of 30 sec of denaturation at 95°C, 30 sec of annealing at 56°C, and 30 sec of extension at 72°C, with a final extension at 72°C for 7 min. The PCR products, 30 μl of PCR reaction per well, were separated on 1.5% agarose gels.
Real-time RT-PCR
Total RNA and the cDNAs were generated as described above. SYBR Green I-based real-time PCR was carried out on a MJ Research DNA Engine Opticon Continuous Fluorescence Detection System (MJ Research Inc., Waltham, MA). All PCR mixtures contained PCR buffer (final concentration, 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 2 mM MgCl2, and 0.1% Triton X-100), 250 μM deoxynucleoside triphosphate (Roche Molecular Biochemicals), 0.5 μM of each PCR primer, 0.5× SYBR Green I (Molecular Probes, Carlsbad, CA), 5% DMSO, and 1 unit of Taq DNA polymerase (Promega) with 2 μl of cDNA in a 25-μl final volume reaction mix. The samples were loaded into wells of Low Profile 96-well microplates. After an initial denaturation step for 3 min at 95°C, conditions for cycling were 39 cycles of 30 sec at 95°C, 30 sec at 56°C, and 1 min at 72°C. After incubation for 5 sec at 79°C following the extension step, the fluorescence signal was measured immediately to eliminate possible primer-dimer detection. At the end of the PCR cycles, a melting curve was generated to identify specificity of the PCR product. For each run, serial dilutions of GAPDH plasmids were used as standards for quantitative measurement of the amount of amplified DNA. For normalization of each sample, GAPDH primers were used to measure the amount of GAPDH cDNA. All samples were run in triplicate, and the data were calculated as the ratio of ESE-1 or COL2A1: GAPDH. The primers used for real time PCR are as follows: human GAPDH, 5′-CAAAGTTGTCATGGATGACC-3′ (sense) and 5′-CCATGGAGAAGGCTGGGG-3′ (antisense); human ESE1, 5′-CTGAGCAAAGAGTACTGGGACTGTC-3′(sense) and 5′-CCATAGTTGGGCCACAGCCTCGGAGC-3′ (antisense); human COL2A1, 5′-CAACACTGCCAACGTCCAGAT-3′ (sense) and 5′-CTGCTTCGTCCAGATAGGCAAT-3′ (antisense).
Luciferase reporter constructs and expression plasmids
The COL2A1 sequence spanning –577 to +3,428 bp (Ryan et al., 1990; Goldring et al., 1994b) was cloned into the pGL2-basic (pGL2B) luciferase reporter gene vector (Promega) to generate the pGL2-COL2 construct containing approximately 4 kb of 5′-flanking sequence. Mutation of the translation start site ATG to CTG in the pGL2-COL2 construct did not increase the activity indicating that splicing occurred correctly and did not interfere with translation of the luciferase reporter. Deletion constructs containing –577/+125 bp, –403/+125 bp, –131/+125 bp, –90/+125 bp were prepared by restriction enzyme digestion of pGL2-COL2 at PstI, NheI, NcoI, ApaI, and PvuII sites, respectively, followed by ligation. Mutations in ETS sites in the pGL2-COL2 construct were generated using the PCR-based QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Luciferase reporter plasmids were prepared for transfection using the EndoFree plasmid maxi kit (Qiagen). The expression vectors encoding ESE-1, ETS-1, other ETS factors, DN-ESE-1, DN-Egr-1, and Runx2 were described previously (Oettgen et al., 1999a,b; Tan et al., 2003; Brown et al., 2004; Wang et al., 2004; Grall et al., 2005; Ijiri et al., 2005). The sequences of all constructs were confirmed by DNA sequencing performed at the Beth Israel Deaconess Medical Center DNA sequencing facility using the ABI Prism BigDye™Primer Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and the Automatic DNA Sequencer Model 373A (Applied Biosystems).
Transient transfections and luciferase assays
Transient transfection experiments were carried out in C-28/I2 cells using LipofectAMINE PLUS™ Reagent (Invitrogen). Cells were seeded in six-well tissue culture plates at 2.0 × 105 cells/well in DMEM/F12 containing 10% FCS and incubated for 2–3 days with medium changes at 24 and 3 h prior to transfection. For each well, 0.8 μg of plasmid DNA (600 ng of COL2A1-luciferase reporter plasmid and up to 200 ng of expression vector plus empty vector DNA), 6 μl of PLUS reagent and 92 μl of serum-free DMEM/F12 were mixed and incubated for 15 min at room temperature. LipofectAMINE+ reagent (4 μl) in 100 μl of serum-free medium was then added to each reaction mixture and incubation was continued for an additional 30 min at room temperature. Finally, the transfection mixture was combined with 800 μl of serum-free medium and the lipid-nucleic acid complex was transferred to the washed cell monolayer in each well. After incubation for 4 h at 37°C, the transfection mix was diluted with an equal volume of DMEM/F12 containing 2% Nutridoma-SP, and incubation was continued for 18 h in the absence or presence of IL-1β. For cotransfections, the cells were incubated for 24 h to permit expression of recombinant proteins prior to treatment with IL-1β for a further 18 h. Cell lysates were prepared by extraction with 200 μl of Reporter Lysis Buffer (Promega) and the protein content was determined using the Coomassie Plus Protein Assay Reagent (Pierce Chemical Company, Rockford, IL). Luciferase activities were determined by chemiluminescence assay using the Autolumat LB953 luminometer (EG&G Berthold, Oak Ridge, TN), normalized to the amount of protein and expressed as relative activities against that of untreated pGL2-COL2 in each experiment. Each experiment was repeated at least three times and each data point was calculated as the mean (3–6 wells per experiment) standard deviation. Cotransfection with pRL-TK Renilla luciferase control vector was used only for determining transfection efficiencies of new plasmid preparations, since many commonly used vectors contain binding sites for ETS and other ubiquitous transcription factors and potential artifacts with this technique have been reported (Farr and Roman, 1992). The relative activity of each preparation of wild-type or mutant construct was checked by the Dual-Luciferase Reporter Assay (Promega) using 600 ng of COL2A1-luciferase vector and 20 ng of the pRL-TK Renilla luciferase control vector. The levels of luciferase activity, expressed as relative light U/μg of protein, ranged from ~1 × 103 to 1 × 105 for COL2A1 promoter activity in untreated C-28/I2 cells.
Preparation of siRNA-ESE1 and siRNA-ETS1
The siRNA oligonucleotides for ESE1 (sense: GCACUCCCUAAUUUAUGUGtt; antisense: CACAUAAAUUAGGGAGUGCtc) were Silencer pre-designed from Ambion (Elf3: Cat# 16704, siRNA ID# 114424). The siRNA oligonucleotides for ETS1, (sense: ACUUGCUACCAUCCCGUAC-dTT; antisense: GUACGGGAUGGUAGCAAGU-dTT) and control siRNAs (sense, AGGAGAUAUUUCGAGGCUU-dTT; antisense, AAGCUCGAAAUAUCUCCU-dTT) were obtained from Invitrogen and annealed as reported by Ito et al. (2004). After 4 h of transfection with reporter gene as described above, 40 nM of siRNA was added in each well for another 24 h. For verification, protein extracts from cells transfected with ESE1-Flag or ETS1-Flag and the respective siRNA were collected and analyzed by Western blotting with anti-Flag antibody.
Electrophoretic mobility shift assay (EMSA) and supershift analysis
EMSAs were performed using 2 μl of in vitro translation product of pCI-ESE1, prepared as described (Oettgen et al., 1997) using the TNT Quick Coupled Transcription/Translation System (Promega), and radiolabeled oligonucleotides encompassing the ESE-1 binding site in the NOS2 promoter (Rudders et al., 2001) or the wild-type COL2A1 promoter or mutant sequences containing putative ETS binding sites, listed in Figure 1. Double-stranded DNA oligonucleotides were end-labeled using T4 polynucleotide kinase and [α-32P]dATP. Binding reactions were carried out for 30 min at room temperature using 0.8 pmol (~10,000 cpm) of labeled probe in a final volume of 20 μl containing 10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM EDTA, 10 mM KCl, 1 mM dithiothreitol (DTT), 10% glycerol, 0.1 μg bovine serum albumin (BSA), and 5 ng poly(dI-dC). Unlabeled wild-type oligonucleotides were added as competitors at 50- or 100-fold excess. For supershift, 1 μl of rabbit polyclonal anti-ESE-1 antibody raised against a glutathione S-transferase fusion protein of the N terminus of human ESE-1 (East Acres Biologicals, Southbridge, MA), as reported previously (Brown et al., 2004) was incubated with the binding reaction mixture for 30 min at room temperature before electrophoresis. The binding reactions were blocked using the rabbit polyclonal antibody against ESE-1 from Orbigen, (catalog #pad-10221) (Grall et al., 2005). The protein-DNA complexes were separated on 4.2% polyacrylamide gels using 0.5× Tris borate-EDTA buffer (TBE) buffer (45 mM Tris borate, pH 8.3, and 1 mM EDTA) and visualized by autoradiography.
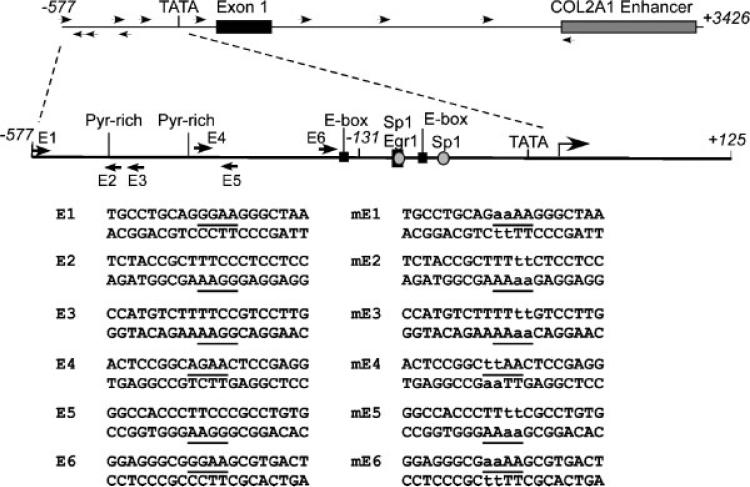
Fig. 1.
Potential ESE-1 binding sites in the COL2A1 promoter. The structure of the COL2A1 regulatory region spanning –577 to +3,426 bp is shown at top with potential ETS binding site indicated with arrows. The previously identified Sp1, Egr-1/Sp1, E-box, and TATA-box sites are represented schematically within the –577/+125 bp promoter. The oligonucleotides that were used for EMSAs are listed below with putative ETS binding sites underlined (left) and the corresponding mutant oligonucleotides (right).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY). Briefly, the C-28/I2 chondrocytes were plated on 150-cm dishes, transfected with either pcDNA3Flag-ESE1 or pcDNA3Flag, incubated for 24 h, or stimulated with IL-1β (0.5 ng/ml) for 6 h. The cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and washed twice with ice-cold PBS containing protease inhibitors (protease inhibitor cocktail tablets from Roche). After the cells were scraped into a conical tube and centrifuged at 700g for 4 min at 4°C, cell pellets were resuspended in 1 ml of cell lysis buffer, 5 mM Pipes (KOH), pH 8.0/85 mM KCl/0.5% NP-40, containing the following protease inhibitors 1 μg/ml leupeptin, 1 μg/ml aprotinin and 1 mM PMSF and incubated on ice for 10 min. Nuclei are pelleted by centrifugation (5,000 rpm for 5 min), resuspended in 400 μl of SDS lysis buffer, and incubated on ice for 10 min. The lysates were sonicated to shear the DNA to lengths of about 600 bp with an optimized condition on power setting 4.5 using a 550 Sonic Dismembrator (Fisher Scientific, Pittsburgh, PA) for 5 sec pulses, repeated 6 times. For immunoprecipitation, the lysates were incubated with anti-Flag antibody or rabbit polyclonal antibody against ESE-1 (Orbigen, San Diego, CA) at 4°C overnight. Following immunoprecipitation, beads were washed once with low-salt immune complex wash buffer, once with high-salt immune complex wash buffer, once with LiCl immune complex wash buffer, and twice with TE buffer. Crosslinked histone-DNA complexes were eluted from the antibody by adding 250 μl of fresh elution buffer (1% SDS, 0.1 M NaHCO3). After centrifugation, the supernatant was collected. Cross-linked histone-DNA complexes were separated by adding 20μl of 5 M NaCl and heating at 65°C for 4 h. DNA was recovered using the MinElute PCR Purification Kit (catalog 28004; Qiagen). The final DNA preparations were subjected to PCR using the following set of primers: 5′-GCTAGCCTGTCTCCAAGATT-3′ (sense) and 5′-TTTCGAGGCTGGCGAACTC-3′ (antisense), to amplify 497 bp of the human COL2A1 promoter.
Immunohistochemistry
Multiple areas (0.5 × 0.5 cm2) of articular cartilage from two asymptomatic hip joints obtained at joint replacement due to bone fracture and from five knee or hip joints obtained at joint replacement because of symptomatic OA were used for immunohistochemical staining. The samples were fixed in 4% paraformaldehyde for 4 h at room temperature and paraffin embedded. Serial sections of 8 μm were cut and every fiftieth section was collected (from lateral to medial sides). Sections were deparaffinized, subjected to microwave antigen retrieval in 10 mM EDTA, pH 7.5, at 93°C for 10 min, and allowed to cool in a running water bath for 15 min. Protein block (DAKO x0909) was applied and immunostaining was performed using a rabbit polyclonal antibody against ESE-1 (1:1,000 dilution; from Orbigen, catalog #pad-10221), biotinylated anti-rabbit IgG secondary antibody, and the Vectastain Elite ABC kit (Vector Laboratories, Inc., Burlingame, CA PK-6101). The sections were counterstained with methyl green (S1962, DAKOCytomation). For negative controls, normal rabbit IgG (sc-2027) was used in place of the primary antibody.
Results
The suppression of COL2A1 gene expression by IL-1β requires NF-κB and is associated with NF-κB-dependent induction of ESE-1 mRNA in human immortalized and primary chondrocytes
Although previous studies have suggested a role for NF-κB in cytokine-mediated suppression of COL2A1 transcription (Murakami et al., 2000; Seguin and Bernier, 2003), direct regulation via a binding site responsive to NF-κB in the COL2A1 promoter has not been demonstrated. The COL2A1 promoter, however, contains several potential binding sites for ETS factors, as shown in Figure 1. We showed previously that IL-1β or TNF-a could stimulate the expression of ESE-1 in human synovial fibroblasts, osteoblasts and monocytes, as well as in several immortalized human chondrocyte cell lines (Grall et al., 2003). Since ESE-1 mediates, at least partially, the induction of IL-1β-responsive genes by a mechanism requiring the activation of NF-κB (Rudders et al., 2001; Grall et al., 2003, 2005), we asked whether a similar mechanism could account for the transcriptional suppression of COL2A1 expression by IL-1β. Subconfluent cultures of C-28/I2 chondrocytes were infected with the adenoviral vector encoding IκBα, which blocks NF-κB activation and translocation to the nucleus, and incubated for 4 h prior to treatment with IL-1β for 18 h. Adenoviral overexpression of IκB blocked the IL-1β-induced suppression of COL2A1 mRNA and the IL-1β-mediated induction of ESE-1 mRNA (Fig. 2A). These results indicate that the suppression of COL2A1 and the induction of ESE-1 gene expression by IL-1β require NF-κB as an intermediate.
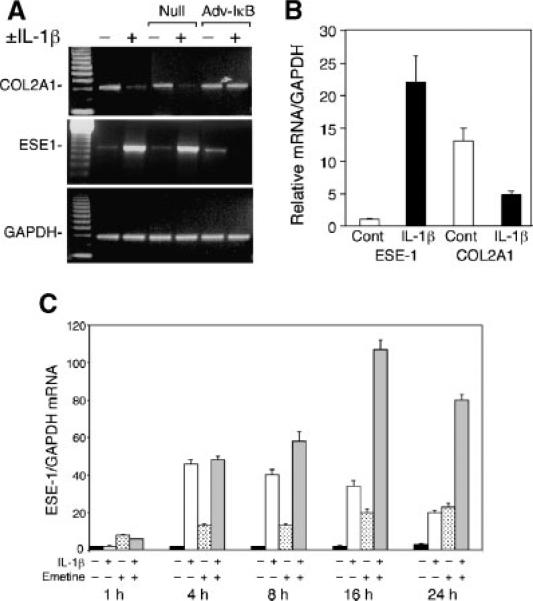
Fig. 2.
The suppression of COL2A1 gene expression by IL-1β requires NF-κB and is associated with NF-κB-dependent induction of ESE-1 mRNA. A: Cultures of the immortalized human chondrocyte cell line, C-28/I2,were infected with adenovirus expressing IκB (adv-IkB) or, as a null control, adv-βgal, incubated for 4 h in serum-containing medium, changed to serum-free medium containing 1% Nutridoma, and treated with IL-1β (500 pg/ml) for 18 h. Total RNA was extracted and analyzed by RT-PCR using specific primers to detect COL2A1, ESE1, and GAPDH mRNAs. B: Primary cultures of human adult articular chondrocytes at day 5 after isolation from the cartilage were treated with IL-1β for 18h, and ESE1 and COL2A1 mRNA were analyzed by real-time RT-PCR. C: The C-28/I2 cells were cultured in serum-containing culture medium for 5 days, and confluent cultures were changed to serum-free medium containing 1% Nutridoma 24 h before treatment with IL-1β for the times indicated. Emetine (10 μg/ml) was also added to the indicated cultures. Total RNA extracts were analyzed for ESE1 mRNA expression by real-time PCR.
To verify the induction of ESE-1 gene expression in chondrocytes isolated from human articular cartilage, we treated chondrocytes at 5–7 days of primary culture with IL-1β for 18 h and analyzed ESE-1 mRNA by real time RT-PCR. As shown in a representative experiment in Figure 2B, ESE1 mRNA was virtually undetectable in untreated primary chondrocytes, but was induced by IL-1β. The increase in ESE1 mRNA was accompanied by the suppression of COL2A1 mRNA. Since the primary chondrocytes were derived from intact cartilage samples from OA patients, these results indicate that the isolated cells do not express high levels of ESE1 mRNA unless stimulated by an inflammatory cytokine. To further examine the mechanism of the IL-1β response, we performed a detailed time course analysis of ESE-1 mRNA expression by real-time PCR using the C-28/I2 chondrocyte cell line. After preincubation in serum-free medium for 24 h, IL-1β was added without medium change and incubation continued for 1, 4, 8, 16, and 24 h (Fig. 2C). Compared to untreated controls at each time point, IL-1β increased the levels of ESE-1 mRNA by 4 h with peak induction between 4 and 8 h, and the IL-1β-stimulated levels declined gradually thereafter without reaching the control level by 24 h. Emetine, a protein synthesis inhibitor without function as a signaling agonist (Edwards and Mahadevan, 1992), enhanced ESE-1 mRNA levels in a gradual, time-dependent manner by more than threefold with sustained expression up to 24 h. When added to some cultures 1 h before the addition of IL-1β, emetine enhanced the IL-1-induced levels of ESE-1 mRNA by 8 h when the IL-1-induced levels had started to decline, with superinduction at 16 h that had begun to decline by 24 h (Fig. 2C). These results indicate that emetine acts as a translational inhibitor to stabilize and superinduce ESE-1 mRNA levels and that the later decrease in the levels of ESE-1 mRNA after the peak induction by IL-1β is due to mRNA instability. In previous studies, we have shown that cycloheximide also superinduces ESE-1 mRNA without reversing the inhibition of COL2A1 mRNA by IL-1β (Grall et al., 2003; Tan et al., 2003). Furthermore, the induction of ESE-1 mRNA by IL-1β precedes the suppression of COL2A1 mRNA but occurs with later and more sustained expression than that of the immediate early gene EGR-1, as described previously (Tan et al., 2003).
ESE-1 decreases COL2A1 promoter activity in a dose-dependent manner and enhances IL-1β-dependent inhibition
We reported previously that IL-1β suppresses COL2A1 promoter activity in both primary and immortalized human chondrocytes in transfection assays using CAT (Goldring et al., 1994a,b) and luciferase (Robbins et al., 2000; Tan et al., 2003) reporter vectors. Note that in previous studies and in the experiments described below, we have consistently observed no more than 50% inhibition by IL-1β of constitutive COL2A1 promoter activity, which represents luciferase activity that has accumulated before the addition of IL-1β. We can enhance the suppression by adding another inhibitory cytokine such as TNFα or IFNγ or by inhibiting IL-1β-induced prostaglandin biosynthesis (Goldring et al., 1994b). IL-1β also induces other factors that may blunt the response.
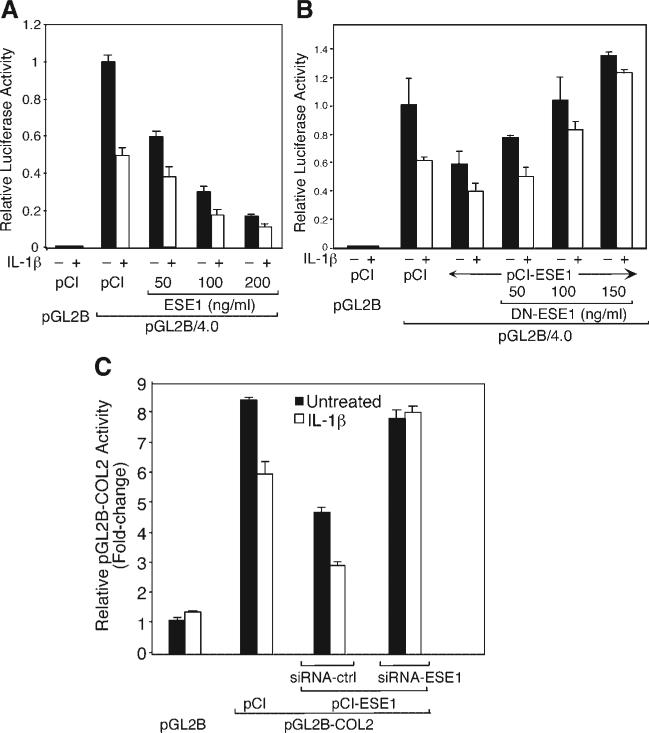
To determine the dependence of the inhibition of COL2A1 promoter activity by IL-1 on ESE-1, we performed cotransfections with pCI-ESE1 in the absence or presence of IL-1β and then determined the extent to which DN-ESE1 could reverse the suppression. As shown in Figure 3A, IL-1β alone suppressed pGL2B-COL2 activity by 50%, and cotransfection with increasing amounts of pCI-ESE1 inhibited COL2A1 promoter activity by up to 80%. In comparison to the suppression by IL-1β in the absence of pCI-ESE-1, the contribution of IL-1β to the inhibition decreased as the amount of ESE-1 increased. This interpretation was further supported by experiments in which co-transfections with pCI-ESE1 at 50 ng/ml, DN-ESE-1 reversed the ESE-1-dependent inhibition in a concentration-dependent manner. DN-ESE1 was less effective against the IL-1β-dependent suppression, approaching complete reversal at 150 ng/ml of the DN vector (Fig. 3B). Finally, siRNA-ESE1 reversed the inhibition of promoter activity in the presence of both IL-l and overexpressed ESE-1 (Fig. 3C). These results show that, by itself, ESE-1 is a potent inhibitor of COL2A1 promoter activity and accounts, at least partially, for the IL-1β-mediated suppression. Although IL-1β induces other transcription factors that suppress COL2A1 promoter activity, high levels of ESE-1 produce such potent inhibition that further effects of other factors may not be observed.
Fig. 3.
ESE-1 decreases COL2A1 promoter activity and enhances IL-1β-dependent inhibition. The C-28/I2 chondrocytes were cotransfected with pGL2B or with pGL2-COL2 together with pCI empty vector or with (A) increasing amounts of pCI-ESE1 (50, 100, and 200 ng/ml), (B) pCI-ESE1 (50 ng/ml) in the absence or presence of increasing amounts of dominant-negative (DN)-ESE1. C: The C-28/I2 cells were cotransfected with pGL2-COL2 and pcDNA3.1 (empty vector) or pcDNA-ESE1 together with siRNA-Ctrl (control) or siRNA-ESE1. Incubations were for 24 h followed by treatment with IL-1β (500 pg/ml) for a further 18 h.
Comparison of ESE-1 and other ETS family members
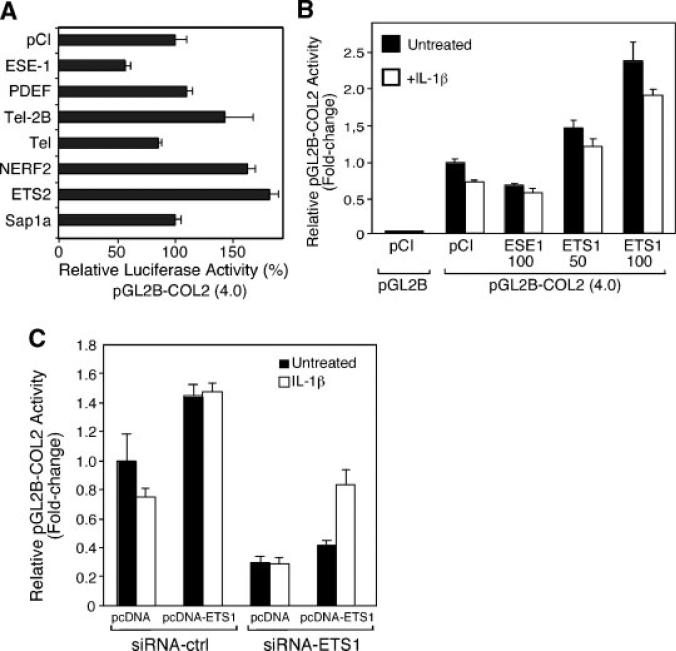
Cotransfection of expression vectors (100 ng/well) encoding various ETS factors showed that ELF-1 (not shown) and TEL produced less than 20% inhibition and PDEF, and SAP-1A had no effect on pGL2-COL2 activity. TEL2B, NERF2, and ETS-2 increased COL2A1 promoter activity, but by less than twofold (Fig. 4A). In additional screens, we had noted that ETS-1 was a more potent inducer of COL2A1 promoter activity than the other ETS factors tested. We, therefore, compared the effects of ETS-1 and ESE-1 in a cotransfection experiment and found that indeed these two ETS factors produced opposite effects. At 100 ng/ml, pCI-ETS1 increased promoter activity by approximately 2.5-fold, and blocked the inhibition by IL-1β compared to the IL-1β-treated pcDNA control (Fig. 4C). Furthermore, in the absence of IL-1β, siRNA-ETS1 decreased promoter activity in either the absence or presence of pcDNA-ETS1 (Fig. 4B). In IL-1-treated cultures, the siRNA-ESE1 further suppressed promoter activity but restored the suppression in the presence of pcDNA-ETS1, compared to the IL-1-treated control (pcDNA + siRNA-Ctrl), suggesting that ETS-1 may be one of the factors blunting the IL-1 inhibitory effect (Fig. 4C). These results indicate that the COL2A1 promoter is a target for transcriptional repression by ESE-1 but that the response may depend upon the balance of intracellular concentrations of other ETS family members in the cell.
Fig. 4.
Comparison of ESE-1 with other ETS family members. C-28/I2 cells were cotransfected with pGL2-COL2 and pCI or expression plasmids encoding (A) various ETS family members (100 ng/well in 12-well plates) or (B) ESE-1 (100 ng/ml) or ETS-1 (50 and 100 ng/ml). C: The C-28/I2 cells were cotransfected with pGL2-COL2 and pcDNA3.1 (empty vector) or pcDNA-ETS1 together with siRNA-Ctrl (control) or siRNA-ETS1. The cells were incubated for 24 h prior to the addition of IL-1β for a further 18 h, cell harvest, and luciferase assay.
IL-1β induces ESE-1 and ESE-3 coordinately with the suppression of COL2A1 mRNA, but ESE-1 is a more potent suppressor of COL2A1 promoter activity than ESE-3
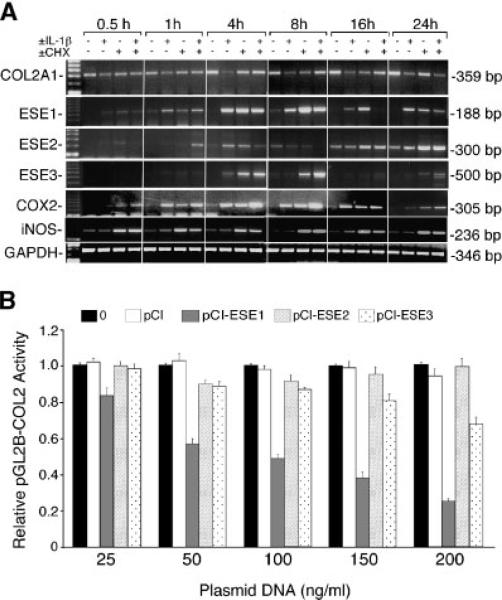
Constitutive levels of COL2A1 mRNA increased with time throughout the 24 h time course. COL2A1 mRNA levels were strongly inhibited by IL-1β by 4 h and the inhibition was maintained up to 24 h (Fig. 5A). Cycloheximide abolished the inhibition at the earlier time points, although it had no effect by 24 h in either the absence or the presence of IL-1β, as we had found previously (Tan et al., 2003; Goldring et al., 1994a,b). Compared to the strong stimulation of ESE-1 mRNA by IL-1β at 4 h that was sustained throughout the 24 h time course, ESE-2 mRNA was not consistently regulated by IL-1β and ESE-3 mRNA was only mildly stimulated at 4 h. Incubation with cycloheximide stabilized mRNA levels at 4 and 8 h for ESE-1 and ESE-3 and at later time points for ESE-2 mRNA. The time courses of stimulation by IL-1β of COX2 and NOS2 mRNAs, which are also inducible by ESE-1 (Rudders et al., 2001; Grall et al., 2005), are shown as positive indicators of the IL-1 response.
Fig. 5.
IL-1β induces ESE-1 and ESE-3 coordinately with the suppression of COL2A1 mRNA but ESE-1 inhibits COL2A1 promoter activity more effectively than ESE-3. A: The C-28/I2 cells were cultured as describe in Figure 2C and treated with IL-1β and cycloheximide alone or together for the times indicated. Total RNA extracts were analyzed by semi-quantitative RT-PCR for COL2A1, ESE1, ESE2, ESE3, and GAPDH mRNAs. COX2 and NOS2 mRNAs were also assayed as positive controls. B: The C-28/I2 cells were cotransfected with pGL2-COL2 together with pCI empty vector or increasing amounts (25, 50, 100, 150, and 200 ng/ml) of pCI-ESE1 or pCI-ESE3.
Since IL-1β increased the levels of ESE-1 and ESE-3 mRNA with a similar time course, we analyzed pGL2-COL2 activity after cotransfection with increasing amounts of plasmid encoding ESE-1 or ESE-3. As shown in Figure 5B, ESE-3 could also inhibit pGL2-COL2 activity, but less effectively than ESE-1. Cotransfection with pCI-ESE1 produced dose-dependent inhibition of pGL2-COL2 similar to that observed in the experiment shown in Figure 3A. Overexpression of ESE-3 also produced dose-dependent inhibition, but to a lesser extent than pCI-ESE1, which produced ~80% inhibition at the highest concentration tested in this experiment (Fig. 5B). Cotransfection of pCI-ESE2 had no significant effect. These results indicate that ESE-1 is a potent suppressor of COL2A1 promoter activity compared to the other ESE isoforms.
The inhibitory responses to IL-1β and ESE-1 colocalize in the COL2A1 promoter region
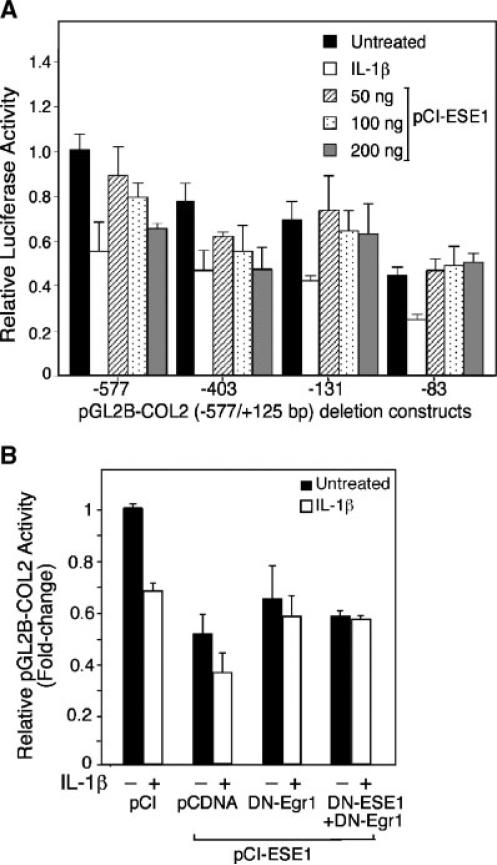
To localize the responses to IL-1β and ESE-1 in the COL2A1 promoter, we made several 5′-deletions of the –577/+127 bp promoter construct, which we had previously determined to be responsive to IL-1β. As shown in Figure 6A, all five promoter constructs responded to IL-1β showing at least 50% inhibition. The activities of the –577/+127 bp and –403/+127 bp constructs were also inhibited in a dose-dependent manner by pCI-ESE1. Cotransfection with the highest amount of pCI-ESE1 used, 200 ng, produced levels of inhibition similar to IL-1β. In contrast, the –131/+127 bp, –83/+127 bp constructs did not respond to pCI-ESE1 either positively or negatively. We showed previously that IL-1β-induced EGR-1 suppresses the activity of the –131 bp promoter region by displacement of Sp1 (Tan et al., 2003). As shown in Figure 6B, DN-Egr1 was unable to reverse the inhibition of pGL2-COL2 activity by pCI-ESE1, but it did reverse the inhibition by IL-1. Note that the 50 ng of DN-ESE1 added together with 50 ng of DN-Egr1 was insufficient to block the suppression by 100 ng of pCI-ESE1. Together, these results indicate that the COL2A1 promoter region upstream of –131 bp contains elements that respond to both IL-1β and ESE-1.
Fig. 6.
ESE-1 decreases activity of the COL2A1 promoter containing the –577/–132 bp region. A: The C-28/I2 cells were cotransfected with pGL2 constructs containing COL2A1 promoter sequences, –577/+125 bp, –403/+125 bp, –131/+125 bp, and –90/+125 bp, together 200 ng of pCI (filled bar) or 50, 100, or 200 ng/ml of pCI-ESE1. B: The cells were cotransfected with pGL2-COL2 and pcDNA3.1 (empty vector) or expression plasmids encoding ESE-1 (100 ng/ml), DN-ESE1 (50 ng/ml), or DN-ESE1 and DN-Egr1, each at 50 ng/ml. The cultures were then incubated for 24 h to permit expression of recombinant proteins. Some cultures were then treated with IL-1β (empty bar) and the incubations continued for a further 18h. All cultures were harvested at the same time for luciferase assay of reporter gene expression. The results show the means ± SD derived from three separate transfection experiments, each done in triplicate.
EMSA and ChIP analysis of binding of ESE-1 to putative ETS sites in the COL2A1 promoter
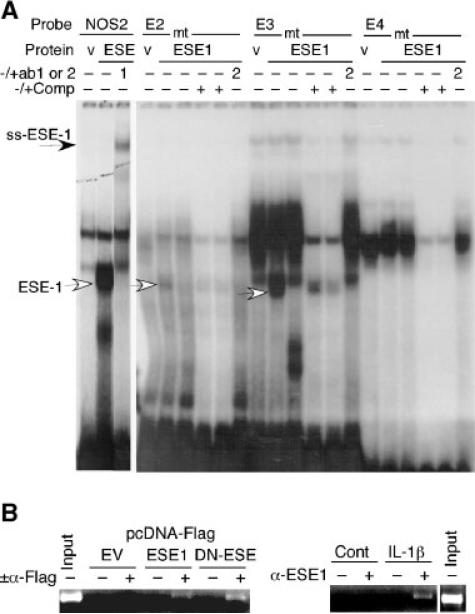
To further define the role of ESE-1 in regulating COL2A1 promoter activity, we evaluated ESE-1 binding to putative ETS sites by gel mobility shift assays. We identified six potential binding sites for ESE-1 in the COL2A1 promoter region spanning –577 to –131 bp at the positions shown in Figure 1. Binding of in vitro-translated ESE-1 protein to radiolabeled oligonucleotides containing the putative ETS sites was compared to binding to the ESE-1 site in the NOS2 probe reported previously (Rudders et al., 2001). Whereas we observed weak binding of ESE-1 to the E2 probe compared to extract made with empty vector (v), ESE-1 bound with high affinity to E3 (Fig. 7A). Furthermore, in vitro-translated ESE-1 did not bind specifically to mutant probes and the addition of 50x or 100x excess unlabeled E2 or E3 oligonucleotide prevented binding to these ESE-1 sites. An ESE-1 antibody (ab), which was different from that used to supershift ESE-1 binding to the NOS2 probe, abolished binding of ESE-1 to the E2 and E3 probes without producing supershifts. However, no ESE-1-specific binding could be observed on the radiolabeled E4 probe (Fig. 7A) or on E1, E5, or E6 (data not shown).
Fig. 7.
ESE-1 binds to ETS sites E2 and E3 within –403/–381 bp. A: End-labeled double-stranded oligonucleotides containing the ESE-1 consensus (NOS2 promoter) or the E2, E3, and E4 oligonucleotides or mutants (see Fig. 1) were incubated with in vitro translated protein made with empty vector (v) or pCI-ESE1 (E). Competitor oligonucleotides (at 50× and 100× in adjacent lanes) and antibody were added as indicated and as described in the Methods. Similar background binding of in vitro-translated vector or ESE-1 has been reported on other promoter sequences (Brown et al., 2004; Wang et al., 2004; Grall et al., 2005). Note that the third lane of each set shows binding to mutant (mt) E2, E3, and E4 probes. B: Chromatin immunoprecipitations (ChIP) were performed using C-28/I2 cells transfected with pcDNA3-FLAG empty vector (EV) or pcDNA3-FLAG-ESE1 or -DN-ESE1 (left), or they were treated with IL-1β for 6 h (right). Chromatin proteins were crosslinked to DNA with formaldehyde and purified nucleoprotein complexes were immunoprecipitated using anti (α)-FLAG or α-ESE1 antibody. The precipitated DNA fractions were analyzed by PCR for the presence of the COL2A1 promoter region encompassing the ESE-1 binding sites. The input DNA was used as a positive control for the PCR and water was used as negative control (not shown).
To confirm ESE-1 binding to the COL2A1 promoter in vivo, a chromatin immunoprecipitation experiment was performed. The C-28/I2 cells were transfected with the pcDNA3-FLAG empty vector (EV), or pcDNA3-ESE1 or DN-ESE1, or they were treated with IL-1β for 6 h. The proteins bound to DNA were cross-linked with formaldehyde and cell extracts were prepared by sonication and immunoprecipitated using antibodies against FLAG or ESE-1. As shown in Figure 7B (left part), when the anti-FLAG antibody was used for the chromatin immunoprecipitation and PCR was performed using oligonucleotide primers encompassing the ESE-1 binding sites, a specific fragment was detected in the precipitate from cells transfected with pcDNA3-FLAG-ESE1 that was not detected in cells transfected with the empty vector. Since the DN-ESE contains the DNA-binding domain, it was also immunoprecipitated with the FLAG antibody.
Furthermore, the ESE-1-specific band corresponded to the size of the DNA fragment recovered from cells treated with IL-1β (Fig. 7B, right part) These results demonstrate that ESE-1 binds directly to the COL2A1 promoter region containing the ESE-1 binding sites and that IL-1β induces binding of endogenous ESE-1 to the same site in vivo.
Immunohistochemical localization of ESE-1 in chondrocytes in vivo in cartilage
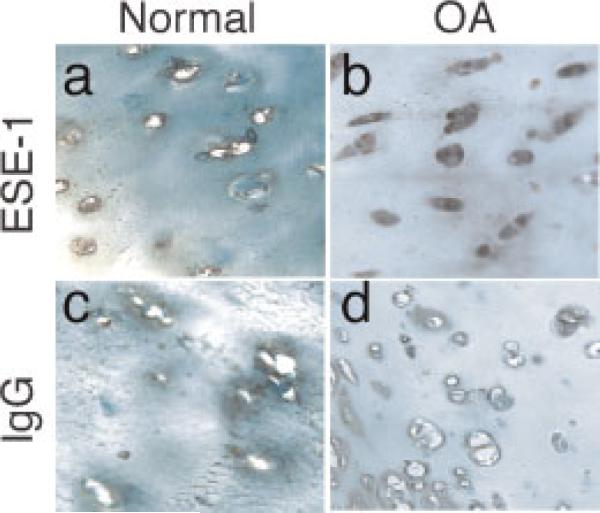
We showed previously that ESE-1 is expressed in rheumatoid synovium and to a lesser extent in synovium from patients with osteoarthritis (OA) showing histological evidence of inflammation (Grall et al., 2003). To determine whether ESE-1 is expressed by chondrocytes in vivo, we analyzed ESE-1 protein in human cartilage by immunohistochemistry, as shown in Figure 8. As shown in a representative human OA cartilage specimen, ESE-1 protein expression was cell-associated in the cytoplasm and nuclei of chondrocytes distributed throughout the cartilage, but predominantly in the deep zone and in the intact superficial and middle zones near the degraded cartilage lesion. In contrast, normal cartilage derived from adult donors with no histological evidence of OA showed no staining.
Fig. 8.
Immunolocalization of ESE-1 in chondrocytes in vivo associated with osteoarthritis but not in normal articular cartilage and induction of ESE-1 mRNA by IL-1β in primary human articular chondrocytes. Samples of articular cartilage from individuals without apparent joint destruction (normal; a and c) and from patients with osteoarthritis (OA; b and d) were subjected to immunohistochemical staining using an antibody against human ESE-1 or isotype-matched IgG as negative control. Positive staining for ESE-1 appears as a brown color in the cytoplasm and nuclei of chondrocytes in this representative sample of OA cartilage. (Original magnifications, 40×). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Staining of serial sections of OA cartilage with isotype control antibody was also negative. Although ESE-1 is restricted to epithelial tissues under normal physiological conditions, these results indicate that this transcription factor is expressed in chondrocytes that have been subjected to inflammation or stress in vivo.
Discussion
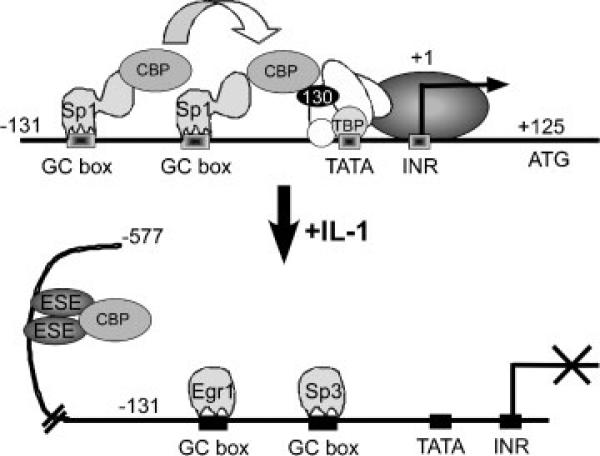
Our results indicate that ESE-1 acts as a direct repressor of COL2A1 promoter activity via binding to at least two tandem ETS sites upstream of –131 bp and accounts, in part, for the sustained suppression by IL-1β. IL-1 treatment of chondrocytes increases the ratio of Sp3/Sp1 binding to the Sp1 sites (Chadjichristos et al., 2003), and we found previously that IL-1β-induced and -activated EGR-1 inhibits COL2A1 transcription by binding to the –131/+125 bp core promoter and displacing Sp1 from at least one of the overlapping GGGCG boxes (Tan et al., 2003). Since overexpression of CBP reverses the inhibition, we proposed that EGR-1, an immediate early response factor, acts by disruption of the interactions among Sp1, CBP, and TATA-binding proteins (Tan et al., 2003). Our study suggested that this early response was transient and that complete transcriptional repression of COL2A1 promoter activity would be dependent upon the binding of other IL-1β-induced factors to upstream promoter sequences. Candidate IL-1-induced transcription factors include C/EBPβ and C/EBPδ, which downregulate the expression of both CD-RAP and COL2A1 (Okazaki et al., 2002), and ESE-1 (Rudders et al., 2001; Grall et al., 2003; Brown et al., 2004), both of which function as activators in the context of cytokine-induced genes such as COX2 (Thomas et al., 2000; Grall et al., 2005). The presence of these factors at low or undetectable concentrations in the cytoplasm, the peak induction of mRNA levels by IL-1β at intermediate or later time points with sustained protein expression, and their ability to interact with CBP suggest involvement in the maintenance of the transcriptional response (Okazaki et al., 2002; Grall et al., 2003; Wang et al., 2004) (Fig. 9).
Fig. 9.
Mechanism of suppression of COL2A1 promoter activity by IL-1β-induced transcription factors.
Early work on gene transcription suggested that NF-κB and AP-1 were primary response factors for the regulation of cytokine-induced genes. Further work showed that AP-1 (c-Jun/c-Fos), one of the first transcription factors studied, was insufficient for IL-1β-induced metalloproteinase (MMP)-1 gene expression by fibroblasts, and that ETS factors played important regulatory roles (Vincenti and Brinckerhoff, 2002). Furthermore, it has not been possible to attribute the regulation of a significant number of cytokine-responsive genes to direct interaction of NFκB or AP-1 with DNA elements, including those in the COL2A1 promoter, which does not contain functional binding sites for these transcription factors. The ETS transcription factors are nuclear effectors of the Ras-Raf-MAPK pathway that require the activation and binding of AP-1 to DNA adjacent to ETS sites on responsive promoters (Wasylyk et al., 1998). Reddy et al. (2003) showed that ESE-1 synergizes with c-Jun/AP-1 to enhance the activity of an epithelial-specific promoter in response phorbol ester. A recent study showed that c-Jun N-terminal kinase and c-Jun/AP-1 mediate IL-1-induced type II collagen mRNA and promoter activity via suppression of Sox9, but direct binding of AP-1 to the COL2A1 promoter was not implicated (Hwang et al., 2005).
Previous studies have shown that IL-1β-induced NF-κB inhibits COL2A1 gene expression by suppressing Sox9 promoter activity (Murakami et al., 2000) or by destabilizing Sox9 mRNA (Sitcheran et al., 2003). Consistent with those findings, adenoviral overexpression of IκB in C-28/I2 chondrocytes blocks the suppression of COL2A1 mRNA by IL-1β. As we showed previously in other cell types (Grall et al., 2003), the induction of ESE-1 mRNA by IL-1β can be blocked in chondrocytes by the overexpression of IkB, and our present results indicate that NF-κB-dependent induction of ESE-1 is involved in the suppression of COL2A1 gene expression by IL-1β. In contrast to the findings of Murakami et al. (2000), however, the constitutive levels of Sox9, L-Sox5, and Sox6 mRNA levels are not suppressed by IL-1β in our model (data not shown), similar to our findings for IFN-γ (Osaki et al., 2003), and overexpression of the three SOX proteins, which bind to the intronic enhancer (Lefebvre et al., 1998), does not reverse the inhibition of COL2A1 activity by IL-1β (manuscript in preparation). Similarly, a recent study also showed that NF-κB is not required for modulation of Sox9-dependent COL2A1 promoter activity by Bcl-2 (Yagi et al., 2005).
ESE-1 has two DNA binding domains, a classical ETS domain, which would bind the ETS sites in the COL2A1 promoter, as shown here, and an A/T hook domain, which is found also in HMG proteins and recognizes the A/T-rich region of double-stranded DNA (Oettgen et al., 1997). The absence of the A/T hook domain in ESE-3 (Kas et al., 2000) could explain why it is a less effective suppressor of COL2A1 promoter activity, even though IL-1β increases the levels of mRNA encoding this ESE family member. Interestingly, ESE-3 decreases MMP-1 promoter activity in lung fibroblasts (Silverman et al., 2002), whereas ESE-1 increases MMP-1 promoter activity in F9 embryonal carcinoma cells in a manner dependent upon the AT-hook and serine-aspartic acid-rich (SAR) domains (Kopp et al., 2007). Our finding that ETS-1 increases COL2A1 promoter activity suggests that ESE-1 may compete for ETS-1 binding sites, but also may block the activities of other ETS factors that maintain constitutive expression. Such differential regulation by ETS factors, such as that observed on the COL11A2 promoter in response to ERG and the Ewing's sarcoma variant EWS/ERG, is dependent upon association with a histone acetylase complex and recruitment to RNA polymerase II (Matsui et al., 2003). ESE-1 stimulates promoter activity of the type II TGFβ receptor gene in differentiated but not undifferentiated F9 cells in a manner distinct from other ETS factors, including ETS-1 (Kopp et al., 2004). The roles played by ETS factors as positive or negative regulators in chondrogenesis, as well as in regulating the expression of MMPs and other collagen genes, suggest that the stage of chondrocyte differentiation, the promoter context, and the relative concentrations of different factors determine the extent of repression or activation of gene transcription by ETS factors (Iwamoto et al., 2000, 2007; Czuwara-Ladykowska et al., 2001; Silverman et al., 2002; Jinnin et al., 2005).
The strong positive staining in the nuclei of chondrocytes in OA but not in normal articular cartilage confirms a pathological role for ESE-1 in non-epithelial tissues, as indicated by our previous studies (Grall et al., 2003; Gravallese et al., 2003). Those studies suggested that ESE-1 is a potent inducer of events associated with inflammation and tissue destruction (Grall et al., 2003; Gravallese et al., 2003; Brown et al., 2004; Grall et al., 2005), whereas Sandell and coworkers (Imamura et al., 2005; Okazaki et al., 2006) have proposed a role for C/EBP proteins as negative regulators of chondrogenesis during skeletal development, where IL-1β is not known to play a role. Our present results indicate that ESE-1 serves a repressor function by binding directly to the COL2A1 promoter and that NF-κB and Egr-1 also participate by indirect and direct mechanisms, respectively. The differential activation of upstream signaling events that result in induction of these transcription factors could explain the synergy and redundancy in cytokine responses. While ESE-1, by itself, is a potent negative regulator of COL2A1 promoter activity, IL-1β also stimulates the production of prostaglandins and bone morphogenetic protein (BMP)-2, which can promote COL2A1 expression and blunt the effects of the negative regulators (Goldring et al., 1994b; Fukui et al., 2003). Thus, in the context of OA cartilage, the initial events that activate chondrocyte synthetic activity probably result in activation of the normally inactive COL2A1 promoter, which would then be susceptible to transcriptional repression depending upon the balance of positive and negative regulators. NF-κB can specifically activate transcription of the gene encoding BMP-2 (Feng et al., 2003), suggesting multiple unexpected feedback regulatory mechanisms. IL-1β also induces the expression of IL-6 that, together with the soluble IL-6 receptor that is not present in sufficient amounts in cultured cells, down-regulates COL2A1, aggrecan, and link protein via the JAK/STAT pathway (Legendre et al., 2003). TGF-β expression is also induced by IL-1β through activation of the bHLH factor AP-4, which binds to a sequence overlapping the AP-1 site (Andriamanalijaona et al., 2003).
Our findings suggest the involvement of multiple IL-1β-induced transcription factors in the suppression of chondrocyte-specific genes. These factors are usually present at low or undetectable concentrations in the cytoplasm and are part of a cascade of cytokine-regulated genes that are induced at intermediate and later time points. Their subsequent sustained expression suggests involvement in maintenance of transcriptional responses. Since these factors also upregulate genes associated with catabolic and inflammatory responses in chondrocytes, including COX2, MMP13, and NOS2, and similar signaling pathways may be induced by adverse mechanical stress, the dissection of the molecular mechanisms involved may lead to an understanding of strategies for blocking destruction and promoting repair of cartilage matrix.
Acknowledgments
We are grateful to Dr. Benjamin Bierbaum, New England Baptist Hospital, for providing human articular cartilage. This study was supported by the National Institutes of Health (R01-AR45378 and R01-AG022021 to M.B.G., R01-AI49527 to T.A.L., and R01-AR47952 to P.O.), Arthritis Foundation, Biomedical Science Grant (to M.B.G.).
Literature Cited
- Andriamanalijaona R, Felisaz N, Kim SJ, King-Jones K, Lehmann M, Pujol JP, Boumediene K. Mediation of interleukin-1b-induced transforming growth factor b1 expression by activator protein 4 transcription factor in primary cultures of bovine articular chondrocytes: Possible cooperation with activator protein 1. Arthritis Rheum. 2003;48:1569–1581. doi: 10.1002/art.11020. [DOI] [PubMed] [Google Scholar]
- Bau B, Gebhard PM, Haag J, Knorr T, Bartnik E, Aigner T. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46:2648–2657. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- Brown C, Gaspar J, Pettit A, Lee R, Gu X, Wang H, Manning C, Voland C, Goldring SR, Goldring MB, Libermann TA, Gravalllese EM, Oettgen P. ESE-1 is a novel transcriptional mediator of angiopoietin-1 expression in the setting of inflammation. J Biol Chem. 2004;279:12794–12803. doi: 10.1074/jbc.M308593200. [DOI] [PubMed] [Google Scholar]
- Chadjichristos C, Ghayor C, Kypriotou M, Martin G, Renard E, Ala-Kokko L, Suske G, de Crombrugghe B, Pujol JP, Galera P. Sp1 and Sp3 transcription factors mediate interleukin-1 b down-regulation of human type II collagen gene expression in articular chondrocytes. J Biol Chem. 2003;278:39762–39772. doi: 10.1074/jbc.M303541200. [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, Trojanowska M. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J Biol Chem. 2001;276:20839–20848. doi: 10.1074/jbc.M010133200. [DOI] [PubMed] [Google Scholar]
- Edwards DR, Mahadevan LC. Protein synthesis inhibitors differentially superinduce c-fos and c-jun by three distinct mechanisms: Lack of evidence for labile repressors. EMBO J. 1992;11:2415–2424. doi: 10.1002/j.1460-2075.1992.tb05306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr A, Roman A. A pitfall of using a second plasmid to determine transfection efficiency. Nucleic Acids Res. 1992;20:920. doi: 10.1093/nar/20.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JQ, Xing L, Zhang JH, Zhao M, Horn D, Chan J, Boyce BF, Harris SE, Mundy GR, Chen D. NF-kB specifically activates BMP-2 gene expression in growth plate chondrocytes in vivo and in a chondrocyte cell line in vitro. J Biol Chem. 2003;278:29130–29135. doi: 10.1074/jbc.M212296200. [DOI] [PubMed] [Google Scholar]
- Foxwell B, Browne K, Bondeson J, Clarke C, de Martin R, Brennan F, Feldmann M. Efficient adenoviral infection with IkB a reveals that macrophage tumor necrosis factor a production in rheumatoid arthritis is NF-kB dependent. Proc Natl Acad Sci USA. 1998;95:8211–8215. doi: 10.1073/pnas.95.14.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui N, Zhu Y, Maloney WJ, Clohisy J, Sandell LJ. Stimulation of BMP-2 expression by pro-inflammatory cytokines IL-1 and TNF-α in normal and osteoarthritic chondrocytes. J Bone Joint Surg Am. 2003;85-A:59–66. doi: 10.2106/00004623-200300003-00011. [DOI] [PubMed] [Google Scholar]
- Goldring MB. Human chondrocyte cultures as models of cartilage-specific gene regulation. Methods Mol Med. 2004;107:69–96. doi: 10.1385/1-59259-861-7:069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB, Birkhead J, Sandell LJ, Kimura T, Krane SM. Interleukin 1 suppresses expression of cartilage-specific types II and IX collagens and increases types I and III collagens in human chondrocytes. J Clin Invest. 1988;82:2026–2037. doi: 10.1172/JCI113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB, Birkhead JR, Suen L-F, Yamin R, Mizuno S, Glowacki J, Arbiser JL, Apperley JF. Interleukin-1b-modulated gene expression in immortalized human chondrocytes. J Clin Invest. 1994a;94:2307–2316. doi: 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring MB, Fukuo K, Birkhead JR, Dudek E, Sandell LJ. Transcriptional suppression by interleukin-1 and interferon-g of type II collagen gene expression in human chondrocytes. J Cell Biochem. 1994b;54:85–99. doi: 10.1002/jcb.240540110. [DOI] [PubMed] [Google Scholar]
- Grall F, Gu X, Tan L, Cho J-Y, Inan MS, Pettit A, Thamrongsak U, Choy BK, Manning C, Akbarali Y, Zerbini L, Rudders S, Goldring SR, Gravallese EM, Oettgen P, Goldring MB, Libermann TA. Responses to the pro-inflammatory cytokines interleukin-1 and tumor necrosis factor a in cells derived from rheumatoid synovium and other joint tissues involve NF kB-mediated induction of the Ets transcription factor ESE-1. Arthritis Rheum. 2003;48:1249–1260. doi: 10.1002/art.10942. [DOI] [PubMed] [Google Scholar]
- Grall FT, Prall WC, Wei W, Gu X, Cho JY, Choy BK, Zerbini LF, Inan MS, Goldring SR, Gravallese EM, Goldring MB, Oettgen P, Libermann TA. The Ets transcription factor ESE-1 mediates induction of the COX-2 gene by LPS in monocytes. FEBS J. 2005;272:1676–1687. doi: 10.1111/j.1742-4658.2005.04592.x. [DOI] [PubMed] [Google Scholar]
- Gravallese EM, Pettit AR, Lee R, Madore R, Manning C, Tsay A, Gaspar J, Goldring MB, Goldring SR, Oettgen P. Angiopoietin-1 is expressed in the synovium of patients with rheumatoid arthritis and is induced by tumour necrosis factor a. Ann Rheum Dis. 2003;62:100–107. doi: 10.1136/ard.62.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, Sawaji Y, Bolton M, Alexander S, Wallace A, Begum S, Wait R, Saklatvala J. Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin bA (activin A), a regulatory molecule for chondrocytes. J Biol Chem. 2004;279:43514–43521. doi: 10.1074/jbc.M407041200. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Yu SS, Poo H, Chun JS. c-Jun/activator protein-1 mediates interleukin-1b-induced dedifferentiation but not cyclooxygenase-2 expression in articular chondrocytes. J Biol Chem. 2005;280:29780–29787. doi: 10.1074/jbc.M411793200. [DOI] [PubMed] [Google Scholar]
- Ijiri K, Zerbini LF, Peng H, Correa RG, Lu B, Walsh N, Zhao Y, Taniguchi N, Huang XL, Otu H, Wang H, Wang JF, Komiya S, Ducy P, Rahman MU, Flavell RA, Gravallese EM, Oettgen P, Libermann TA, Goldring MB. A novel role for GADD45b as a mediator of MMP-13 gene expression during chondrocyte terminal differentiation. J Biol Chem. 2005;280:38544–38555. doi: 10.1074/jbc.M504202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Imamura C, Iwamoto Y, Sandell LJ. Transcriptional co-activators CREB-binding Protein/p300 increase chondrocyte Cd-rap gene expression by multiple mechanisms including sequestration of the repressor CCAAT/enhancer-binding protein. J Biol Chem. 2005;280:16625–16634. doi: 10.1074/jbc.M411469200. [DOI] [PubMed] [Google Scholar]
- Ito H, Duxbury M, Benoit E, Clancy TE, Zinner MJ, Ashley SW, Whang EE. Prostaglandin E2 enhances pancreatic cancer invasiveness through an Ets-1-dependent induction of matrix metalloproteinase-2. Cancer Res. 2004;64:7439–7446. doi: 10.1158/0008-5472.CAN-04-1177. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Higuchi Y, Koyama E, Enomoto-Iwamoto M, Kurisu K, Yeh H, Abrams WR, Rosenbloom J, Pacifici M. Transcription factor ERG variants and functional diversification of chondrocytes during limb long bone development. J Cell Biol. 2000;150:27–40. doi: 10.1083/jcb.150.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Tamamura Y, Koyama E, Komori T, Takeshita N, Williams JA, Nakamura T, Enomoto-Iwamoto M, Pacifici M. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol. 2007;305:40–51. doi: 10.1016/j.ydbio.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Yamane K, Mimura Y, Asano Y, Tamaki K. a2(I) collagen gene regulation by protein kinase C signaling in human dermal fibroblasts. Nucleic Acids Res. 2005;33:1337–1351. doi: 10.1093/nar/gki275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas K, Finger E, Grall F, Gu X, Akbarali Y, Boltax J, Weiss A, Oettgen P, Kapeller R, Libermann TA. ESE-3, a novel member of an epithelium-specific ets transcription factor subfamily, demonstrates different target gene specificity from ESE-1. J Biol Chem. 2000;275:2986–2998. doi: 10.1074/jbc.275.4.2986. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Squires GR, Mousa A, Tanzer M, Zukor DJ, Antoniou J, Feige U, Poole AR. Role of interleukin-1 and tumor necrosis factor a in matrix degradation of human osteoarthritic cartilage. Arthritis Rheum. 2005;52:128–135. doi: 10.1002/art.20776. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Wilder PJ, Desler M, Kim JH, Hou J, Nowling T, Rizzino A. Unique and selective effects of five Ets family members, Elf3, Ets1, Ets2, PEA3, and PU.1, on the promoter of the type II transforming growth factor-b receptor gene. J Biol Chem. 2004;279:19407–19420. doi: 10.1074/jbc.M314115200. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Wilder PJ, Desler M, Kinarsky L, Rizzino A. Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J Biol Chem. 2007;282:3027–3041. doi: 10.1074/jbc.M609907200. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre F, Dudhia J, Pujol JP, Bogdanowicz P. JAK/STAT but not ERK1/ERK2 pathway mediates interleukin (IL)-6/soluble IL-6R down-regulation of Type II collagen, aggrecan core, and link protein transcription in articular chondrocytes. Association with a down-regulation of SOX9 expression. J Biol Chem. 2003;278:2903–2912. doi: 10.1074/jbc.M110773200. [DOI] [PubMed] [Google Scholar]
- Lubberts E, van den Berg WB. Cytokines in the pathogenesis of rheumatoid arthritis and collagen-induced arthritis. Adv Exp Med Biol. 2003;520:194–202. doi: 10.1007/978-1-4615-0171-8_11. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Chansky HA, Barahmand-Pour F, Zielinska-Kwiatkowska A, Tsumaki N, Myoui A, Yoshikawa H, Yang L, Eyre DR. COL11A2 collagen gene transcription is differentially regulated by EWS/ERG sarcoma fusion protein and wild-type ERG. J Biol Chem. 2003;278:11369–11375. doi: 10.1074/jbc.M300164200. [DOI] [PubMed] [Google Scholar]
- Mengshol JA, Vincenti MP, Brinckerhoff CE. IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells: Requirement for Runx-2 and activation by p38 MAPK and JNK pathways. Nucleic Acids Res. 2001;29:4361–4372. doi: 10.1093/nar/29.21.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Lefebvre V, de Crombrugghe B. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-a. J Biol Chem. 2000;275:3687–3692. doi: 10.1074/jbc.275.5.3687. [DOI] [PubMed] [Google Scholar]
- Oettgen P, Alani RM, Barcinski MA, Brown L, Akbarali Y, Boltax J, Kunsch C, Munger K, Libermann TA. Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family. Mol Cell Biol. 1997;17:4419–4433. doi: 10.1128/mcb.17.8.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettgen P, Barcinski M, Boltax J, Stolt P, Akbarali Y, Libermann TA. Genomic organization of the human ELF3 (ESE-1/ESX) gene, a member of the Ets transcription factor family, and identification of a functional promoter. Genomics. 1999a;55:358–362. doi: 10.1006/geno.1998.5681. [DOI] [PubMed] [Google Scholar]
- Oettgen P, Kas K, Dube A, Gu X, Grall F, Thamrongsak U, Akbarali Y, Finger E, Boltax J, Endress G, Munger K, Kunsch C, Libermann TA. Characterization of ESE-2, a novel ESE-1-related Ets transcription factor that is restricted to glandular epithelium and differentiated keratinocytes. J Biol Chem. 1999b;274:29439–29452. doi: 10.1074/jbc.274.41.29439. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Li J, Yu H, Fukui N, Sandell LJ. CCAAT/enhancer-binding proteins b and d mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein Induced by interleukin-1b. J Biol Chem. 2002;270:31526–31533. doi: 10.1074/jbc.M202815200. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Yu H, Davies SR, Imamura T, Sandell LJ. A promoter element of the CD-RAP gene is required for repression of gene expression in non-cartilage tissues in vitro and in vivo. J Cell Biochem. 2006;97:857–868. doi: 10.1002/jcb.20648. [DOI] [PubMed] [Google Scholar]
- Osaki M, Tan L, Choy BK, Yoshida Y, Cheah KS, Auron PE, Goldring MB. The TATA-containing core promoter of the type II collagen gene (COL2A1) is the target of interferong-mediated inhibition in human chondrocytes: Requirement for Stat1 a, Jak1 and Jak2. Biochem J. 2003;369:103–115. doi: 10.1042/BJ20020928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AR. Cartilage in health and disease. In: Koopman W, editor. Arthritis and allied conditions: A rextbook of rheumatology. 15 edition. Lippincott, Williams, and Wilkins; Philadelphia: 2005. pp. 223–269. [Google Scholar]
- Reddy SP, Vuong H, Adiseshaiah P. Interplay between proximal and distal promoter elements is required for squamous differentiation marker induction in the bronchial epithelium: Role for ESE-1. Sp1, and AP-1 proteins. J Biol Chem. 2003;278:21378–21387. doi: 10.1074/jbc.M212258200. [DOI] [PubMed] [Google Scholar]
- Robbins JR, Thomas B, Tan L, Choy B, Arbiser JL, Berenbaum F, Goldring MB. Immortalized human adult articular chondrocytes maintain cartilage-specific phenotype and responses to interleukin-1b. Arthritis Rheum. 2000;43:2189–2201. doi: 10.1002/1529-0131(200010)43:10<2189::AID-ANR6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Rudders S, Gaspar J, Madore R, Voland C, Grall F, Patel A, Pellacani A, Perrella MA, Libermann TA, Oettgen P. ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-kB to regulate the inducible nitric-oxide synthase gene. J Biol Chem. 2001;276:3302–3309. doi: 10.1074/jbc.M006507200. [DOI] [PubMed] [Google Scholar]
- Ryan MC, Sieraski M, Sandell LJ. The human type II procollagen gene: Identification of an additional protein-coding domain and location of potential regulatory sequences in the promoter and first intron. Genomics. 1990;8:41–48. doi: 10.1016/0888-7543(90)90224-i. [DOI] [PubMed] [Google Scholar]
- Sandell LJ, Aigner T. Articular cartilage and changes in arthritis. An introduction: Cell biology of osteoarthritis. Arthritis Res. 2001;3:107–113. doi: 10.1186/ar148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin CA, Bernier SM. TNFa suppresses link protein and type II collagen expression in chondrocytes: Role of MEK1/2 and NF-κB signaling pathways. J Cell Physiol. 2003;197:356–369. doi: 10.1002/jcp.10371. [DOI] [PubMed] [Google Scholar]
- Silverman ES, Baron RM, Palmer LJ, Le L, Hallock A, Subramaniam V, Riese RJ, McKenna MD, Gu X, Libermann TA, Tugores A, Haley KJ, Shore S, Drazen JM, Weiss ST. Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am J Respir Cell Mol Biol. 2002;27:697–704. doi: 10.1165/rcmb.2002-0011OC. [DOI] [PubMed] [Google Scholar]
- Sitcheran R, Cogswell PC, Baldwin AS., Jr NF-κB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes Dev. 2003;17:2368–2373. doi: 10.1101/gad.1114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1b. J Biol Chem. 2003;278:17688–17700. doi: 10.1074/jbc.M301676200. [DOI] [PubMed] [Google Scholar]
- Tchetina EV, Antoniou J, Tanzer M, Zukor DJ, Poole AR. Transforming growth factor-b2 suppresses collagen cleavage in cultured human osteoarthritic cartilage, reduces expression of genes associated with chondrocyte hypertrophy and degradation, and increases prostaglandin E(2) production. Am J Pathol. 2006;168:131–140. doi: 10.2353/ajpath.2006.050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow LC, Adlam DJ, Woolley DE. Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage. Arthritis Rheum. 2001;44:585–594. doi: 10.1002/1529-0131(200103)44:3<585::AID-ANR107>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Thomas B, Berenbaum F, Humbert L, Bian H, Béréziat G, Crofford L, Olivier JL. Critical role of C/EBPd and C/EBPb factors in the stimulation of cyclooxygenase-2 gene transcription by interleukin-1b in articular chondrocytes. Eur J Biochem. 2000;267:1–13. doi: 10.1046/j.1432-1033.2000.01778.x. [DOI] [PubMed] [Google Scholar]
- Tower GB, Coon CI, Belguise K, Chalbos D, Brinckerhoff CE. Fra-1 targets the AP-1 site/2G single nucleotide polymorphism (ETS site) in the MMP-1 promoter. Eur J Biochem. 2003;270:4216–4225. doi: 10.1046/j.1432-1033.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: Integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 2002;4:157–164. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fang R, Cho JY, Libermann TA, Oettgen P. Positive and negative modulation of the transcriptional activity of the ETS factor ESE-1 through interaction with p300, CREB-binding protein, and Ku 70/86. J Biol Chem. 2004;279:25241–25250. doi: 10.1074/jbc.M401356200. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: Nuclear effectors of the Ras-Map-kinase signaling pathway. TIBS. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Yagi R, McBurney D, Horton WE., Jr Bcl-2 positively regulates Sox9-dependent chondrocyte gene expression by suppressing the MEK-ERK1/2 signaling pathway. J Biol Chem. 2005;280:30517–30525. doi: 10.1074/jbc.M502751200. [DOI] [PubMed] [Google Scholar]