Abstract
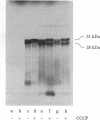
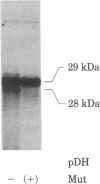
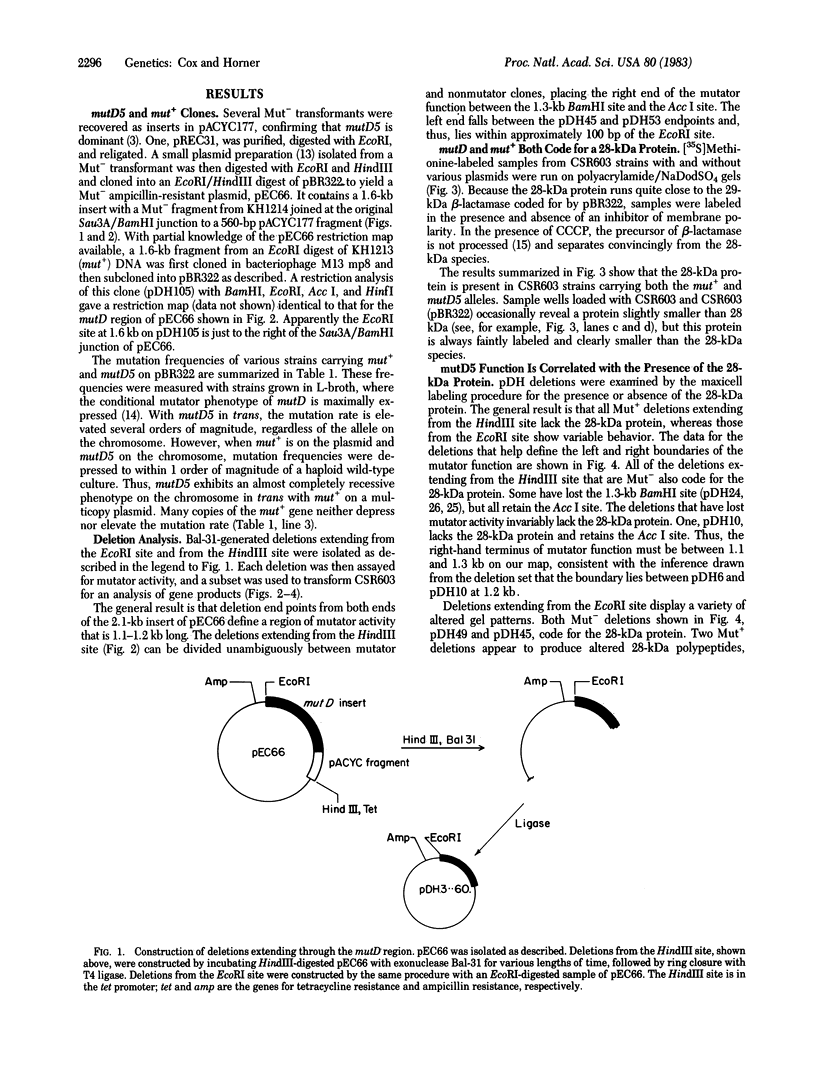
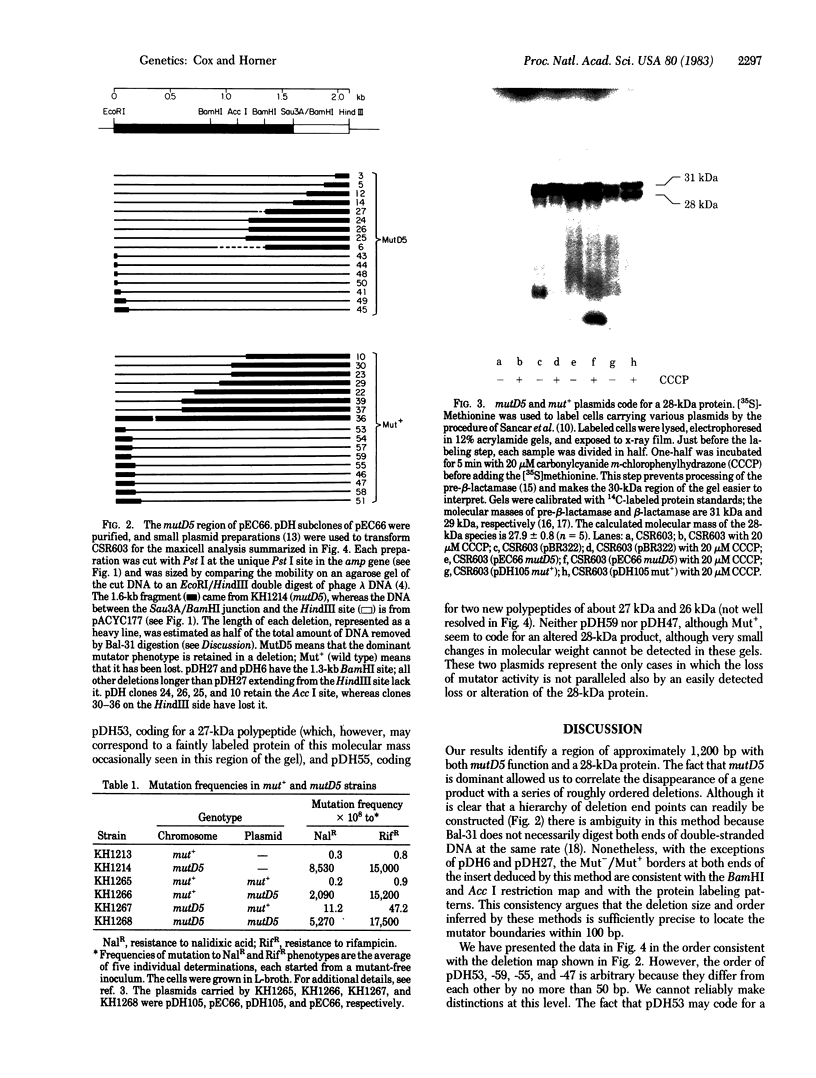
The region of the Escherichia coli chromosome coding for the mutD gene was cloned, mutD5 function resides on a 1.2-kilobase fragment coding for a 28-kilodalton (kDa) protein. A deletion end-point analysis shows that the presence of the 28-kDa protein is required for mutD5 function and suggests that the mutD functional region has sufficient capacity to code for a second polypeptide of approximately 20 kDa. Plasmids carrying the mutD5 and mut+ alleles both produce the 28-kDa species. The product of mutD5 is dominant when carried by single and multicopy plasmids. The product of mut+ is dominant only on multicopy plasmids. Thus, mutD5 exhibits negative complementation. We suggest that the 28-kDa protein participates in a multimeric structure, perhaps at the replication fork.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Scott G. K. Partial amino acid sequence of penicillinase coded by Escherichia coli plasmid R6K. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3732–3736. doi: 10.1073/pnas.75.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. Dominant mutators in Escherichia coli. Genetics. 1982 Jan;100(1):7–18. doi: 10.1093/genetics/100.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C. J., Bole D. G., Quay S. C., Oxender D. L. Role for membrane potential in the secretion of protein into the periplasm of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5396–5400. doi: 10.1073/pnas.78.9.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974 Feb;117(2):477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H. A., Cox E. C. Interaction of an Escherichia coli mutator gene with a deoxyribonucleotide effector. Mol Gen Genet. 1980;178(3):703–708. doi: 10.1007/BF00337881. [DOI] [PubMed] [Google Scholar]
- Fowler R. G., Degnen G. E., Cox E. C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133(3):179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Winston T. P., Hodnett J. L., Legerski R. J., Nees D. W., Wei C. F., Robberson D. L. The extracellular nuclease from Alteromonas espejiana: an enzyme highly specific for nonduplex structure in nominally duplex DNA. Gene Amplif Anal. 1981;2:169–203. [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Maruyama M., Sekiguchi M. Identification of the dnaQ gene product and location of the structural gene for RNase H of Escherichia coli by cloning of the genes. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3770–3774. doi: 10.1073/pnas.78.6.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol Gen Genet. 1978 Jul 25;163(3):277–283. doi: 10.1007/BF00271956. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Maki H., Sekiguchi M. Conditional lethality of Escherichia coli strains carrying dnaE and dnaQ mutations. Mol Gen Genet. 1981;181(1):24–28. doi: 10.1007/BF00339000. [DOI] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci U S A. 1968 Apr;59(4):1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Wharton R. P., Seltzer S., Kacinski B. M., Clarke N. D., Rupp W. D. Identification of the uvrA gene product. J Mol Biol. 1981 May 5;148(1):45–62. doi: 10.1016/0022-2836(81)90234-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of small plasmids in extracts of Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):273–280. doi: 10.1007/BF00325823. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]