Abstract
Methylation of cytosines in the mammalian genome represents a key epigenetic modification and is dynamically regulated during development. Compelling evidence now suggests that dynamic regulation of DNA methylation is mainly achieved through a cyclic enzymatic cascade comprised of cytosine methylation, iterative oxidation of methyl group by TET dioxygenases, and restoration of unmodified cytosines by either replication-dependent dilution or DNA glycosylase-initiated base excision repair. In this review, we discuss the mechanism and function of DNA demethylation in mammalian genomes, focusing particularly on how developmental modulation of the cytosine-modifying pathway is coupled to active reversal of DNA methylation in diverse biological processes.
Introduction
It has long been recognized that cytosines in the genome, as part of the genetic code, also carry epigenetic information through chemical modification of its pyrimidine ring (Holliday and Pugh, 1975; Riggs, 1975). The dual functions associated with cytosines provide a means by which developmental stage- and cell-type-specific epigenetic memory can be directly deposited onto DNA itself (Bird, 2002). Methylation of the fifth position of cytosine (5-methylcytosine, 5mC) is a highly conserved epigenetic modification of DNA found in most plant, animal and fungal models (Law and Jacobsen, 2010), and has a profound impact on genome stability, gene expression, and development (Jaenisch and Bird, 2003; Smith and Meissner, 2013).
In mammals, new DNA methylation pattern is established by de novo DNA methyltransferases, DNMT3A and DNMT3B (Okano et al., 1999; Okano et al., 1998) (Figure 1A–B). Their activity can be modulated by a catalytically inactive family member, DNMT3L (Goll and Bestor, 2005). In somatic cells, 5mC is primarily restricted to palindromic CpG dinucleotides, which are typically methylated in a symmetric manner (de novo methylation in Figure 2A). Methylation of cytosine in non-CpG context (CpH, H=A, T, C) is prevalent in plants (Law and Jacobsen, 2010), but is rare in most mammalian cell-types. Recent work suggests that non-CpG methylation is relatively abundant in oocytes, pluripotent embryonic stem cells (ESCs) and mature neurons (Lister et al., 2013; Lister et al., 2009; Shirane et al., 2013; Xie et al., 2012), but the function of mammalian non-CpG methylation remains unclear. Of the roughly 28 million CpGs in the human genome, 60–80% are methylated in somatic cells (Smith and Meissner, 2013). During mitosis, the global CpG methylation pattern is faithfully maintained in daughter cells through the action of maintenance DNA methyltransferase DNMT1 and its obligate partner, the ubiquitin-like plant homeodomain and RING finger domain 1 (UHRF1), which preferentially recognizes hemi-methylated CpGs (Bostick et al., 2007; Hermann et al., 2004; Sharif et al., 2007) (maintenance methylation in Figure 1A and 2A). Such inheritability of CpG methylation suggests a role for 5mC in long-term epigenetic regulation required for diverse biological processes, such as stable silencing of gene expression, maintenance of genome stability and establishment of genomic imprinting (Bird, 2002).
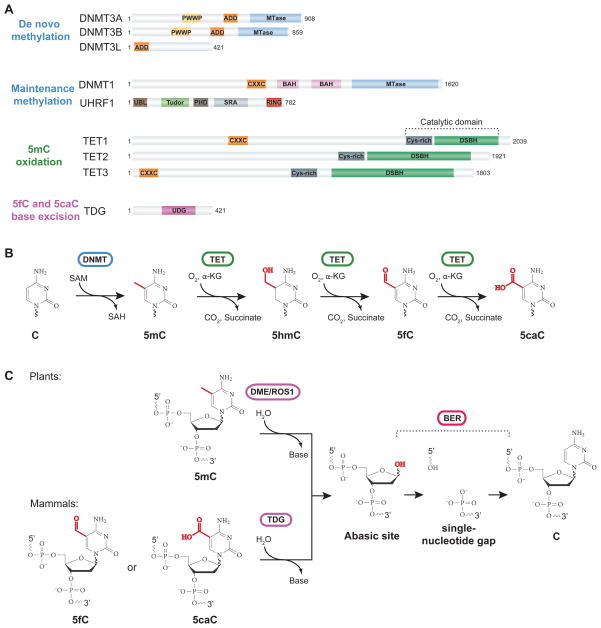
Figure 1. Domain architecture and enzymatic activities of cytosine methylation and demethylation machineries.
(A) Schematic diagrams of predicted functional domains in proteins involved in de novo methylation, maintenance methylation, 5mC oxidation, and 5fC/5caC excision. All TET proteins contain a C-terminal catalytic domain that includes a Cys-rich insert and a large double-stranded β-helix (DSBH) domain. TET1 and TET3 proteins also contain a N-terminal CXXC domain. TDG contains a uracil DNA glycosylase (UDG) domain. The number of amino acids of the full-length isoform is indicated, and the numbering corresponds to mouse proteins. ADD, ATRX-DNMT3-DNMT3L; PWWP, proline-tryptophan-tryptophan-proline motif; MTase, DNA methyltransferase domain; BAH, bromo-adjacent homology; CXXC, zinc-finger Cystien-X-X-Cystine; SRA, SET and RING associated; RING, Really interesting new gene; PHD, Plant homeodomain; UBL, ubiquitin-like domain.
(B) The step-wise cytosine modification pathway that includes cytosine methylation by DNMTs and iterative oxidation of 5mC by TET proteins. DNMTs use S-adenosylmethionine (SAM) as a methyl donor to catalyze methylation at the 5-position of cytosine, yielding S-adenosylhomocysteine (SAH). As Fe(II)/α-ketoglutarate (α-KG)-dependnet dioxygenases, TET proteins use a base-flipping mechanism to flip the target base out of the duplex DNA into the catalytic site. The active site Fe(II) is bound by conserved His-His-Asp residues in TETs and coordinates water and α-KG. TET enzymes use molecular oxygen as a substrate to catalyze oxidative decarboxylation of α−KG, yielding CO2, enzyme-bound succinate, and a reactive high-valent Fe(IV)-oxo intermediate. The enzyme-bound Fe(IV)-oxo intermediate reacts with 5mC/5hmC/5fC to generate 5hmC/5fC/5caC, with a net oxidative transfer of the single oxygen atom to the substrate, resulting in regeneration of the Fe(II) species.
(C) Base excision repair of 5mC (in plants) or 5fC/5caC (in mammals) to complete DNA demethylation. In plants, the DME/ROS1 family of bifunctional DNA glycosylases is capable of catalyzing the release of 5mC base from DNA by cleavage of the N-glycosidic bond, generating an abasic [apurinic and apyrimidinic (AP)] site. The DNA backbone is then nicked by AP lyase activity of DME/ROS1 enzymes. The 3′ sugar group is then cleaved by an AP endonuclease and the resulting single-nucleotide gap is filled in with an unmodified C by DNA polymerase and ligase activities. In mammals, oxidized 5mC bases (5fC/5caC), could be excised by TDG. TDG is a monofunctional DNA glycosylase, so other proteins are required to provide the AP lyase activity to remove the sugar ring to generate a single-nucleotide gap.
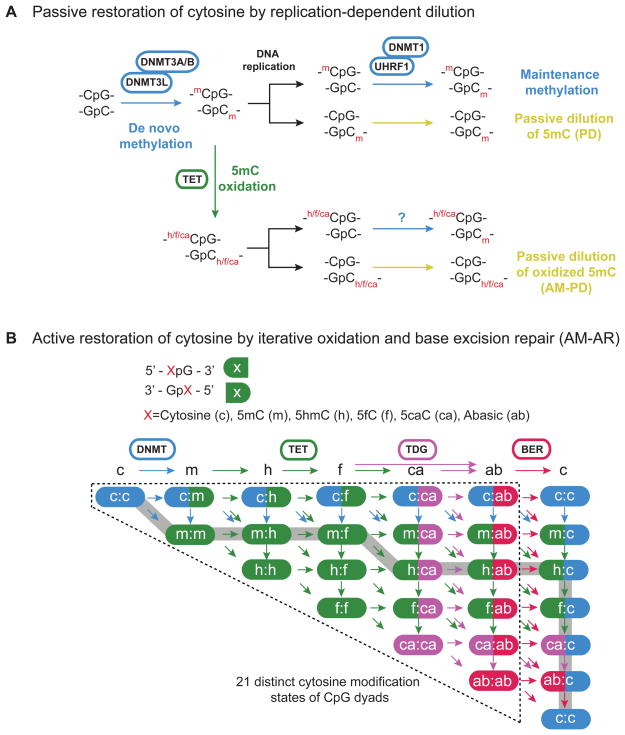
Figure 2. Mechanisms of passive and active reversal of CpG DNA methylation.
(A) Schematic diagrams of replication-dependent passive loss of 5mC or oxidized 5mC within CpG dyads. Replication-dependent passive dilution (PD) of 5mC occurs in the absence of the DNA methylation maintenance machinery (DNMT1/UHRF1). By contrast, 5mC oxidation derivatives, 5hmC (h) [potentially 5fC (f) and 5caC (ca)], may facilitate passive demethylation as hemi-hydroxymethylated CpGs is an inefficient substrate for DNMT1. This form of active DNA demethylation is termed as active modification (AM) followed by passive dilution (AM-PD). Whether DNMT1 or DNMT3 can efficiently methylate CpG dyads hemi-modified with 5fC/5caC is unclear.
(B) Schematic diagrams of replication-independent DNA demethylation within CpG dyads. TET and TDG mediate sequential 5mC oxidation and 5fC/5caC excision. The resulting abasic site is repaired by BER to regenerate unmodified cytosines. All possible intermediate products (a total of 21 distinct cytosine modification states for a single CpG dyad) of the DNMT/TET/TDG/BER enzymatic cascade are depicted. For each state, the left half represents the top strand of CpG dyad, whereas the right half indicates the bottom strand. The arrows represent enzymatic reactions and are color-coded according to that of the corresponding enzyme. One of many potential combinations of intermediate steps for the TET/TDG-mediated active DNA demethylation process is highlighted in gray. Of note, 5fC can be directly excised by TDG to generate abasic site (e.g. f:ca -> ab:ab). For clarity, all f->ab reactions are not depicted.
Although DNA methylation pattern in somatic cells is stably maintained, genome-wide loss of 5mC, or DNA demethylation, has been observed in specific developmental stages such as pre-implantation embryos and developing primordial germ cells (PGCs) (Hajkova et al., 2002; Mayer et al., 2000; Oswald et al., 2000; Sasaki and Matsui, 2008). Global DNA demethylation is important for setting up pluripotent states in early embryos and for erasing parental-origin-specific imprints in developing PGCs (Feng et al., 2010b). Mounting evidence indicates that the rapid erasure of 5mC during these two major waves of epigenetic reprogramming could not be fully explained by replication-dependent passive loss of 5mC, suggesting the existence of enzymatic activities capable of actively removing or modifying methyl groups on cytosines (Wu and Zhang, 2010). However, a unifying mechanistic understanding of active DNA demethylation processes in mammalian cells does not emerge until recently. As we will discuss below, the transformative discovery of Ten-eleven translocation (TET) proteins as 5mC oxidase has provided major insights into mechanisms of active DNA demethylation. The biochemical basis of TET enzymes in oxidative modification of 5mC has recently been reviewed elsewhere (Kohli and Zhang, 2013; Pastor et al., 2013). In this review, we focus on an integrated understanding of mechanisms, genomics and biological functions of mammalian DNA demethylation process. First, we summarize the current mechanistic understanding of passive and active DNA demethylation pathways. Second, we examine the recent advances in development of genomic mapping technologies for 5mC oxidation derivatives as well as the understanding of potential regulatory functions of oxidized cytosine bases in the mammalian genome. Finally, we discuss how distinct modes of DNA demethylation dynamics can be achieved through modulation of the cytosine modifying enzymatic pathway to satisfy the needs of diverse biological processes.
DNA methylation and demethylation in mammals: a cyclic enzymatic cascade
DNA demethylation can take place either passively or actively. Upon DNA replication, the maintenance machinery DNMT1/UHRF1 restores the symmetrical CpG methylation pattern by methylating the unmodified cytosine in the nascent DNA strand (Bostick et al., 2007; Sharif et al., 2007). Passive loss of 5mC can therefore be achieved through successive cycles of DNA replication in the absence of functional DNMT1/UHRF1 [passive dilution of 5mC (PD) in Figure 2A]. One example of replication-dependent passive loss of 5mC is the global erasure of 5mC in the maternal genome during mouse pre-implantation development (see below). Compelling evidence indicates that 5mC can also be rapidly erased independent of DNA replication in specific biological settings in both plants and mammals (Law and Jacobsen, 2010; Wu and Zhang, 2010). Such change involves active DNA demethylation mechanisms, which require one or more enzymatic reactions to modify or remove 5mC. In flowing plants (e.g. Arabidopsis thaliana), active erasure of 5mC is mediated by the Demeter (DME)/repressor of silencing 1 (ROS1) family of DNA glycosylases and base excision repair (BER) machinery (Zhu, 2009) (Figure 1C). However, mammalian orthologs of DME/ROS1 enzymes have not been identified. Mammalian cells may therefore achieve active DNA demethylation using different mechanisms. Early efforts focused on testing various known DNA modifying proteins, such as DNA repair enzymes, cytosine deaminases, and DNA glycosylases (Box 1A). However, experimental evidence for these proposed candidates is often restricted to very specific biological settings, and in many cases the activity cannot be substantiated by follow-up studies (Ooi and Bestor, 2008; Wu and Zhang, 2010). Thus, these studies did not reveal a consensus mechanistic framework for active DNA demethylation in mammals.
Box 1. Known and putative mechanisms of active erasure of 5-methylcytosines.
A. 5mC oxidation-independent active DNA demethylation mechanisms
1. Enzymatic removal of the methyl group from 5mC
The most straightforward way to achieve DNA demethylation is direct removal of the methyl-group. However, this is a thermodynamically unfavorable reaction. One study proposed that methyl-CpG-binding domain protein 2 (MBD2) can catalyze such a reaction leading to the release of methanol, but other laboratories could not reproduce this finding and active erasure of paternal 5mC occurs normally in fertilized oocytes lacking Mbd2 (Wu and Zhang, 2010). To date, no compelling evidence supports that this biochemical reaction could take place under physiological conditions.
2. Nucleotide excision repair to erase 5mC
Excision repair of short genomic regions that contain methylated cytosine nucleotides can indirectly result in erasure of 5mC. There are two major DNA excision repair mechanisms to repair single-stranded DNA damage: base excision repair (BER) and nucleotide excision repair (NER). BER is used to correct damaged bases or mismatched base pairs (e.g. G:T mismatch), whereas NER can repair bulky DNA lesions formed by exposure to radiation or chemicals. Once the bulky DNA lesion is recognized, specific enzymatic activities introduce dual incisions flanking the damage region. The resulting single-stranded gap (usually 24–32 nucleotides) is then filled in by DNA repair polymerases and ligases. The Gadd45 (growth arrest and DNA-damage-inducible protein 45) family of proteins was reported to stimulate active DNA demethylation via NER (Barreto et al., 2007). While some evidence suggests that Gadd45 may induce active DNA demethylation in Xenopus laevis embryos and mammalian cells (Ma et al., 2009; Rai et al., 2008; Schmitz et al., 2009), conflicting results were also reported (Jin et al., 2008). Furthermore, Gadd45a- and Gadd45b-null mice are grossly normal and have no global alteration in DNA methylation levels (Engel et al., 2009; Ma et al., 2009). Thus, the role of Gadd45 in DNA demethylation remains uncertain.
3. Direct base excision repair of 5mC by DNA glycosylases
Active DNA demethylation can be achieved by a DNA glycosylase that efficiently excises 5mC base from DNA. Strong genetic and biochemical evidence supports the use of this mechanism in plants (Zhu, 2009). Mammalian DNA glycosylases, including TDG and methyl-CpG binding domain-containing protein 4 (MBD4), were also proposed to have excision activity against 5mC (Zhu et al., 2000). However, 5mC excision activity of these enzymes is significantly (30–40 fold) lower than their activity for T or U in T•G or U•G mismatches. In addition, Mbd4-null mice are viable and have normal DNA methylation pattern during development. Interestingly, Tdg-null mice die around E12.5 and exhibit modest increase in level of 5mC at some CpG-rich gene promoters (Cortazar et al., 2011; Cortellino et al., 2011). Because TDG, but not MBD4, has robust activity towards 5fC and 5caC, it is likely that 5fC/5caC excision activity, instead of the marginal 5mC excision activity of TDG, is responsible for methylation defects observed in Tdg-deficient cells (see main text).
4. Deamination and repair of 5mC base
Deamination of 5mC generates thymine and the resulting G•T mismatch can be repaired by multiple DNA glycosylases (e.g. TDG) and BER to replace the mismatched T with C. Activation-induced deaminase (AID)/APOBEC-family of cytidine deaminases are known to function in generating mutations in DNA and RNA, which are biologically important for antibody diversification in B cells and for RNA editing (Conticello, 2008). In vitro biochemical analysis suggests that AID/APOBEC could also deaminate 5mC to T (Morgan et al., 2004). However, AID/APOBEC only efficiently acts on single-strand but not double-strand DNA (Bransteitter et al., 2003), and exhibit substantially lower activity on 5mC (10–20 fold) relative to cytosine, their canonical substrate (Nabel et al., 2012). In addition, AID- or APOBEC1-knockout mice exhibit the expected B cell and immunological defects, but are viable and fertile (Wu and Zhang, 2010). Nevertheless, studies in zebrafish embryos (Rai et al., 2008), mouse PGCs (Popp et al., 2010), mouse ESC/human fibroblast fused heterokaryons (Bhutani et al., 2010), and iPSCs (Kumar et al., 2013), support a potential role of AID/APOBEC-mediated 5mC deamination in DNA demethylation. Further studies are needed to clarify the exact biochemical mechanism and function of AID/APOBEC proteins in DNA demethylation. Moreover, De novo DNMTs have been reported to possess in vitro 5mC deaminase activity in the absence of the methyl-donor S-adenosylmethionine (SAM) (Metivier et al., 2008). Given that SAM is relatively abundant in all living cells and is a general methyl-group donor for many essential enzymes, the in vivo relevance of DNMT-mediated deamination of 5mC remains unknown (Ooi and Bestor, 2008).
5. Radical SAM mechanisms
Elongator complex protein 3 (ELP3) was reported to promote paternal genome demethylation in mouse zygotes (Okada et al., 2010). While it is proposed that the Fe-S radical SAM domain of ELP3 may be involved in the demethylation process (Wu and Zhang, 2010), direct biochemical evidence demonstrating the demethylation activity of ELP3 is currently lacking.
B. 5mC oxidation-dependent active DNA demethylation mechanisms
1. Replication-dependent passive dilution of oxidized 5mC
Iterative oxidation of 5mC by TET proteins generates oxidized cytosine bases (5hmC/5fC/5caC). These cytosine analogs may facilitate DNA demethylation by impairing the binding and/or activity of maintenance methylation machinery (DNMT1/UHRF1) during DNA replication. Compelling evidence suggests that this mechanism is used in the erasure of paternal genome methylation during pre-implantation development as well as in the complete demethylation of imprinting control regions in developing PGCs (see main text).
2. DNA repair-mediated excision of modified 5mC
TDG-mediated excision of 5fC/5caC followed by BER
TET proteins further oxidize 5hmC to 5fC and 5caC, which can be efficiently excised by TDG and repaired by BER pathway. Multiple studies have biochemically validated this pathway (see main text). Compelling genetic and genomic evidence suggests that TET/TDG-mediated active DNA demethylation takes place in mouse ESCs, but how extensive this mechanism is utilized in vivo and in other cell-types is currently unknown.
AID/APOBEC-mediated deamination of 5hmC followed by BER
Similar to 5mC deamination, AID/APOBEC proteins may theoretically deaminate 5hmC to generate 5-hydroxyuracil (5hmU). One study has shown that 5hmC deamination by AID/APOBEC may take place in cultured cells and in mouse brain (Guo et al., 2011). The resulting 5hmU•G mismatch can be repaired by DNA glycosylases such as TDG and SMUG1 (Cortellino et al., 2011). However, subsequent studies show that AID/APOBEC proteins exhibit no detectable deaminase activity against 5hmC in in vitro biochemical assays or in cells overexpressing these enzymes (Nabel et al., 2012), challenging the biochemical plausibility of this proposed mechanism. One potential explanation for the negligible activity of AID/APOBEC on 5hmC is that the relatively bulky steric size of 5-position substituent (cytosine [H-] < 5mC [CH3-] < 5hmC [OHCH2-]) may have a significant negative impact on deaminase activity (Nabel et al., 2012).
3. Direct removal of the oxidized 5-position substitute
Dehydroxymethylation by DNMTs
Although direct removal of a methyl group from 5mC is thermodynamically challenging, the removal of the oxidized methyl group is more feasible. In vitro biochemical studies suggest that DNMT3A/3B can remove the hydroxymethyl group of 5hmC to generate unmodified cytosine in the absence of SAM and under oxidizing conditions (Chen et al., 2012; Liutkeviciute et al., 2009). Given that SAM is present at relatively high levels in in all cell types, the physiological relevance of this reaction remains unclear.
Decarboxylation of 5caC
In the thymine salvage pathway, T-to-U conversion is completed by iso-orotate decarboxylase-mediated decarboxylation of 5-carboxyuracil (Smiley et al., 2005). Given the chemical similarity between THase and TET enzymes, it has been proposed that decarboxylation activity may exist in mammalian cells to directly convert 5caC to unmodified cytosine (Wu and Zhang, 2010). One study suggests that 5caC decarboxylase activity may exist in mouse ESC lysates (Schiesser et al., 2012), but the responsible enzyme awaits to be identified.
The breakthrough came in 2009 when the presence of 5-hydroxymethylcytosine (5hmC), an oxidized derivative of 5mC, was unambiguously demonstrated in the mammalian genome and TET family of DNA dioxygenases were identified as the enzyme that convert 5mC to 5hmC (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Subsequent studies have revealed that TET proteins can further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) (He et al., 2011; Ito et al., 2011; Pfaffeneder et al., 2011). This series of discoveries have established the biochemical basis for understanding the molecular mechanism of active DNA demethylation in mammals.
Iterative oxidation of 5mC by TET proteins
The discovery of enzymatic activity modifying 5mC through oxidation was motivated by two known pathways involved in oxidative modification of thymine (T). The first involves the biosynthesis of “base J” (β-D-glucosyl-hydroxymethymethyluracil), a modified base present in the genome of the parasite Trypanosome brucei that causes African sleeping sickness (Borst and Sabatini, 2008). Base J biosynthesis is achieved by a two-step process. J-binding protein (JBP) 1 and 2 first oxidize T in the DNA to 5-hydroxymethyluracil (5hmU); then a glycosyltransferase completes the synthesis of base J by adding glucose groups to 5hmU (Borst and Sabatini, 2008). JBP1/2 proteins are members of the Fe(II)/α-ketoglutarate (α-KG)-dependent dioxygenase family (Loenarz and Schofield, 2011). Systematic bioinformatic analysis identified the human gene, Ten-eleven translocation (TET) 1, as a homolog of JBP1/2 (Iyer et al., 2009). TET1, initially discovered as a fusion partner of the histone H3 Lys4 methyltransferase mixed-lineage leukaemia 1 (MLL1) [t(10;11)(q22;q23)] in certain acute myeloid leukemia (AML) (Lorsbach et al., 2003; Ono et al., 2002), contains a C-terminal catalytic domain consisting of a Cys-rich region and a double-stranded β-helix (DSBH) fold (Figure 1A). Overexpression of TET1 in cultured cells results in reduction of genomic 5mC level, and recombinant TET1 proteins can oxidize 5mC in vitro and the resulting product was determined as 5hmC (Tahiliani et al., 2009) (Figure 1B). Similar enzymatic activity was also demonstrated in all three TET proteins in mouse (TET1-3) (Ito et al., 2010). Supporting the physiological relevance of TET-mediated 5mC oxidation, 5hmC can be readily detected in most cell-types (1–5% of 5mC) and is particularly enriched in the genome of adult neurons (15–40% of 5mC) (Globisch et al., 2010; Kriaucionis and Heintz, 2009).
The second pathway that provides clues on enzymatic activity of TET proteins involves T-to-uracil (U) conversion in the pyrimidine salvage pathway. In this process, T is first converted by thymine hydroxylase (THase), also a Fe(II)/α-KG-dependent dioxygenase, to 5hmU, 5-formyluracil (5fU), and 5-carboxyluracil (5caU) through three successive oxidation reactions; then isoorotate decarboxylase completes the T-to-U conversion through decarboxylation of 5caU (Smiley et al., 2005). The similarity in chemistry between T-to-U conversion and oxidative modification of 5mC suggests the possibility that TET enzymes may function similarly to THase and can mediate interative oxidation of 5mC (Wu and Zhang, 2010). With improved chromatographic separation conditions, TET proteins have been shown to be capable of further oxidizing 5hmC to 5fC and 5caC (He et al., 2011a; Ito et al., 2011) (Figure 1B). Mass spectrometry analysis has shown that 5fC is present in a wide-range of cell types and 5caC is detectable in mouse ESCs, although their levels are at least an order of magnitude less than that of 5hmC (He et al., 2011; Ito et al., 2011; Pfaffeneder et al., 2011). The low abundance of 5fC and 5caC is probably due to the presence of enzymatic activity capable of efficiently excising these highly oxidized bases from DNA (see below). Thus, TET-mediated 5mC oxidation reactions extend the previously recognized binary state of cytosine modification (C/5mC) to five distinct states (C/5mC/5hmC/5fC/5caC) in the mammalian genome, raising the possibility that these new cytosine analogs may play a role in dynamic regulation of 5mC.
Passive restoration of unmodified cytosines by replication-dependent dilution of oxidized 5mC
The presence of oxidized 5mC bases at CpG sites may facilitate replication-dependent passive loss of 5mC. While there are conflicting results regarding whether UHRF1 can efficiently recognize hemi-hydroxymethylated CpG sites [hCG:GC] (Frauer et al., 2011; Hashimoto et al., 2012), independent studies have established that DNMT1 is significantly less efficient (10~60-fold) in methylating hCG:GC than hemi-methylated [mCG:GC] sites in vitro (Hashimoto et al., 2012; Valinluck et al., 2004). Whether hemi-modified CpGs carrying 5fC or 5caC [XG:GC, X=fC or caC] also impair DNMT1 functions is unclear. In summary, TET proteins may initiate a two-step demethylation process in proliferating cells that involves initial active modification (AM) of 5mC through oxidation and subsequent replication-dependent passive dilution (PD) of 5hmC and potentially 5fC/5caC (AM-PD in Figure 2A). This mode of active DNA demethylation is distinct from simple passive dilution of 5mC (PD in Figure 2A) (Kohli and Zhang, 2013), as AM-PD may be effective even in the presence of functional methylation maintenance machinery. Replication-dependent DNA demethylation in the presence of DNMT1/UHRF1 has been reported during erythropoiesis (Shearstone et al., 2011), it will be interesting to examine whether 5mC oxidation may contribute to DNA demethylation in this system.
Interestingly, DNMT3A/B has comparable in vitro methylase activity towards mCG:GC and hCG:GC sites (Hashimoto et al., 2012; Otani et al., 2013b), suggesting that DNMT3 may be involved in maintaining 5mC levels in dividing cells with high TET activity. In support of this notion, mouse ESCs deficient in both DNMT3A and DNMT3B become globally hypomethylated after prolonged culturing (Chen et al., 2003). Because 5hmC/5fC/5caC in the paternal genome can be passively diluted in the presence of maternal DNMT3A proteins during early pre-implantation development (Inoue et al., 2011; Inoue and Zhang, 2011), DNMT3A alone may not be sufficient to effectively methylate hemi-modified sites [XG:GC, X= hC, fC, or caC]. Further studies are needed to elucidate the interplay between 5mC oxidation and replication-dependent demethylation.
Active restoration of unmodified cytosines by enzymatic removal of oxidized 5mC
In addition to passive dilution of modified cytosines, several TET initiated but replication-independent active demethylation mechanisms have been proposed (Box 1B). These active DNA demethylation processes also involve initial oxidative modification of 5mC but complete the demethylation through replication-independent active restoration (AR) of unmodified C. First, removal of oxidized 5-position substituent, through either dehydroxymethylation of 5hmC by DNMTs (Chen et al., 2012; Liutkeviciute et al., 2009) or decarboxylation of 5caC by a putative decarboxylase (Schiesser et al., 2012; Wu and Zhang, 2010), may directly revert the oxidized 5mC base to unmodified cytosine. Second, DNA glycosylase-mediated excision of oxidized 5mC bases, including removal of 5hmU [deamination product of 5hmC] (Guo et al., 2011) or 5fC/5caC (He et al., 2011; Maiti and Drohat, 2011), may result in abasic sites that are further repaired by BER to restore unmethylated C.
Among these proposed TET-dependent AM-AR demethylation pathways, the mechanism that involves TDG-mediated excision of 5fC/5caC has received the most experimental support. As a member of uracil DNA glycosylase (UDG) superfamily (Figure 1A), TDG is known to use a base-flipping mechanism to remove the pyrimidine base from T•G or U•G mismatches to initiate BER (Stivers and Jiang, 2003). Given the precedent of DME/ROS1-mediated excision repair of 5mC in plants (Figure 1C), DNA glycosylases have long been suspected in playing a role in active DNA demethyation in mammals. TDG is a particularly attractive candidate as it can act as a scaffold protein to bridge transcriptional coactivator CBP/p300 to numerous transcription factors, supporting a gene regulatory role of TDG (Dalton and Bellacosa, 2012). Early studies suggested that TDG might promote DNA demethylation through either direct excision of 5mC or T (the deamination product of 5mC). Although biochemically feasible, these mechanisms probably play a limited role in DNA demethylation given the marginal 5mC excision activity of TDG and AID/APOBEC deaminases’ selectivity for single-stranded DNA (Box 1A). The role of TDG in DNA demethylation was revisited after additional oxidized cytosine analogs were discovered in the genome. While TDG cannot efficiently remove 5mC or 5hmC, it exhibits robust in vitro excision activity towards 5fC or 5caC [5fC•G or 5caC•G pairs] in duplex DNA (He et al., 2011; Maiti and Drohat, 2011). In fact, TDG has a higher activity for 5fC•G compared to T•G mismatch, its cognate substrate (Maiti and Drohat, 2011). The molecular basis for excision of 5fC/5caC by TDG stems from the effect of 5fC and 5caC in destabilizing the base-sugar linkage (N-glycosidic bond) as C bases with a weakened N-glycosidic bond can be efficiently excised by DNA glycosylases (Bennett et al., 2006). Further biophysical analysis indicates that the active site of TDG may possess specific structural features to mediate the recognition of oxidized 5mC bases (Zhang et al., 2012). Supporting the physiological relevance of this mechanism, overexpression of TET and TDG in the HEK293 cells rapidly depleted 5fC and 5caC (Nabel et al., 2012), whereas Tdg-deficiency in mouse ESCs caused a 5–10 fold increase in levels of 5fC and 5caC (He et al., 2011; Shen et al., 2013; Song et al., 2013a). Thus, methylation, iterative oxidation and excision repair offer a complete cytosine-modifying cascade that permits replication-independent erasure of DNA methylation (Figure 1C and 2B).
While other members of the UDG family of DNA glycosylases (UNG, MBD4, and SMUG1) are dispensable for embryonic development (Kemmerich et al., 2012; Millar et al., 2002), Tdg-null mouse embryos exhibit a wide range of developmental defects and die around embryonic day (E) 12.5 (Cortazar et al., 2011; Cortellino et al., 2011). Interestingly, among all UDG DNA glycosylases, only TDG can efficiently excise 5fC/5caC (He et al., 2011; Maiti and Drohat, 2011), suggesting a unique role of TDG’s 5fC/5caC excision activity in embryonic development. Indeed, mutant embryos carrying the glycosylase activity defective mutation (N151A in mouse TDG) phenocopy developmental defects of Tdg-null embryos (Cortellino et al., 2011). Given its robust excision repair activity for mismatch mutations, TDG may potentially ensure proper development by reducing genetic mutations; however, Tdg-deficiency induced embryonic lethality phenotype was fully penetrant, which is incompatible with an anti-mutagenic DNA repair defect that would exhibit highly variable phenotypes due to stochastic mutations in different genes. Thus, inactivation of TDG may primarily result in epigenetic rather than genetic defects in the genome. Dys-regulation of C/5mC/5hmC levels and/or accumulation of ectopic 5fC/5caC at specific gene regulatory sequences in Tdg-deficient mice may potentially affect transcription and cause developmental defects. Further in vivo analysis is needed to clarify how extensive TET/TDG-mediated DNA demethylation pathway is utilized in vivo and to elucidate the exact mechanism by which TDG activity contributes to transcription and development.
Intermediate steps of TET/TDG-mediated replication-independent DNA demethylation
Considering the cytosine methylation and demethylation process in the context of TET/TDG-mediated step-wise enzymatic cascade prompts key mechanistic questions: What are the preferred intermediate steps through which TET and TDG enzymatically revert 5mC to unmodified cytosine? Does TDG-mediated excision of 5fC/5caC preferentially generate single strand break (SSB) or double strand break (DSB)? Base-resolution mapping studies showed that steady-state 5hmC is almost exclusively found in the CpG context (>99% in CpGs), even in mouse ESCs and neurons where non-CpG methylation is prevalent (Lister et al., 2013; Yu et al., 2012). Thus, oxidative modification of 5mC by TET proteins may occur mostly in the CpG context. In line with this, recent structural analysis of TET2 catalytic domain in complex with methylated DNA indicates that TET2 has a stronger preference to oxidize 5mC in CpG sites than in non-CpG context (Hu et al., 2013). Alternatively, iterative oxidation of 5mC at non-CpG sites could occur at a much faster rate such that 5hmC at these sites is only transiently generated and rapidly converted to 5fC/5caC, which are removed by TDG/BER. Base-resolution mapping of 5fC/5caC in Tdg-mutant cells can potentially capture these short-lived oxidized 5mC bases within non-CpG sites.
Within each CpG dyad, the cytosine-modifying pathway (DNMT-TET-TDG/BER) can theoretically generate 21 distinct cytosine modification states [XG:GX, X=C, mC, hmC, fC, caC or abasic], six of which are in symmetric form (Figure 2B). Base-resolution mapping of 5hmC in mouse ESCs indicates that less than 20% of 5hmC is potentially present in symmetrically modified form [hmCG:GChm] (Yu et al., 2012). Thus, hemi-modified CpG dyads may be the predominant pool of intermediates generated from 5mC oxidation reactions. Strand asymmetry associated with 5hmC (or 5fC and 5caC) implies that TET/TDG-mediated active demethylation may frequently generate single abasic site at methylated CpGs, which typically leads to formation of SSB. Reducing the frequency of symmetrically oxidized CpGs [XG:GX, X=fC or caC] may be physiologically relevant as two abasic sites in a single CpG dyad [XG:GX, X=abasic] could frequently generate DSB, increasing the chance of introducing deleterious mutations. Curiously, in vitro biochemical assay showed that TDG has higher excision activity for hemi-carboxylmethylated CpG site [caCG:GC] than for fully carboxylmethylated one [caCG:GCca] (He et al., 2011), which may also help reduce frequency of generating DSB. Sequencing technology that allows simultaneous mapping of 5fC and 5caC at base-resolution may help determine the relative frequency of SSBs versus DSBs introduced during TET/TDG-mediated active demethylation process.
Regulatory mechanisms of TET-dependent DNA demethylation
Maintaining proper gene expression and cellular identity requires precise control of the DNA methylation pattern (Ji et al., 2010; Ziller et al., 2013). 5mC is likely to be the default epigenetic state of CpGs (except for those within CpG-rich promoters) in the mammalian genome, thus direct regulation of DNA methylation machineries generally leads to global changes in 5mC levels; however, regulatory mechanisms that modulate substrate accessibility, enzymatic activity, expression levels, and genomic targeting of TET enzymes may provide more precise control of the pattern of 5mC (or oxidized 5mC) at specific loci in the genome.
Regulation by substrate preference and accessibility of TET enzymes
Quantitative mass spectrometry analysis of mouse ESCs suggests that 5hmC (~5% of 5mC) is significantly more prevalent than 5fC (0.06–0.6% of 5mC) and 5caC (0.01% of 5mC) (Ito et al., 2011; Pfaffeneder et al., 2011), suggesting that TET enzymes tend to stall at 5hmC. The processivity of 5mC oxidation may be regulated by substrate preference and/or the accessibility of TET enzymes. In vitro biochemical analysis suggests that the initial reaction rate of TET enzymes for oxidizing 5mC to 5hmC is 5–8 fold higher than that of oxidizing 5hmC or 5fC (Ito et al., 2011), indicating that TET proteins may be more efficient in utilizing 5mC (as compared to 5hmC or 5fC) as a substrate. Structural analysis of TET2 catalytic domain shows that there is no binding difference between methylated and unmethylated CpG sites (Hu et al., 2013), implying that the methyl group on the 5-position does not impact DNA binding. Thus, it remains to be determined whether the active site of TET enzymes has distinct affinity for 5hmC and 5fC. Interestingly, a SRA domain containing protein, UHRF2, has been recently identified as a 5hmC-binding protein and reported to be able to enhance the processivity of TET enzymes (Spruijt et al., 2013), potentially via its ability to flip modified cytosine from DNA duplex and increase the substrate accessibility of TET proteins.
Regulation of enzymatic activity by metabolites and co-factors
Emerging evidence indicates that changes of metabolic states in cells may affect the relative concentration of enzymatic cofactors and in turn influence the activity of DNA and histone modifying enzymes (Kaelin and McKnight, 2013). TET enzymes require oxygen, Fe (II) and α-KG to catalyze 5mC oxidation reactions. α-KG is primarily produced from isocitrate in the tricarboxylic acid (TCA) cycle by isocitrate dehydrogenases 2 and 3 (IDH2 and IDH3) in the mitochondria. Cytosolic IDH1 can provide an alternative source of α-KG. Interestingly, inactivating mutations in the TCA cycle enzymes, fumarate hydratase (FH) and succinate dehydrogenase (SDH), or tumor-associated missense mutations in IDH1/2 result in accumulation of high concentrations of fumarate, succinate, and the oncometabolite 2-hydroxyglutarate (2-HG), all of which share structural similarity with α-KG and competitively inhibit Fe(II)/α-KG-dependent dioxygenases (Kaelin and McKnight, 2013), including TET enzymes and JmjC-domain containing histone demethylases (Klose et al., 2006).
Vitamin C (also known as ascorbic acid), a micronutrient known for its anti-scurvy activity in humans, is also implicated in modulation of TET activity. Recent studies have supported a role of vitamin C in stimulating the catalytic activity of TET proteins (Blaschke et al., 2013; Minor et al., 2013; Yin et al., 2013). Vitamin C may directly interact with the catalytic domain of TET proteins and act as a positive modulator of TET activity by facilitating conformational changes and/or recycling of the cofactor Fe (II) (Yin et al., 2013). In mouse ESCs, vitamin C significantly increases levels of all 5mC oxidation products, particularly 5fC and 5caC (>10-fold). Moreover, ground-state mouse ESCs cultured in the presence of vitamin C adopt a methylome more resembling that of blastocysts in vivo (Blaschke et al., 2013).
Regulation of genomic targeting of TET enzymes
In addition to the C-terminal catalytic domain, TET1 and TET3 encode a zinc finger Cystein-X-X-Cystein (CXXC) domain at their N-termini (Figure 1A). The CXXC domain is present in many chromatin associated proteins, and has a strong preference for unmethylated CpGs (Long et al., 2013). Although TET2 lacks a CXXC domain, a neighboring gene IDAX (inhibition of the Dvl and axin complex; also known as CXXC4) encodes a CXXC domain highly similar to those in TET1 (also known as CXXC6) and TET3 (CXXC10). It is possible that IDAX was originally part of an ancestral TET2 gene that underwent a chromosomal inversion, which separated the CXXC domain from the catalytic domain (Iyer et al., 2009; Ko et al., 2013). Genome-wide occupancy analysis suggests that TET1 is preferentially localized to unmethylated CpG-rich promoters and genic regions in mouse ESCs (Williams et al., 2011; Wu et al., 2011b), and IDAX is also enriched at unmethylated CpG sequences and directly interacts with TET2 (Ko et al., 2013). Further biochemical and structural analyses indicate that TET1 CXXC domain may also recognize methylated cytosines (Xu et al., 2011b; Zhang et al., 2010), and the TET3 CXXC domain can target unmethylated cytosines within both CpG and non-CpG contexts (Xu et al., 2012). Thus, unlike other CXXC domains that specifically recognize unmethylated CpG sites, CXXC domains of TET1/3 and IDAX may have increased flexibility in sequence selectivity and play a role in facilitating the recruitment of TET enzymes to their specific genomic targets.
Transcriptional and post-transcriptional regulation of TET levels
Three TET genes exhibit different expression patterns in a developmental-stage- and cell-type-specific manner (Ito et al., 2010; Szwagierczak et al., 2010). A high-level of TET1 is specifically detected in mouse ESCs, the inner cell mass of blastocysts, and developing PGCs (E9.5-12.5), whereas TET2 and TET3 are broadly expressed in various mouse adult tissues (Ito et al., 2010; Yamaguchi et al., 2012). Interestingly, TET3 is the only TET enzyme present in mouse oocytes and one-cell zygotes (Iqbal et al., 2011; Wossidlo et al., 2011). Transcriptional control of TET genes is likely mediated by cell-type specific transcription factors. For instance, the upstream promoter region of the TET1 gene contains a large cluster of binding sites for pluripotency-related transcription factors (TFs) (Ficz et al., 2011), suggesting a role of pluripotency-related TFs in maintaining TET1 expression. Indeed, TET1 is highly expressed in mouse ESCs, but its level is rapidly down-regulated upon differentiation (Ito et al., 2010; Koh et al., 2011).
Given the relatively large size of mRNA and 3′ untranslated regions (UTR) of TET1-3 genes, TET mRNAs may also be regulated post-transcriptionally by microRNAs. Indeed, two recent studies have reported a role of oncogenic microRNA miR-22 in negatively regulating TET protein levels in the context of breast cancer development and hematopoietic stem cell transformation (Song et al., 2013b; Song et al., 2013c). In addition, IDAX may target TET2 proteins for degradation via a caspase-dependent mechanism; similarly, the CXXC domain of TET3 may negatively regulate its own protein stability (Ko et al., 2013), suggesting an auto-inhibitory mechanism of regulating TET protein levels.
Genomic distribution of oxidized methylcytosines and their potential roles in transcription regulation
Genome-wide mapping of oxidized 5mC bases in the mammalian genome represents an unbiased approach to investigate the potential roles of TET-mediated demethylation pathway and these oxidized cytosines in epigenome function and transcriptional regulation. In principle, DNMT and TET enzymes can modify each of the approximately 28 million CpGs in the diploid mammalian genome to adopt one of the five epigenetic states (C/5mC/5hmC/5fC/5caC).
Genome-wide mapping technologies for oxidized 5mC bases
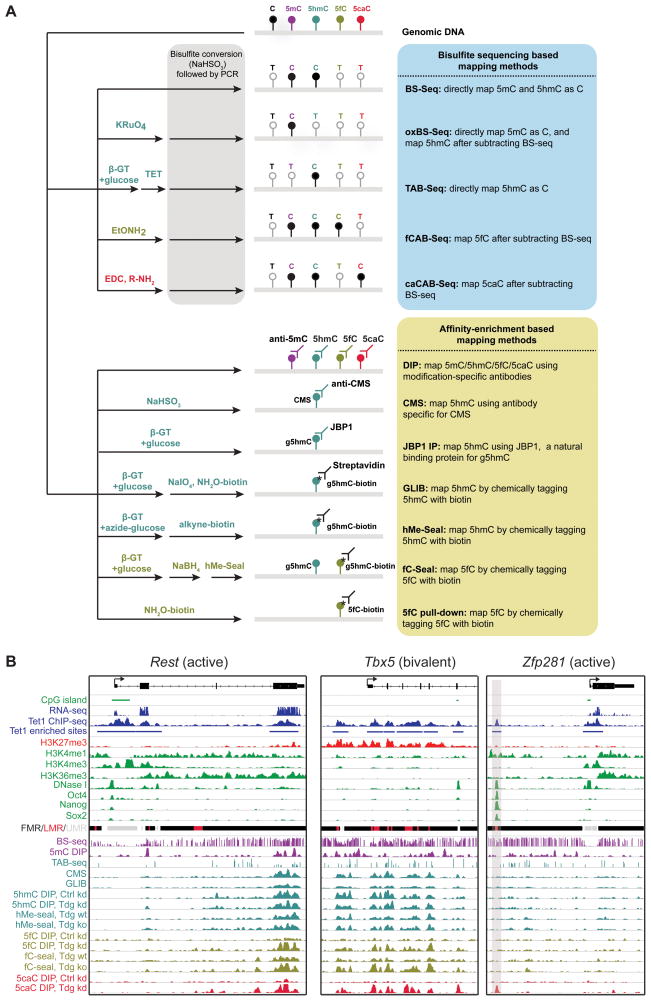
Multiple methods exploiting specific biochemical or biophysical properties of modified cytosine bases have been developed to map their distribution across the genome (Figure 3A). These methods can be categorized into two groups: first, affinity-enrichment based approaches, which utilize modification-specific antibodies or selective chemical labeling of modified cytosines (Box 2A); second, base-resolution mapping methods such as bisulfite sequencing (BS-Seq) (Box 2B). The affinity- based methods are generally straightforward to perform and more cost-effective. However, the resolution of these methods is limited by the size of DNA fragments (several hundred base pairs), and affinity-enrichment methodology identifies regions enriched for modified bases by comparing signals [normalized sequencing read counts] in pull-down and those in negative control (IgG or input), thereby providing a measurement of relative fold-enrichment [pull-down/control]. By contrast, base-resolution mapping of modified bases can determine which cytosine (including exact position and strand) is modified and typically measures the percentage of modification level [(number of modified cytosines/number of all cytosines) × 100%] at a given cytosine. The probability that a cytosine can be confidently identified as modified is governed by the sequencing depth and abundance of the modification at that cytosine in a specific cell type. Given the low abundance of oxidized methylcytosines, base-resolution methods require a relatively high sequencing depth to detect them. It has been estimated that at a normal sequencing depth (20–30x for BS-Seq), base-resolution mapping of 5hmC (TAB-Seq in Figure 3A and Box 2B) can confidently detect cytosines with an average 20% or higher level of hydroxymethylation regardless of the local density of cytosine substrates (Yu et al., 2012). As a result, base-resolution methods have enhanced the sensitivity for detecting individual cytosine that is modified at medium-to-high level but sparsely distributed (that is, not clustered with other 5hmC). Indeed, while 80–90% of 5hmC peaks identified by affinity-based methods (e.g. 5hmC antibody IP and GLIB in Figure 3A) contain at least one of the approximately 2.06 million high-confidence 5hmC-marked cytosines detected by TAB-Seq in mouse ESCs, 5hmC peaks from affinity-based methods only cover 30–40% of high-confidence 5hmC identified by TAB-Seq (Yu et al., 2012). Because affinity-based methods integrate signals from multiple cytosines within a given genomic region, they are capable of detecting closely clustered cytosines with low level of modification, which may otherwise be missed by base-resolution methods. Further increasing the sequencing depth of base-resolution mapping experiments may help identify cytosines carrying low-level modification, although it is unclear whether these infrequently modified cytosines are biologically relevant. In summary, both affinity-based and single-base mapping methods can consistently identify regions enriched for cytosines that are frequently modified (medium-to-high level), but their results may differ at sparsely distributed or infrequently modified cytosines. Discrepancies between published genome-wide mapping studies may be due to the use of different methodologies and data analysis strategies as well as varying definition of a given genomic feature. Here we focus on recent advances on mapping of oxidized 5mC bases including 5fC/5caC in stem cells and their potential gene regulatory roles at various genomic features.
Figure 3. Genome-wide mapping methods for cytosine modifications.
(A) Summary of BS-Seq or affinity-enrichment based genome-wide mapping methods of DNA cytosine modifications. In conjunction with various chemical and enzymatic pre-treatment, BS-Seq or affinity-enrichment (antibody or chemical tagging) methods have been developed to map 5mC (BS-Seq and oxBS-Seq; 5mC DIP), 5hmC (oxBS-seq and TAB-Seq; 5hmC DIP, CMS IP, JBP1 IP, GLIB, and hMe-Seal), 5fC (fCAB-Seq; 5fC DIP, fC-Seal, and 5fC pull-down), and 5caC (caCAB-Seq; 5caC DIP) (see Box 2). Notably, oxBS-Seq, fCAB-Seq, and caCAB-Seq require subtracting signals of conventional BS-Seq from those of modified BS-Seq to indirectly determine the position and abundance of oxidized 5mC bases. 5fC and 5caC have not been systematically mapped at base-resolution, as they are relatively rare in the wild-type cells and require unusually high sequencing coverage to confidently determine their position and abundance.
(B) Shown are genomic distributions of oxidized 5mC bases at representative loci in mouse ESCs. Genomic distribution of 5hmC/5fC/5caC in control (wt: wild-type; Ctrl kd: control knockdown) and Tdg-depleted (ko: knockout; Tdg kd: Tdg knockdown) mouse ESCs are shown for three representative loci. 5hmC distribution was measured by both affinity-based method [5hmC DIP, CMS, 5hmC-seal, and GLIB] (Pastor et al., 2011; Shen et al., 2013; Song et al., 2013a) and base-resolution method [TAB-Seq, scale: 0–50%] (Yu et al., 2012). Distributions of 5fC and 5caC were determined by antibody [5fC/5caC DIP] (Shen et al., 2013) or chemical tagging [5fC-seal] (Song et al., 2013a) methods. DNA methylation levels were estimated by 5mC DIP (Shen et al., 2013) or conventional BS-Seq (5mC+5hmC, scale: 0–100%) in wild-type mouse ESCs (Stadler et al., 2011). Based on local methylation levels, the genome can be categorized into FMRs (Fully methylated regions, 50–100% methylation), LMRs (low methylated regions, 10–50%), and UMRs (unmethylated regions, <10%) (Stadler et al., 2011). Also shown are other genomic datasets in wild-type mouse ESCs, including TET1 ChIP-seq (Wu et al., 2011b), RNA-seq (Ficz et al., 2011), major histone modifications (Mikkelsen et al., 2007), DNase I hypersensitivity site (modENCODE project) and binding sites of pluripotency TFs (Whyte et al., 2012). A region located upstream of Zfp281 gene is highlighted in gray, where Tdg-deficiency induced ectopic 5caC is overlapped with chromatin features of active enhancer (DNase I+/H3K4me1+/H3K4me3−) and binding sites of pluripotency factors (Oct4/Nanog/Sox2).
Box 2. Genome-wide mapping technologies for 5mC oxidation products.
A. Affinity-enrichment methods
Affinity-enrichment is achieved by using either modification-specific antibodies or chemical tagging specific 5mC oxidation derivatives (Figure 3A).
1. Antibody-based immunoprecipitation (IP) of 5hmC/5fC/5caC
Modification-specific antibodies are available for all three oxidized 5mC bases and the IP procedure is simple (Song et al., 2012b; Wu and Zhang, 2011a). However, there is evidence that antibody-based method may pull-down certain type of simple repeats (e.g. poly-CA) non-specifically (Matarese et al., 2011). Subtracting IgG control signals from IP signals can partially alleviate this problem (Shen et al., 2013).
2. CMS antibody-based IP of 5hmC
This method is based on sodium bisulfite-mediated conversion of 5hmC to cytosine methylene sulphonate (CMS). Highly specific antibody against CMS can be generated and used to enrich 5hmC containing DNA fragment (Huang et al., 2012). Anti-CMS serum may have lower background compared to anti-5hmC serum (Pastor et al., 2011). However, the presence of bulky CMS moiety in DNA can introduce PCR biases.
3. JBP1 IP of 5hmC
The method involves initial conversion of 5hmC to glycosylated 5hmC (g5hmC) using β-glucosyltransferase (β-GT) from T4 bacteriophage. JBP1, a natural g5hmC-binding protein, is then used to enrich g5hmC-containing genomic DNA (Robertson et al., 2011). However, a recent study suggests that JBP1 IP generally produces weaker signals compared to 5hmC antibody IP or hMe-Seal (Thomson et al., 2013).
4. Selective chemical labeling 5hmC with biotin (hMe-Seal or GLIB)
By exploiting the naturally occurring glucosylation process of 5hmC by β-GT, several methods were developed to chemically and enzymatically label 5hmC with the biotin tag (Pastor et al., 2011; Song et al., 2011; Szwagierczak et al., 2010). The highly selective and strong interaction between biotin and streptavidin allows efficient pull-down of 5hmC-containing DNA. In the first method, termed hMe-Seal (Song et al., 2011), purified β-GT is used to transfer an azide-modified glucose analogue (UDP-6-N3-glucose) to 5hmC. Then, a biotin group can be attached to g5hmC with alkyne-biotin probe via click chemistry (Song et al., 2011). The second approach, named GLIB (glucosylation, periodate oxidation, biotinylation), first uses β-GT to transfer an unmodified glucose to 5hmC. Next, sodium periodate (NaIO4) is used to oxidize the vicinal hydroxyl group in the glucose to aldehyde group, which is further modified with aldehyde-reactive probe (aminooxy-biotin or NH2O-biotin) to add biotin tag to g5hmC (Pastor et al., 2011). These chemical labeling methods have enhanced sensitivity and lower background as compared to the antibody-based method (Song et al., 2012b). However, similar to 5hmC antibody IP, these methods still exhibit some degree of density-dependency and tend to more efficiently pull-down DNA fragment containing multiple modified bases (Shen et al., 2013).
5. Selective chemical labeling 5fC with biotin (5fC-Seal or 5fC pull-down)
A strategy for direct labeling 5fC with biotin tag using aminooxy-biotin probes has been suggested (Pfaffeneder et al., 2011) and was used to generate the genome-wide distribution map of 5fC (Raiber et al., 2012). However, this method may have side reactions with other cytosines or abasic sites, resulting in relatively high background. Based on hMe-Seal, an indirect 5fC labeling method (termed fC-Seal) has been developed (Song et al., 2013a). In fC-Seal, β-GT is first used to block endogenous 5hmC in the genome with unmodified glucose. Next, sodium borohydride (NaBH4) is used to reduce 5fC to 5hmC, which is further converted to g5hmC-biotin (derived from endogenous 5fC) as in hMe-Seal.
B. Single-nucleotide resolution methods
Base-resolution mapping of 5mC oxidation derivatives involves the use of bisulfite sequencing (BS-Seq) or third-generation sequencing platforms.
1. BS-Seq (5mC+5hmC)
In standard BS-Seq, 5mC and 5hmC are resistant to deamination and consequently be read out as C after PCR amplification (Clark et al., 1994). In contrast, unmodified C, 5fC and 5caC are deaminated in bisulfite treatment and read out as T in sequencing. Thus, methylation signals (C) in standard BS-Seq represent the sum of 5mC and 5hmC, complicating the interpretation of BS-Seq results (Huang et al., 2010). By exploiting distinct chemical properties of 5hmC/5fC/5caC in bisulfite treatment (NaHSO3), traditional BS-Seq can be adapted to map oxidized 5mC (Figure 3A).
2. oxBS-Seq (5hmC)
In oxidative bisulfite sequencing (oxBS-Seq) (Booth et al., 2012), potassium perruthenate (KRuO4) was used to specifically oxidize 5hmC to 5fC, which can be deaminated by bisulfite treatment. After the DNA sample is treated with KRuO4 and NaHSO3, 5hmC/5fC/5caC is detected as T in sequencing, whereas 5mC remains intact and is read as C. Therefore, subtracting signals of oxBS-Seq maps from standard BS-Seq signals would reveal the location and abundance of 5hmC (Booth et al., 2012).
3. TAB-Seq (5hmC)
In TET-assisted bisulfite sequencing (TAB-Seq) (Yu et al., 2012), 5hmC is first protected from TET-mediated oxidation by glucosylation using β-GT. Next, 5mC and 5fC are oxidized to 5caC in the presence of highly active recombinant TET proteins.
4. fCAB-Seq (5fC)
In this chemical modification assisted BS-Seq (CAB) method, O-ethylhydroxylamine (EtONH2) is used to prevent 5fC from bisulfite-mediated deamination (Song et al., 2013a). Subtracting standard BS-Seq signals from signals of fCAB-Seq will reveal the location and abundance of 5fC.
5. caCAB-Seq (5caC)
Similar to fCAB-Seq, 5caC can be modified by 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC) to resist deamination (Lu et al., 2013). However, this method may have side reactions with the genomic DNA.
6. Third-generation sequencing technology
These single-molecule sequencing technologies directly sequence DNA molecules without the need of PCR amplification. In single-molecule real-time (SMRT) sequencing, real-time recording of nucleotide incorporations generates not only the sequence identity but also DNA polymerase kinetics. The polymerase replication rate at and around modified cytosines is slower compared to unmodified bases. When combined with chemical labeling or enzymatic modification (adding a glucosyl group to 5hmC; oxidizing 5mC to 5caC) to increase kinetic differences, SMRT can be used to detect strand-specific 5mC and oxidized bases at base-resolution with high confidence (Clark et al., 2013; Song et al., 2012a). Nanopore sequencing monitors the current signal when individual DNA molecular pass through nanoscale protein pores and has also shown the potential to detect oxidized 5mC bases (Li et al., 2013; Wanunu et al., 2011). While third generation sequencing technologies have the potential to directly map various cytosine modifications, significant technological advances are required to further improve the detection accuracy and to increase the sequencing throughput for analyzing complex mammalian genomes.
Oxidized methylcytosines at gene promoters
Approximately 70% of mammalian promoters are associated with short CpG-dense sequences [termed CpG islands (CGIs), on average 1 kilobase (kb) long] (Deaton and Bird, 2011). CpG-rich promoters [also known as high-CpG promoters (HCPs)] are generally unmethylated and transcriptionally active as H3K4 methylation and histone variant H2A.Z enriched at these HCPs antagonize DNMT binding (Ooi et al., 2007; Zilberman et al., 2008). The lack of 5mC at HCPs predicts that TET-mediated oxidative modification of 5mC is minimal at these promoters. Indeed, the current consensus view from both affinity-enrichment and base-resolution mapping studies suggests that proximal regions surrounding the transcriptional start site (TSS) of active promoters (mostly HCPs) are generally devoid of 5hmC/5fC/5caC in both wild-type and Tdg-depleted mouse ESCs [exemplified by promoters of Rest and Zfp281 in Figure 3B, overlapping with unmethylated regions (UMR)] (Shen et al., 2013; Song et al., 2013a; Szulwach et al., 2011a; Wu et al., 2011a; Yu et al., 2012). TET proteins enriched at CpG-rich active promoters may therefore facilitate steady-state transcription through catalytic activity-independent functions such as stabilizing or recruiting other chromatin modifying complexes (Box 3).
Box 3. Catalytic activity-independent role of TET proteins in transcriptional regulation.
Genomic mapping of TET proteins in ESCs, HEK293T cells and bone marrow tissues suggest that all three TET proteins are preferentially localized to unmethylated CpG-rich promoters (Chen et al., 2013b; Deplus et al., 2013; Vella et al., 2013; Williams et al., 2011; Wu et al., 2011b). In mouse ESCs, TET1 is highly enriched at actively transcribed CpG-rich promoters as well as bivalent gene promoters, which are repressed by Polycomb repression complex 2 (PRC2). The preference for CGIs is likely due to its N-terminal CXXC domains. Depletion of TET1 causes transcriptional activation and repression of many direct TET1 targets (Williams et al., 2011; Wu et al., 2011b; Xu et al., 2011b). These results suggest that TET proteins may have dual functions in transcriptional regulation via distinct mechanisms. The transcriptional activation role of TET1 may be dependent on its 5mC oxidation activity to maintain unmethylated states at active promoters or distal enhancers (Ficz et al., 2011; Shen et al., 2013; Wu et al., 2011b); in contrast, a role of TET1 in transcriptional repression may be related to its physical interaction with the SIN3A repressor complex (Williams et al., 2011), or its ability to facilitate the recruitment of the MBD3/NURD (Yildirim et al., 2011) and PRC2 (Wu et al., 2011b) to chromatin. TET2 and TET3 (to a lesser extent for TET1), can directly interact with O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT), and recruit OGT to CpG-rich gene promoters in mouse ESCs and differentiated cells (Chen et al., 2013b; Deplus et al., 2013; Vella et al., 2013). Although this physical interaction appears not to regulate enzymatic activity of TET proteins, depletion of TET proteins reduces OGT binding to chromatin and its activity to glycosylates histone H2B (Chen et al., 2013b) and other chromatin regulators such as HCF1 (host cell factor 1) (Deplus et al., 2013), a component of the H3K4 methyltransferase SET1/COMPASS complexes. Thus, TET proteins may promote transcription of target genes by enhancing histone modifications such as O-GlcNAcylation at Ser 112 of H2B and trimethylation of Lys 4 on H3 (H3K4me3), both of which are associated with gene activation. Together, these results suggest that TET proteins possess catalytic activity-independent gene regulatory functions.
Affinity-based methods showed that 5hmC is preferentially enriched at promoters of genes expressed at medium-to-low levels in mouse and human ESCs (Pastor et al., 2011; Szulwach et al., 2011a; Wu et al., 2011a; Xu et al., 2011b), and these 5hmC-enriched promoters are generally associated with intermediate or low CpG-density [intermediate-CpG promoter (ICP) or low-CpG promoter (LCP)]. Base-resolution mapping of 5hmC also indicated that 5hmC is more abundant at ICPs/LCPs than at HCPs (Yu et al., 2012). Recent studies of 5fC and 5caC showed that 5fC/5caC tend to accumulate at transcriptionally inactive or poised promoters in the absence of TDG in mouse ESCs (Shen et al., 2013; Song et al., 2013a). In fact, some of the highest levels of 5hmC/5fC/5caC in Tdg-depleted mouse ESCs are found at promoters and flanking regions of lineage-specific transcription factor (mostly ICPs) [Tbx5 in Figure 3B] (Shen et al., 2013), indicating that TET/TDG-mediated active demethylation occurs at these loci. 5hmC/5fC/5caC peaks at or surrounding these ICPs of lineage-specific genes are frequently overlapped with low-methylation regions (LMRs) that are previously identified by standard BS-Seq as partially methylated regions (Stadler et al., 2011), as well as bivalent domains, which contain H3K4me3 [active histone mark] and H3K27me3 [polycomb repression complex 2 (PRC2)-deposited repressive mark] (Bernstein et al., 2006). TET proteins and oxidized methylcytosines may contribute to the establishment and/or maintenance of transcriptionally poised/inactive state at lineage-specific promoters through several mechanisms, including maintenance of partially methylation state by mediating active demethylation, recruitment of TET-interacting co-repressor complexes such as SIN3A (Box 3), and attracting/repelling specific reader proteins for oxidized 5mC bases (see below). Further studies are needed to clarify the relative importance of these distinct mechanisms in gene regulation.
Oxidized methylcytosines within gene bodies
Enrichment of 5hmC within intragenic regions, particularly at 3′ portion of gene bodies, is found to be a hallmark of active transcription in virtually all cell-types investigated, including mouse/human ESCs (Ficz et al., 2011; Pastor et al., 2011; Szulwach et al., 2011a; Williams et al., 2011; Wu et al., 2011a; Xu et al., 2011b) (Rest in Figure 3B), mouse cerebellum (Mellen et al., 2012; Song et al., 2011), and mouse/human fontal cortex (Lister et al., 2013). Despite their scarcity in the bulk genome, 5fC and 5caC also exhibit a tendency to accumulate within actively transcribed gene bodies in the absence of TDG (Shen et al., 2013; Song et al., 2013a) (Rest in Figure 3B). The exact role of intragenic 5hmC/5fC/5caC in transcriptional regulation is currently unclear. One possible function of intragenic 5fC/5caC is to regulate the rate of RNA polymerase II transcription (Kellinger et al., 2012). In mouse ESCs, 5hmC/5fC/5caC is also more enriched at exons relative to introns, raising the possibility that 5mC oxidation may regulate mRNA processing events such as splicing.
Oxidized methylcytosines at distal cis-regulatory elements
Both affinity enrichment-based and base-resolution mapping analyses indicate that 5hmC is highly enriched at active distal-regulatory elements (Stroud et al., 2011; Yu et al., 2012), which are marked by DNase I hypersensitivity, enhancer-associating histone modifications (H3K4me1 and H3K27ac) (Shen et al., 2012) and LMRs. Interestingly, base-resolution mapping indicates that 5hmC is actually depleted at the binding motif of transcription factors or CTCF insulators, but enriched at immediately adjacent regions (Lister et al., 2013; Yu et al., 2012). Furthermore, base-resolution mapping of 5mC and 5hmC during human and mouse fontal cortex development showed that for adult brain-specific enhancers (active in adult brains but inactive in fetal brains), both 5mC and 5hmC are enriched at the center regions of fetal brains, but are lost in adult brains (Lister et al., 2013), supporting a role of TET-mediated 5mC oxidation in enhancer activation in postnatal brains.
In mouse ESCs, 5hmC and 5fC tend to enrich at poised tissue-specific enhancers (H3K4me1-positive, but H3K27ac-negative) (Shen et al., 2013; Song et al., 2013a), indicating that these poised enhancers are pre-marked by 5hmC/5fC for subsequent demethylation during later development. Furthermore, 5caC (to a lesser extent for 5fC) is preferentially accumulated at distally located active enhancers in Tdg-depleted mouse ESCs (Shen et al., 2013; Song et al., 2013a). For instance, 5caC is enriched at a cohort of distal pluripotency TF (Oct4/Nanog/Sox2) binding sites in Tdg-depleted mouse ESCs (Zfp281 in Figure 3B). These results suggest that TET/TDG-mediated active demethylation is preferentially targeted to these active distal-regulatory elements. DNA binding proteins are capable of inducing localized DNA demethylation (Stadler et al., 2011), but it is currently unclear how TET/TDG activity is recruited to these cis-regulatory elements. TET and TDG may be recruited directly by transcription factors or co-activators (CBP/p300). Alternatively, transcription factor binding may first facilitate promoter-enhancer interaction via looping, which allows promoter-bound TET/TDG proteins to access distal-regulatory elements.
Oxidized 5mC bases as potential epigenetic marks
It is known that specific protein domains exist to recognize unmodified, hemi- or fully methylated CpG sites. For example, unmethylated CpG sites [CG:GC] can be specifically recognized by CXXC domain containing proteins (Long et al., 2013), whereas the SRA domain in UHRF1 and methyl-CpG-binding domains (MBD) can specifically recognize hemi-methylated [mCG:GC] and symmetrically methylated [mCG:GCm] CpG dyads, respectively (Hashimoto et al., 2010). Besides acting as intermediates of active DNA demethylation, growing evidence suggests that 5hmC is relatively stable in the genome and may potentially influence transcription by attracting or repelling specific DNA binding proteins. There is evidence that several MBD proteins, including MBD3 (Yildirim et al., 2011), MeCP2 (Mellen et al., 2012), and MBD4 (Otani et al., 2013a), may also bind to hemi- or fully hydroxymethylated CpG sites, suggesting that these proteins have dual binding capacity for 5mC and 5hmC. Recently, mass spectrometry-based unbiased analyses of nuclear proteins in mouse ESCs, neural progenitor cells (NPC) and adult brains have identified a large number of candidate proteins bound selectively to or repelled by 5hmC-containing oligonucleotides (Spruijt et al., 2013). While some 5hmC interacting protein candidates (e.g. WDR76, THY28) are ubiquitously expressed, many of them are expressed in a cell-type specific manner. For instance, 5hmC-specific reader protein UHRF2 and THAP11 are preferentially enriched in NPCs and adult brains, respectively, suggesting that 5hmC may mediate cell-type specific functions by recruiting distinct reader proteins.
Despite their scarcity in the genome, 5fC and 5caC possess unique chemical groups for protein recognition, raising the possibility that they may act as dynamic/transient epigenetic marks (Song and He, 2013). For example, TDG and a number of DNA repair proteins were found to specifically bind to 5fC- and 5caC-containing DNA oligonucleotides (Spruijt et al., 2013). Another study also identified many candidate proteins from mouse ESC nuclear extract that specifically bind to 5fC, including transcription factors, DNA repair proteins and chromatin regulators (Iurlaro et al., 2013). However, these studies cannot distinguish between proteins that bind directly and indirectly to 5fC/5caC, and some candidates may simply bind to formyl or carboxyl groups non-specifically. In addition to the possibility of acting as epigenetic marks, 5fC/5caC may contribute to transcriptional regulation through eliciting “scheduled” DNA repair response at specific gene regulatory elements (Fong et al., 2013).
Biological functions of DNA demethylation in diverse biological processes
Global erasure of 5mC is known to occur in specific stages of mammalian development, including early pre-implantation embryos and developing germ cells. A better understanding of the biochemical basis of DNA demethylation pathway and emergence of new technologies for mapping various cytosine modifications have opened new avenues to study the role of DNA demethylation in these biological processes. Intriguingly, the broad expression patterns of TET enzymes and other components of active DNA demethylation pathway in somatic tissues such as brains imply that dynamic DNA demethylation may be more prevalent than previously thought. Below, we review recent advances in our understanding on biological functions of DNA demethylation in various biological and pathological settings.
DNA demethylation dynamics in pre-implantation development
After the sperm fertilizes the oocyte and before two pronuclei merge, the paternal genome goes through a complex epigenetic remodeling process that includes global erasure of DNA methylation. The sperm undergoes a rapid loss of 5mC before the onset of first DNA replication as measured by immunostaining and locus-specific bisulfite sequencing (Mayer et al., 2000; Oswald et al., 2000); by contrast, the maternal genome is not affected in one-cell zygote, but become gradually demethylated during subsequent cleavage divisions. Recent studies indicate that the loss of paternal DNA methylation coincides with a rapid increase in 5hmC (Inoue and Zhang, 2011; Iqbal et al., 2011; Wossidlo et al., 2011) and 5fC/5caC (Inoue et al., 2011), suggesting that TET-mediated 5mC oxidation is involved in the erasure of 5mC. Knockdown or deletion of TET3, the only 5mC oxidase present at this stage, abolished the loss of 5mC and 5hmC generation in the male pronucleus (Gu et al., 2011; Wossidlo et al., 2011). Tet3-deficient oocytes fertilized with wild-type sperms can develop normally to blastocysts, but many of these heterozygous embryos exhibit developmental failure (Gu et al., 2011), indicating that TET3-mediated rapid and specific oxidation of 5mC from paternal genome is biologically relevant. It will be interesting to identify which regions in the paternal genome are preferentially targeted by TET3 and to determine why 5mC oxidation of paternal genome is functionally important for early embryogenesis.
The striking asymmetry of epigenetic reprogramming between paternal and maternal genomes in one-cell zygote can potentially be achieved through either specific targeting of TET3 to paternal genome or active protection of maternal genome from TET3-mediated 5mC oxidation. A recent study indicates that Stella (also known as PGC7 or Dppa3), a maternal factor essential for early development (Payer et al., 2003), is recruited to oocyte-derived genome by dimethylation of Lys9 on H3 (H3K9me2), a histone mark that is preferentially enriched on maternal chromatin (Nakamura et al., 2012). In the absence of Stella, TET3 is localized to both paternal and maternal genomes (Nakamura et al., 2012; Wossidlo et al., 2011), and the maternal genome also undergoes 5mC oxidation (Nakamura et al., 2007; Nakamura et al., 2012). Locus-specific bisulfite sequencing analysis further demonstrated that Stella is required for protecting a few imprinting control regions (ICRs) in the paternal genome (Nakamura et al., 2007), which are also associated with H3K9me2 (Nakamura et al., 2012). However, the exact mechanism by which Stella inhibits TET3 function is unknown.
After paternal and maternal pronuclei fuse, bulk 5hmC/5fC/5caC signals in sperm-derived chromosomes (AM-PD in Figure 2A) and 5mC in oocyte-derived chromosomes (PD in Figure 2A) are lost in a replication-dependent manner (Inoue et al., 2011; Inoue and Zhang, 2011; Mayer et al., 2000). The passive dilution of maternal 5mC is believed to be ensured by active exclusion of DNMT1o (an oocyte-derived variant of DNMT1) and likely UHRF1 from the nucleus in early embryos (Seisenberger et al., 2013) (Figure 4A). Despite this wave of global DNA demethylation, a subset of maternally derived methylated CpG-rich sequences, including many maternal ICRs and some retroviral repeats such as the intracisternal A particle (IAP) type, remain fully or partially methylated (Lane et al., 2003; Smallwood et al., 2011; Smith et al., 2012). Recent evidence suggests that maintenance of 5mC at ICRs is perhaps conferred through targeting DNMT1 and/or DNMT3A to these regions by zinc finger transcription factor ZFP57 and its partner, KAP1 (also known as TRIM28) (Li et al., 2008; Messerschmidt et al., 2012; Quenneville et al., 2011). In line with the role of this complex in maintenance of 5mC at imprinting loci, ZFP57 prefers to bind to its target sequence (TGCmCGC, present in all known murine ICRs) when it is methylated, but has reduced binding affinity when its target sequence is unmodified or hydroxymethylated DNA (Liu et al., 2012).
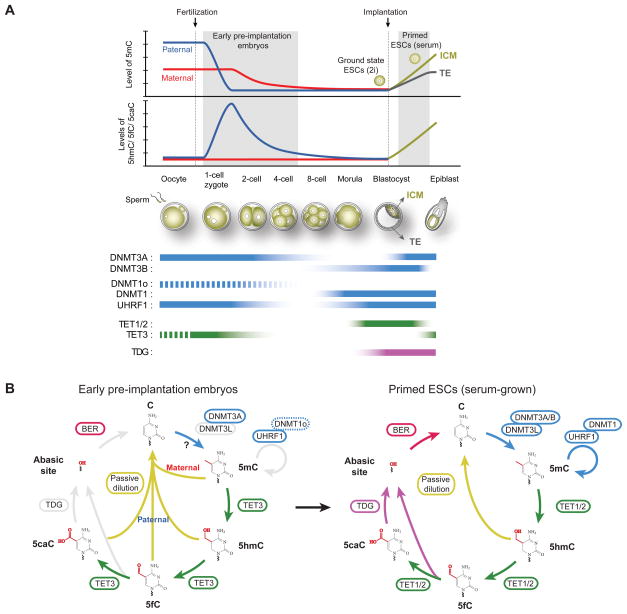
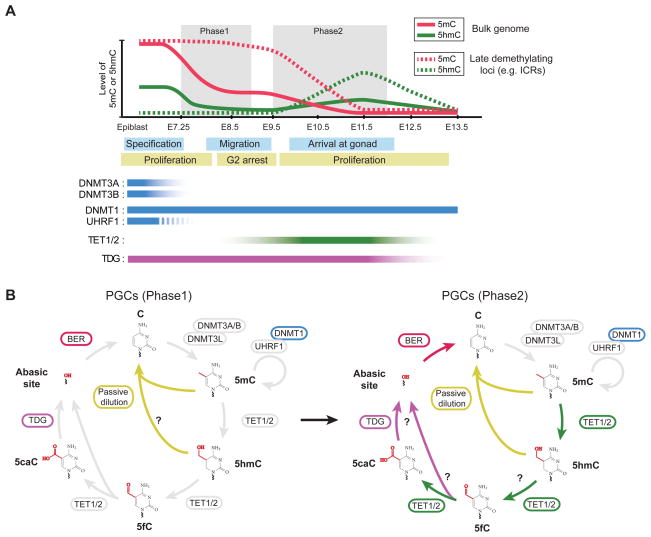
Figure 4. DNA methylation dynamics during mouse preimplantation development.
(A) Dynamic changes in cytosine modifications and relevant enzymes during preimplantation development. Immediately after fertilization, paternal 5mC is rapidly oxidized to 5hmC/5fC/5caC by TET3 proteins. In early pre-implantation embryos, oocyte-derived DNMT1o is largely excluded from nucleus (dash line) and consequently maintenance methylation is inefficient. Oxidized 5mC bases in the paternal genome and 5mC in the maternal genome are therefore passively diluted. The global 5mC level reaches the lowest point around blastocyst stage (E3.5). After implantation, DNA methylation pattern is reestablished by DNMT3A/3B in inner cell mass (ICM) cells, but not in trophectoderm (TE) cells. The genome of ground-state ESC (2i-condition) is hypomethylated (5mC: 1% of all C) and is more similar to the methylome of pre-implantation ICM cells, whereas primed ESCs (serum) possess a methylome (5mC: 4% of all C) that recapitulates overall methylation pattern in epiblast cells.
(B) Developmental stage-specific usage of the cyclic cytosine modifying enzymatic cascade during early embryonic development. Upon fertilization, oocyte-derived TET3 proteins specifically localize to paternal nucleus and convert sperm-derived 5mC to 5hmC/5fC/5caC. DNA replication drives passive loss of 5hmC/5fC/5caC in the paternal genome and 5mC in the maternal genome (left panel). In primed ESCs and cells during ICM-to-epiblast transition in vivo, all enzymes of the cytosine modifying cascade are expressed at relatively high levels. Dnmt3A/3B/3L and DNMT1/UHRF1 establish and maintain 5mC patterns. At some genomic regions (e.g. active enhancers and bivalent promoters) in these cells, TET1/2 oxidize 5mC to 5hmC/5fC/5caC, and TDG/BER excises 5fC/5caC to regenerate unmodified cytosine.
If inefficient maintenance of DNA methylation is a major driver for the genome-wide erasure of 5mC in early embryos (Figure 4B), why the paternal genome needs to be oxidized first? One possible reason might be to rapidly equalize 5mC levels of the two parental genomes as the CpG methylation level differs significantly between sperms (~90%) and oocytes (~40%) before fertilization (Kobayashi et al., 2012). Thus, oxidized 5mC bases in paternal genome may accelerate the demethylation process of paternal genome. 5fC and 5caC generated from further oxidization of 5hmC provide a substrate for TDG-mediated excision repair pathway (Inoue et al., 2011), and there is some evidence supporting a role of BER in paternal genome demethylation (Hajkova et al., 2010). However, bulk 5fC and 5caC signals in the paternal genome remain detectable by immunostaining in 4-cell stage and also exhibit replication-dependent dilution (Inoue et al., 2011), suggesting that 5fC/5caC are not rapidly excised. In support of this view, single-cell RNA-seq analysis suggests that the Tdg gene is expressed at extremely low levels in oocytes and during pre-implantation development (Tang et al., 2011) (Figure 4A), indicating that TDG may only play a limited role in paternal genome demethylation (e.g. locus-specific demethylation). Alternatively, after DNA replication, CpG sites with hemi-modified state [XG:GC, X= hC, fC, caC] in the paternal genome may be more resistant to enzymatic activity of maternal DNMT3A or residual DNMT1o in the nucleus than hemi-methylated CpG sites [mCG:GC], thereby ensuring more efficient paternal demethylation.
Overall, DNA demethylation in early embryos is a highly orchestrated process, requiring both active and passive demethylation. In addition, specific factors, such as Stella and ZFP57/KAP1, may protect 5mC patterns at selective loci (e.g. ICRs) from either active (TET3-mediated 5mC oxidation) or passive (inefficient methylation maintenance) demethylation. Interestingly, in contrast to the situation in mice, recent methylome analyses in Zebrafish suggest that paternal methylation pattern is maintained during early embryogenesis; instead, the relatively hypomethylated maternal genome is progressively reprogrammed to a pattern similar to that of the sperm methylome (Jiang et al., 2013; Potok et al., 2013). Furthermore, 5mC oxidation is not detected during Zebrafish early embryo development (Jiang et al., 2013). Thus, 5mC oxidation-mediated paternal genome demethylation seems to be a unique regulatory mechanism for mammalian embryogenesis. The evolutionary significance of this mechanism awaits further investigation.
DNA demethylation in pluripotent stem cells and reprogramming
As the zygote develops into the blastocyst (E3.5), the genome of inner cell mass (ICM) cells becomes hypomethylated (~21% CpG methylation) as compared to sperm (90%) or oocyte (40%) methylome after replication-dependent dilution of maternal 5mC and paternal 5hmC/5fC/5caC (Kobayashi et al., 2012). Thereafter, DNMT3A/3B mediated de novo methylation takes places during the developmental transition from blastocyst to epiblast stage (70%) by E6.5 (Seisenberger et al., 2012; Smith et al., 2012) (Figure 4A). ESCs are initially derived from the ICM of blastocysts, and these pluripotent stem cells possess unique properties of self-renewing while maintaining potential to differentiate into any somatic cell lineages (Hanna et al., 2010). Traditionally, mouse ESCs are cultured in serum supplemented with leukemia inhibitory factor (LIF) and their methylomes resemble those of somatic cells (70% CpG methylation). Mouse ESCs grown in serum exhibit greater heterogeneity and consist of at least two distinguishable populations, one with a transcriptome similar to the naïve ICM-like state and the other similar to the primed epibalst-like state.
Inhibition of the mitogen-activated protein (MAPK) pathway and the glycogen synthase kinase (GSK3) pathway, via two kinase inhibitors (PD0325901 for MAPK and CHIR99021 for GSK3, known as 2i condition), is sufficient to maintain mouse ESCs in the naïve ICM-like state (Ying et al., 2008). Mouse ESCs grown in the 2i condition appear to be a homogeneous population that has a transcriptome and histone modification profile distinct from that of serum-grown mouse ESCs (Marks et al., 2012). Using mass spectrometry or whole-genome BS-Seq (WGBS), recent studies showed that 2i-grown mouse ESCs adopt a globally hypomethylated genome recapitulating that of ICM cells in vivo (Ficz et al., 2013; Habibi et al., 2013; Leitch et al., 2013). Strikingly, switching from serum to 2i-condition induces genome-wide DNA demethylation in mouse ESCs, which is on a scale and pattern similar to the early embryos. While the maintenance methylation system (DNMT1/UHRF1) and TET1/2 are expressed at comparable levels between ground state (2i) and primed state (serum) mouse ESCs, PR-domain-containing 14 (PRDM14) is up-regulated in 2i-grown mouse ESCs and mediates rapid transcriptional down-regulation of the de novo DNA methylation machinery (DNMT3A/3B/3L) (Yamaji et al., 2013). Available evidence suggests that global demethylation during the serum-to-2i transition is primarily induced by the down-regulation of DNMT3A/3B/3L and TET1/2-mediated transient 5mC oxidation mediate (Blaschke et al., 2013; Ficz et al., 2013; Habibi et al., 2013). Overall, globally hypomethylated genomes observed in ground state mouse ESCs (2i-condition) closely resemble the methylome of ICM cells in pre-implantation blastocysts, whereas serum-grown mouse ESCs have a hypermethylated genome similar to that of post-implantation embryos and somatic cells.
In serum-grown mouse ESCs, all major components of the cytosine-modifying cascade (DNMT, TET, TDG, and BER proteins) are expressed (Figure 4B). Thus, ESCs in primed state provide a model system for understanding DNA demethylation dynamics and its regulatory mechanisms. Single or combined depletion of Tet1 and Tet2 in mouse ESCs, by short-hairpin RNA (shRNA)-mediated knockdown or genetic inactivation, produced variable in vitro phenotypes (Dawlaty et al., 2013; Dawlaty et al., 2011; Ficz et al., 2011; Freudenberg et al., 2012; Ito et al., 2010; Koh et al., 2011; Williams et al., 2011). Multiple studies confirmed that depletion of TET1 alone or in combination with TET2 skews in vitro differentiation towards trophectoderm and primitive endoderm, and teratomas derived from Tet1−/− or Tet1/2−/− ESCs are enriched for trophoblast cells. In support of a role of TET1 in regulating early lineage commitment, TET1 negatively regulates the expression of key regulators of trophectoderm lineage, such as Cdx2 and Eomes, possibly by facilitating binding of repressor complexes to target gene promoters (Williams et al., 2011; Wu et al., 2011b). However, there are conflicting results concerning the role of TET1 in the maintenance of mouse ESCs (Dawlaty et al., 2011; Freudenberg et al., 2012; Ito et al., 2010; Williams et al., 2011; Wu and Zhang, 2011b). The use of different cell lines and culturing conditions may contribute to these discrepancies, although potential off-target effects of specific shRNAs cannot be ruled out. Because all these studies focused on serum-grown ESCs, fluctuating epigenetic state associated with this heterogeneous ESC population may also contribute to the variable phenotype in Tet1-depleted mouse ESCs. Overall, current data indicate that, despite the presence of relatively high level of TET1/2 proteins and oxidized methylcytosines in ESCs, TET enzymes are largely dispensable for ESC maintenance but may play a role in guiding ESCs to properly differentiate into specific lineages and/or regulating later development of the embryos. Indeed, while Tet1−/− and Tet2−/− single mutant mice are viable, roughly half of Tet1−/−Tet2−/− double knockout (DKO) mice from intercrossing double-heterozygous animals die perinatally with severe developmental abnormalities, indicating a role of TET1/2 proteins in regulating embryonic development (Dawlaty et al., 2013). The partially penetrant perinatal lethality of TET1/2 DKO mutants suggests TET3 may compensate for the loss of TET1/2 during development. In line with this notion, all Tet3−/− single mutants die perinatally (Gu et al., 2011).
While additional studies are needed to clarify the exact function of TET proteins in mouse ESCs and embryonic development, recent studies support a role for TET enzymes in transcription factor (Oct4/Klf4/Sox2/c-Myc, referred to as OKSM)-mediated reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) (Hanna et al., 2010). One study showed that TET2 is recruited to the Nanog and Esrrb loci (both are pluripotency-related TFs) to facilitate their transcription in the early stage of reprogramming (Doege et al., 2012). In addition, TET1 and TET2 may interact with Nanog protein and promote iPSC generation in a TET catalytic activity-dependent manner (Costa et al., 2013). More strikingly, TET1 overexpression not only promotes transcriptional reactivation of Oct4, but can also replace Oct4 in the reprogramming OKSM cocktail (Gao et al., 2013). A recent study also implicates a role of vitamin C in modulating TET functions during iPSC generation (Chen et al., 2013a).
In addition to TET proteins, TDG may also play a role in stem cell functions and embryonic development. TDG is broadly expressed during early development and in adult tissues. Depletion of Tdg by shRNA or genetic inactivation in mouse ESCs does not cause any discernible phenotype in ESC proliferation or early differentiation (Cortazar et al., 2011; Shen et al., 2013; Song et al., 2013a), suggesting that TDG may regulate later steps of embryonic development. Indeed, Tdg-deficiency in mice causes widespread development deficits and embryonic lethality (Cortazar et al., 2011; Cortellino et al., 2011). Further studies are needed to clarify the exact mechanism by which TDG contributes to developmental gene regulation.
DNA demethylation dynamics during germ cell development
Around E6.5, a subset of posterior epiblast cells (~40 cells) are instructed by specific factors such as BLIMP1 (also known as PRDM1) and PRDM14 to become PGCs (Sasaki and Matsui, 2008). These committed PGCs suppress somatic transcriptional program and re-activate expression of pluripotency-associated factors. Around E7.5, PGCs start to migrate through the hindgut to the developing gonad, where they colonize by E10.5 to initiate epigenetic reprogramming (Figure 5A) (Saitou et al., 2012). Early PGCs (E7.25) are thought to inherit a methylation pattern similar to that of epiblast cells (~70% CpG methylation), but they undergo global erasure of DNA methylation and become nearly completely demethylated in gonadal PGCs around E13.5 (7–14%) (Seisenberger et al., 2012).
Figure 5. Global reprogramming of DNA methylation patterns during early PGC development.
(A) Dynamic changes in cytosine modifications and expression levels of relevant enzymes during early stage of mouse PGC development. Global erasure of DNA methylation in developing PGCs goes through two phases. In the first phase (E7.25 to E9.0), both de novo and maintenance methylation machineries are functionally impaired due to the repression of DNMT3A/3B/3L and UHRF1. There is also evidence that remaining UHRF1 is excluded from nucleus (dash line). Thus, loss of bulk genomic DNA methylation (solid line, inherited from epiblast cell stage) takes place in a replication-dependent manner in phase 1. However, a portion of the genome (dash line) including ICRs and germline-specific genes are partially or fully protected from the first wave of passive demethylation. In the second phase (E9.5 to E12.0), TET1 (possibly TET2) are up-regulated and oxidize remaining 5mC to 5hmC.
(B) Developmental stage-specific usage of the cytosine modifying enzymatic cascade during early stage of PGC development. In phase 1 of PGC demethylation, bulk of the genome undergoes global demethylation in the absence of de novo and maintenance DNA methylation. In phase 2, TET1/2 proteins oxidize remaining 5mC at late demethylating loci. Replication-dependent loss of 5hmC completes the global demethylation process. 5fC/5caC is not specifically enriched in developing PGCs as compared to surrounding somatic cells, so they may be actively excised by TDG/BER pathway that is present in PGCs.
Recent genome-wide DNA methylation studies suggest that genome-wide erasure of DNA methylation in PGCs takes place in two distinct phases (Seisenberger et al., 2012; Vincent et al., 2013). The first phase of demethylatioin (phase 1 in Figure 5A) occurs before or during PGC migration to genital ridge and appears to primarily involve replication-dependent passive dilution of 5mC. Upon PGC specification (E7.25), all components of de novo methylation systems are transcriptionally down-regulated (Kagiwada et al., 2013), possibly due to PRDM14-mediated repression. UHRF1 is also acutely down-regulated in PGCs (Kagiwada et al., 2013; Seisenberger et al., 2012), which prevents localization of DNMT1 to replication foci. In the absence of efficient de novo and maintenance methylation, global erasure of DNA methylation occurs indiscriminately at nearly all genomic features (promoters, exons, introns, repeats and intergenic regions) in early replicating PGCs. By E9.5, the overall CpG methylation level reduces to ~30% in PGCs (Seisenberger et al., 2012). In line with this model, recent evidence suggests that TET proteins are largely dispensable for the initial global DNA demethylation (Figure 5B). Mass spectrometry and WGBS analysis of PGC-like cells differentiated in vitro from Tet1/2-depleted mouse ESCs (shRNA-mediated depletion of Tet1 in Tet2−/− cells) indicate that global loss of 5mC in early PGCs still occurs in the absence of TET1/2 (Vincent et al., 2013). Furthermore, WGBS analysis of Tet1 mutant PGCs derived from gene-trap female mice indicates that Tet1-deficiency does not affect global demethylation (Yamaguchi et al., 2012).
Methylation levels at some genomic regions, including ICRs, CpG-rich promoters of germ cell specific genes, CGIs on X-chromosome genes, and specific type of repeats (e.g. IAPs), are largely maintained during the first phase of global demethylation in PGCs. Demethylation of these sequences is only completed once PGCs settle in gonad and is most likely initiated by TET1/2-mediated 5mC oxidation (phase 2 in Figure 5A). Expression of TET1/2 is up-regulated in gonadal PGCs (relative to early PGCs and surround somatic cells) (Hackett et al., 2013; Yamaguchi et al., 2012), and antibody-based genome-wide mapping of 5mC and 5hmC in PGCs indicates that 5mC is oxidized to 5hmC at many late demethylating loci between E9.5 to E11.5 (Hackett et al., 2013). Time-course analysis suggests that both 5mC and 5hmC are lost at a rate consistent with replication-dependent dilution during this period (Hackett et al., 2013; Kagiwada et al., 2013). Given that TDG and BER components are expressed throughout the PGC development (Kagiwada et al., 2013) (Figure 5B), it is likely that TDG/BER may play a role in removing 5fC/5caC at specific loci during phase 2. Integrated genomic analysis showed that TET1 is specifically required for promoter demethylation and transcriptional activation of germline-specific genes such as meiosis-related genes (e.g. Mae1, Sycp1, and Sycp3) in developing female PGCs (Yamaguchi et al., 2012). The locus-specific demethylation at meiotic gene promoters by TET1 in female PGCs is biologically important, as half of the Tet1-deficient female gametes exhibits a defect in meiotic synapsis and become developmentally arrested later (Yamaguchi et al., 2012).
Conversion of 5mC to 5hmC at ICRs in late PGCs suggests that TET proteins are also required for the complete 5mC erasure at parental imprints (Hackett et al., 2013). Analysis of methylated ICRs of somatic cells fused with embryonic germ cells (EGCs) suggests that TET1 can induce 5mC oxidation specifically at ICRs and mediate eventual erasure of imprints in this in vitro system (Piccolo et al., 2013). Locus-specific methylation analysis in progenies of Tet1−/−Tet2−/− mice revealed aberrant methylation pattern at some ICRs, further supporting a mechanistic link between TET proteins and erasure of parental imprints in PGCs (Dawlaty et al., 2013). More definitive evidence comes from analysis of progenies derived from crossing Tet1−/− male with wild-type female (Yamaguchi et al., 2013). Despite their identical genotype, these heterozygous progenies exhibit a number of variable phenotypes including placental, fetal and postnatal growth defects, and early embryonic lethality. Several lines of evidence suggest that these developmental defects can be largely explained by the Tet1-deficiency induced epigenetic dysregulation of imprinted genes in PGCs. First, many dysregulated imprinting genes, such as Peg10 and Peg3, exhibit aberrant hypermethylation in the paternal allele of differential methylated regions (DMRs) in progenies of Tet1−/− male. Second, RNA-Seq analysis reveals extensive dysregulation of imprinted genes in these heterozygous progenies. Lastly, reduced representation bisulfite sequencing (RRBS) analysis of E13.5 PGCs and sperms of Tet1−/− male mice reveals that aberrantly hypermethylated ICRs observed in Tet1−/− sperms can be traced back to PGCs. Overall, these data suggest that TET1 plays a critical role in resetting parental origin-specific genomic imprinting in PGCs by facilitating the complete erasure of methylation at ICRs (phase 2 in Figure 5). Curiously, it is current unclear why and how late demethylating loci such as promoters of meiotic genes and ICRs of imprinting genes are protected from the initial phase of global demethylation in early PGCs (phase 1 in Figure 5).
In summary, both passive and active demethylation mechanisms are critical to complete genome-wide erasure of DNA methylation in developing PGCs. This is reminiscent of global demethylation in early embryos. In both cases, passive dilution is facilitated by inefficient maintenance methylation systems (exclusion of Dnmt1o in early embryos and transcriptional repression of Uhrf1 in developing PGCs). Meanwhile, 5mC oxidation by TET3 in zygotes and TET1 (potentially TET2) in PGCs is indispensable for achieving efficient and complete erasure of 5mC in paternal genome of early embryos and late demethylating loci (e.g. meiotic genes and ICRs) in PGCs, respectively.
Role of DNA demethylation in neural development
The development of mammalian brain is a spatiotemporally orchestrated process that requires precisely controlled gene regulatory program to generate diverse, functionally distinct populations of neurons and glia. Inactivation of DNMTs in the nervous system results in neurodevelopmental deficits and impaired neuronal functions (Fan et al., 2001; Fan et al., 2005; Feng et al., 2010a; LaPlant et al., 2010; Nguyen et al., 2007; Wu et al., 2010), suggesting that precise control of DNA methylation pattern is required for proper brain development and maturation. Interestingly, distinct parts of mammalian brains, including frontal cortex (Lister et al., 2013), hippocampus (Munzel et al., 2010), and cerebellum (Song et al., 2011; Szulwach et al., 2011b), all exhibit age-dependent acquisition of 5hmC (e.g. 0.2% of cytosines in fetal cortex versus 0.9% of cytosines in adult cortex) (Lister et al., 2013). In fact, the genome of mature neurons in adult central nervous system contains the highest level of 5hmC of any mammalian cell-type (~40% as abundant as 5mC in Purkinje neurons in cerebellum) (Kriaucionis and Heintz, 2009). These observations indicate that 5mC oxidation and potentially DNA demethylation may be functionally important for neuronal differentiation and maturation processes.
All three Tet genes are expressed at detectable levels in developing mouse brains (Hahn et al., 2013; Szwagierczak et al., 2010). In embryonic cortex, an increase in expression levels of Tet2 and Tet3 is observed when the Nestin-positive neural progenitor cells (NPCs) differentiate into Tubb3-expressing neurons, coinciding with an elevation of 5hmC level during neuronal differentiation (Hahn et al., 2013). Knockdown of Tet2/3 in developing embryonic cortex via in utero electroporation results in neuronal differentiation deficits; in contrast, overexpression of Tet2/3 seems to promote embryonic cortical neurogenesis (Hahn et al., 2013). In addition, depletion of tet3 in Xenopus (X. laevis) embryos by antisense morpholinos causes defects in early eye and neural development. Interestingly, intact CXXC domain and catalytic activity of TET3 protein are both essential for its role in target gene regulation and normal head development (Xu et al., 2012). Thus, these results support a conserved role of specific TET proteins in regulating early stages of brain development.
Growing evidence suggests that TET proteins may also regulate adult brain functions. While Tet1−/− mice have no detectable defects in embryonic development, a recent study demonstrates that TET1 may promote adult hippocampal neurogenesis by regulating neural progenitor proliferation and Tet1-deficiency may cause defects in spatial learning as well as short-term memory (Zhang et al., 2013). Analysis of DNA methylation by RRBS and gene expression in Tet1-deficient NPCs isolated from adult dentate gyrus (DG) of hippocampus identified a cohort of genes that become promoter hypermethylated and down-regulated in the absence of TET1(Zhang et al., 2013). Another study of adult Tet1−/− mice revealed that multiple neuronal activity-regulated genes, including Npas4, c-Fos, and Arc, are transcriptionally down-regulated, and promoter of Npas4 becomes hypermethylated in absence of TET1 (Rudenko et al., 2013). This study also demonstrated that Tet1−/− mice are associated with abnormal hippocampal long-term depression (LTD) and impaired memory extinction. Interestingly, viral-mediated overexpression of TET1 or a catalytically inactive form of TET1 in adult hippocampus resulted in transcriptional up-regulation of several memory-associated genes and impaired contextual fear memory (Kaas et al., 2013), suggesting a 5mC oxidase-independent role of TET1 in adult brains. Given that TET2 and TET3 are also expressed in adult mouse brains, it will be interesting to examine the overlapping and distinct function of different TET proteins in adult brains.
The high abundance of 5hmC in adult brains raises the possibility that 5hmC may act as an epigenetic mark. Indeed, current evidence suggests that 5hmC may recruit MeCP2 (Mellen et al., 2012) and other putative binding protein candidates (Spruijt et al., 2013) to facilitate transcription in postnatal brains. Interestingly, a Rett syndrome mutation, R133C, preferentially disrupts the ability of MeCP2 to bind to 5hmC, but not 5mC (Mellen et al., 2012), implicating impairment of 5hmC binding of MeCP2 in this debilitating autism spectrum disorder. Comparative analysis of 5mC, 5hmC and gene expression in specific cell types in cerebellum or olfactory sensory neurons suggest that enrichment of 5hmC and depletion of 5mC within intragenic regions positively correlate with transcription and chromatin accessibility (Colquitt et al., 2013; Mellen et al., 2012). However, the functional significance of the interplay between 5hmC and its potential binding proteins remains unclear.
While most studies focused on 5mC in CpG context, non-CpG methylation is relatively prevalent in adult mouse brains (Xie et al., 2012). Recently, a comprehensive base-resolution analyses of 5mC and 5hmC in mammalian frontal cortex in both fetal and adult stages indicate that non-CpG methylation (mCH) and CpG hydroxymethylation (hCG) drastically build up in cortical neurons after birth (Lister et al., 2013), coinciding with the peak of synaptogenesis and synaptic pruning in the cortex. Intriguingly, this study demonstrated that mCH could become a dominant form of cytosine modifications in adult brains, accounting for 53% in adult human cortical neuronal genome. Furthermore, the genomic positions of mCH in adult brains are highly conserved between individuals, indicating a precisely controlled process catalyzing non-CpG methylation in adult brain. Current evidence suggests that the developmental acquisition of neuronal mCH is mediated by DNMT3A, whose expression is transiently spiked during the same period. In mature neurons, intragenic mCH is preferentially enriched at inactive non-neuronal lineage-specific genes, indicating a role in negative regulation of the associated transcripts. By contrast, genic hCG is positively correlated with gene expression levels. In addition to the 5mC/5hmC dynamics detected in genic regions, distal cis-regulatory regions are also associated with developmental change in cytosine modification states. In fetal brains, hCG and mCG are preferentially enriched at a large cohort of poised distal-regulatory regions that are subsequently demethylated (converting to unmodified CG) and activated in the adult brain. Tet2-deficiency partially blocks the demethylation process at these hCG-marked poised regulatory elements. However, it is currently unclear whether this TET-mediated demethylation process at poised neuronal enhancers primarily occurs in proliferating neuronal progenitors or post-mitotic neurons. Publically accessible RNA-seq datasets indicate that Tdg gene is expressed in both fetal and adult mouse cortex, suggesting that TET/TDG-active demethylation may take place in both progenitors and neurons. Further studies are needed to reveal the mechanism and biological significance of widespread methylome reconfiguration in the neuronal genome during the maturation process of brain.
DNA demethylation and cancer
Aberrant DNA methylation is one of the hallmarks associated with cancer cells (Baylin and Jones, 2011; Esteller, 2007). Growing evidence suggests that impairment in TET-mediated DNA demethylation may contribute to tumorigenesis (Cimmino et al., 2011; Wu and Zhang, 2011a). The initial evidence implicating TET proteins in cancer development came from the identification of human TET1 as a rare fusion partner of MLL in patients with acute myeloid leukemia (AML) (Ono et al., 2002). Subsequent studies reveal a link between recurrent mutations in the TET2 gene and myelodysplastic disorders (Delhommeau et al., 2009; Langemeijer et al., 2009). Mouse genetic studies have demonstrated that Tet2-deficiency causes enhanced self-renew of hematopoietic stem cells (Ko et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011), supporting a role of TET2 as tumor suppressor for leukemia. TET2 mutations associated with myeloid malignancies impair enzymatic activity and cause consistent loss of 5hmC in patient bone marrows (Ko et al., 2010).
Interestingly, TET2 mutations are found to be mutually exclusive with neomorphic mutations in IDH1/2 (R132 of IDH1 and R140/172 of IDH2) in AML (Dang et al., 2009; Ward et al., 2010). Wild-type IDH1/2 converts isocitrate to α-KG, whereas mutant IDH enzymes produce 2-hydroxyglutarate (2-HG), an oncometabolite that competitively inhibit many α-KG dependent dioxygenases, including TET proteins (Figueroa et al., 2010; Xu et al., 2011a). Similarly, inactivation mutations in SDH and FH, which have been identified in a subset of cancers, lead to marked accumulation of succinate and fumarate, both of which can inhibit α-KG dependent dioxygenases (Xiao et al., 2012). Thus, mutations in these TCA cycle enzymes may lead to inactivation of TET proteins and alteration of methylome in cancer cells.
Recent studies suggest that transcriptional down-regulation of all three TET genes and IDH1/2 is observed in many solid cancers that are not known to be associated with mutations in these enzymes, and global loss of 5hmC may be broadly used as a diagnostic biomarker for human melanoma, breast, liver, lung pancreatic and prostate cancers (Hsu et al., 2012; Lian et al., 2012; Yang et al., 2013). Overexpression of oncogenic miRNAs that target TET mRNAs may be one possible mechanism to down-regulate TET proteins and 5hmC levels (Song et al., 2013c). In summary, multiple mechanisms may cause inactivation of TET-mediated 5mC oxidation, but relevant molecular targets of TET proteins that are causally linked to cancer development are not systematically investigated. It is also unclear whether TDG and 5fC/5caC may play a role in cancer.
Conclusion and Perspective
The discovery of oxidative modification of 5mC and related repair mechanism has provided a new perspective on how heritable DNA methylation pattern may be dynamically regulated in the mammalian genome. The realization that epigenetic information can be reversibly “written” and “erased” on mammalian DNA through the cyclic cytosine-modifying cascade raises new mechanistic and biological questions: Which 5-methylcytosines in the genome are selectively targeted by TET and TDG-mediated demethylation activity? Can oxidized 5mC bases (5hmC/5fC/5caC) act as epigenetic marks by recruiting or repelling specific reader proteins? How does targeted excision repair of 5fC/5caC at distal cis-regulatory elements contribute to gene regulation? How is TET-mediated active erasure of 5mC regulated in response to developmental signaling, environmental stimuli and changes in metabolic states? How does this dynamic epigenetic regulatory process functionally contribute to development and diseases in vivo? New technologies such as TAL effectors and CRISPR/Cas9 that allow manipulation of the epigenetic state of cytosine modification at specific loci (Maeder et al., 2013) as well as base-resolution mapping of all five cytosine modification states promise to advance this exciting field substantially in the coming years.
Acknowledgments
We thank Shinpei Yamaguchi and Jifang Tao for critical reading of the manuscript. We apologize to colleagues whose work cannot be cited owing to space constrains. This work was supported by NIH grants GM68804 and U01DK089565 (to Y.Z.). H.W. is supported by a Jane Coffin Childs postdoctoral fellowship. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez Ja, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013:1–7. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annual review of microbiology. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem. 2012;287:33116–33121. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H, Wang X, Hu X, Gu T, Zhou Z, et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013a doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013b;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET Family Proteins and Their Role in Stem Cell Differentiation and Transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Lu X, Luong K, Dai Q, Boitano M, Turner SW, He C, Korlach J. Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC biology. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquitt BM, Allen WE, Barnea G, Lomvardas S. Alteration of genic 5-hydroxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1302759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4:459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Developmental cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson Ma, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, Tront JS, Erinle T, Nguyen N, Latham KE, Sapienza C, Hoffman B, Liebermann DA. Conserved DNA methylation in Gadd45a(−/−) mice. Epigenetics. 2009:4. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010a;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. FGF Signaling Inhibition in ESCs Drives Rapid Genome-wide Demethylation to the Epigenetic Ground State of Pluripotency. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg JM, Ghosh S, Lackford BL, Yellaboina S, Zheng X, Li R, Cuddapah S, Wade PA, Hu G, Jothi R. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012;40:3364–3377. doi: 10.1093/nar/gkr1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell stem cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011 doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett Ja, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down Ta, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science (New York, NY) 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell reports. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell. 2013 doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor Wa, Zepeda-Martínez Ja, Rao A. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nature protocols. 2012;7:1897–1908. doi: 10.1038/nprot.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ, Andrews S, Balasubramanian S, Reik W. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. Embo J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerich K, Dingler FA, Rada C, Neuberger MS. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung−/−Msh2−/− mice. Nucleic Acids Res. 2012;40:6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, DiMenna L, Schrode N, Liu TC, Franck P, Munoz-Descalzo S, Hadjantonakis AK, Zarrin AA, Chaudhuri J, Elemento O, et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013;500:89–92. doi: 10.1038/nature12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WW, Gong L, Bayley H. Single-molecule detection of 5-hydroxymethylcytosine in DNA through chemical modification and nanopore analysis. Angew Chem Int Ed Engl. 2013;52:4350–4355. doi: 10.1002/anie.201300413. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai C, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011 doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during Mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nature chemical biology. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochemical Society transactions. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- Lu X, Song CX, Szulwach K, Wang Z, Weidenbacher P, Jin P, He C. Chemical Modification-Assisted Bisulfite Sequencing (CAB-Seq) for 5-Carboxylcytosine Detection in DNA. J Am Chem Soc. 2013;135:9315–9317. doi: 10.1021/ja4044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013 doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011 doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese F, Carrillo-de Santa Pau E, Stunnenberg HG. 5-Hydroxymethylcytosine: a new kid on the epigenetic block? Molecular systems biology. 2011;7:562. doi: 10.1038/msb.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nature chemical biology. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Otani J, Arita K, Kato T, Kinoshita M, Kimura H, Suetake I, Tajima S, Ariyoshi M, Shirakawa M. Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J Biol Chem. 2013a;288:6351–6362. doi: 10.1074/jbc.M112.431098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Kimura H, Sharif J, Endo TA, Mishima Y, Kawakami T, Koseki H, Shirakawa M, Suetake I, Tajima S. Cell cycle-dependent turnover of 5-hydroxymethyl Cytosine in mouse embryonic stem cells. PLoS One. 2013b;8:e82961. doi: 10.1371/journal.pone.0082961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA. stella Is a Maternal Effect Gene Required for Normal Early Development in Mice. Current Biology. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153:759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenetics and cell genetics. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schiesser S, Hackner B, Pfaffeneder T, Muller M, Hagemeier C, Truss M, Carell T. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl. 2012;51:6516–6520. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Current opinion in cell biology. 2013 doi: 10.1016/j.ceb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, Kono T, Sasaki H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr, Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochimica et biophysica acta. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012 doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013a;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, He C, Korlach J. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature methods. 2012a;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem Sci. 2013 doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012b;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Su J, Ito K, Ala U, Kats L, Webster K, Sun Su M, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan David E, et al. The Oncogenic MicroRNA miR-22 Targets the TET2 Tumor Suppressor to Promote Hematopoietic Stem Cell Self-Renewal and Transformation. Cell Stem Cell. 2013b;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-Antagonism Regulates Breast Cancer Stemness and Metastasis via TET-Family-Dependent Chromatin Remodeling. Cell. 2013c;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PWTC, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chemical reviews. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Bao S, Lee C, Wang X, Tuch BB, Heard E, Lao K, Surani MA. Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS One. 2011;6:e21208. doi: 10.1371/journal.pone.0021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Hunter JM, Nestor CE, Dunican DS, Terranova R, Moggs JG, Meehan RR. Comparative analysis of affinity-based 5-hydroxymethylation enrichment techniques. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Molecular cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, Lee SA, Le T, Yoon AJ, Faull K, et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011a;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011b;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011a;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011b:10. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nature reviews Molecular cell biology. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011a;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011b;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Mao S-Q, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, et al. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. Journal of the American Chemical Society. 2013 doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nature chemical biology. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci U S A. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]