Abstract
The classic view is challenged that the migration of the repressor of photomorphogenesis COP1 from the nucleus to the cytoplasm is too slow to participate in light-mediated developmental events.
In darkness, the E3 ligase COSTITUTIVE PHOTOMORPHOGENESIS1 (COP1) targets to degradation ELONGATED HYPOCOTYL5 (HY5) and other proteins required for photomorphogenesis (Osterlund et al., 2000a; Saijo et al., 2003; Seo et al., 2003). Light inactivates COP1, allowing photomorphogenesis to proceed (Lau and Deng, 2012). Pioneer studies have demonstrated that biologically active COP1-GUS fusion proteins migrate from the nucleus to the cytoplasm (von Arnim and Deng, 1994) but this shift is too slow to be the mechanism of COP1 inactivation involved in rapid light-induced responses (von Arnim et al., 1997; Lau and Deng, 2012). Here, we re-examine the latter conclusion by using YFP-COP1 and comparing the kinetics to that of nuclear HY5.
The shoot of young seedlings experiences full darkness when the seeds germinate buried in the soil. The absence of light impairs photosynthesis, forcing the seedling to live heterotrophically on its reserves, and induces skotomorphogenesis, a specialized developmental pattern that optimizes the chances of rapid shoot emergence from the soil. This pattern is characterized in Arabidopsis (Arabidopsis thaliana) by the fast growth of the hypocotyl, the restricted growth of the cotyledons, the formation of apical hook, and the presence of a rudimentary photosynthetic apparatus (Kami et al., 2010). Upon light exposure, the growth of the hypocotyl is restricted, the cotyledons expand and unfold, the apical hook disappears, and the synthesis of a fully functional photosynthetic apparatus and photoprotective pigments is stimulated. Light triggers the shift between skoto- and photomorphogenesis (deetiolation).
COP1 is a key repressor of photomorphogenesis, and the cop1 mutants show photomorphogenesis in darkness (Deng et al., 1992). COP1 is a ubiquitin E3 ligase that targets to degradation proteins required for photomorphogenesis, including the basic Leu zipper transcription factors HY5 (Osterlund et al., 2000a, 2000b; Saijo et al., 2003) and HY5-HOMOLOG (Holm et al., 2002), the R2R3-MYB transcription factor LONG AFTER FAR-RED LIGHT1 (LAF1; Seo et al., 2003), and the basic helix-loop-helix protein LONG HYPOCOTYL IN FAR-RED REDUCED PHYTOCHROME SIGNALING1 (Duek et al., 2004; Jang et al., 2005; Yang et al., 2005), among others (Lau and Deng, 2012). In darkness, COP1 forms a complex with SUPPRESSOR OF PHYTOCHROME A-105 1 (SPA1; Hoecker and Quail, 2001; Saijo et al., 2003; Seo et al., 2003), and mutations at spa1 enhance the abundance of HY5, suggesting that SPA1 enhances COP1 activity (Saijo et al., 2003; Zhu et al., 2008).
Light activates phytochrome (Li et al., 2011) and cryptochrome (Yu et al., 2010) photoreceptors and inactivates COP1, allowing the accumulation of its targets (Osterlund et al., 2000a, 2000b; Saijo et al., 2003; Duek et al., 2004; Jang et al., 2005; Yang et al., 2005). Light induces the dissociation of the COP1-SPA1 complex (Saijo et al., 2003). Under blue light, cryptochrome 1 interacts with SPA1, reducing SPA1 interaction with COP1 (Lian et al., 2011; Liu et al., 2011). Because, in darkness, SPA1 cooperates with COP1 to reduce HY5 abundance (Saijo et al., 2003; Zhu et al., 2008), light perceived by cryptochrome 1 would lower COP1 activity by reducing COP1-SPA1 interaction (Fankhauser and Ulm, 2011; Lian et al., 2011; Liu et al., 2011). It is not clear how the disruption of the complex with SPA1 affects COP1 activity toward its nuclear targets; it could be via changes in COP1 E3-ligase activity, but SPA1 enhances in vitro COP1-mediated LAF1 ubiquitylation only at low COP1 concentrations (Seo et al., 2003) and actually inhibits in vitro ubiquitylation of HY5 (Saijo et al., 2003). Whether phytochromes affect the COP1-SPA1 complex remains to be elucidated.
In darkness, COP1 forms nuclear speckles (von Arnim et al., 1998). Deficient translocation to the nucleus (Stacey et al., 2000) or formation of nuclear speckles (Nakagawa and Komeda, 2004) correlates with impaired biological activity of mutant COP1 proteins. Both phytochromes and cryptochromes shift COP1 from the nucleus to the cytoplasm as indicated by the analysis of COP1-GUS fusion proteins (Osterlund and Deng, 1998). Mutations at the nuclear-exclusion motifs indicate that nuclear exclusion of COP1 protein is important to regulate its biological activity and nuclear abundance in the light (Subramanian et al., 2004), although a contribution of differential subcellular stability cannot be excluded. The kinetics of COP1-GUS migration to the cytoplasm is very slow, and changes are not detectable during the first 12 h after the dark-to-light transition, suggesting that relocalization is important to stably maintain a committed photomorphogenic fate rather than in causing such a commitment (von Arnim et al., 1997). This idea has persisted to the present day (Chen and Chory, 2011; Lau and Deng, 2012), and for this reason, COP1 inactivation by light has recently been modeled without incorporating COP1 migration to the cytoplasm (Pokhilko et al., 2011).
A slow kinetics of COP1 remobilization would help to commit the seedling to photomorphogenesis by buffering the fluctuations in light signal input under day-night cycles (von Arnim et al., 1997). However, under day-night cycles and even in response to shade, yellow fluorescent protein (YFP)-COP1 shows rapid remobilization from or toward the nucleus (Pacín et al., 2013). The latter prompted us to reexamine the kinetics of nuclear COP1 during deetiolation to elucidate whether either COP1 subcellular partitioning becomes more dynamic after deetiolation or the depletion of nuclear COP1 during deetiolation is faster than originally revealed by the studies involving GUS staining. This issue is central to our understanding of the mechanisms involved in deetiolation because a rapid decline in nuclear COP1 would frame this process within the time window when COP1 is inactivated and its target proteins involved in photomorphogenesis accumulate upon light exposure.
RAPID LIGHT-INDUCED DECLINE IN NUCLEAR YFP-COP1
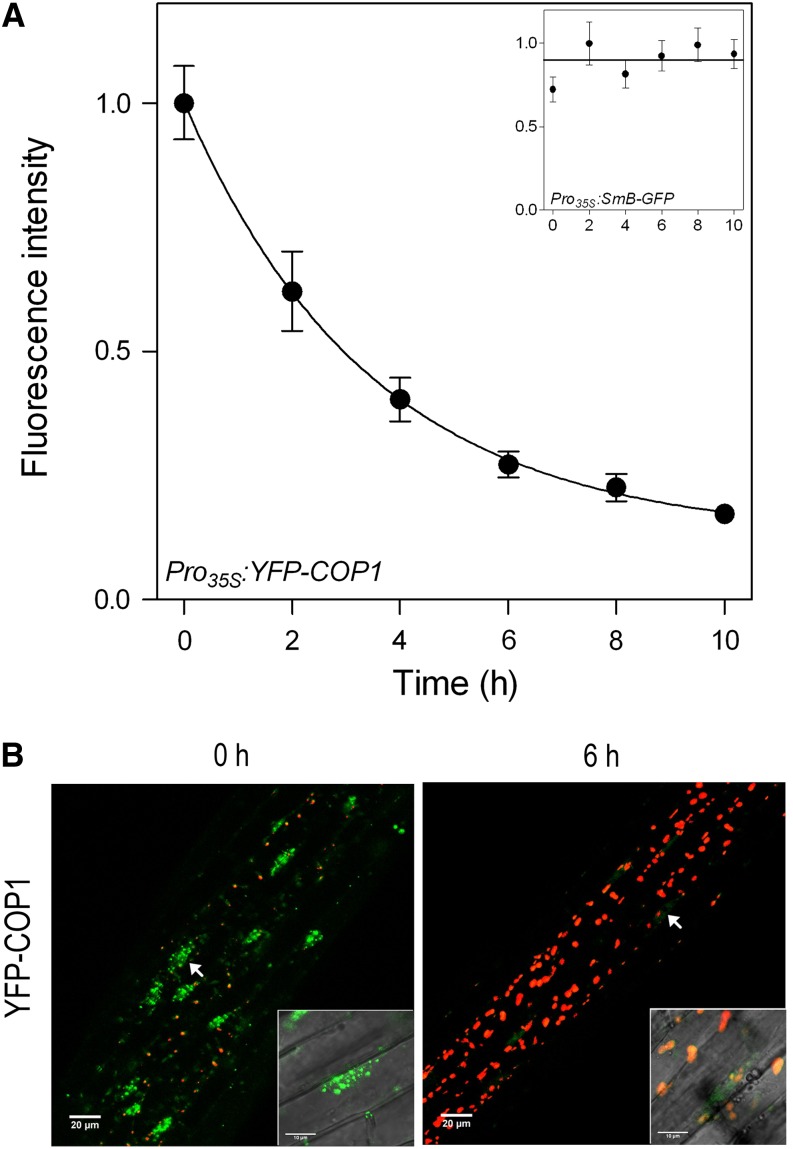
Seedlings of Arabidopsis where the cop1-4 mutation is complemented by Pro 35S:YFP-COP1 (Oravecz et al., 2006) were grown in full darkness for 3 d and then transferred to 100 µmol m–2 s–1 white light with a red/far-red ratio of 1.1 (growth conditions and light spectrum were as described [Pacín et al., 2013]). The nuclear fluorescence of YFP-COP1 was measured by using confocal microscopy as described (Karayekov et al., 2013). The fluorescence signal caused by nuclear YFP-COP1 followed an exponential decay with a half-life of 2.4 ± 0.5 h (mean ± se; Fig. 1). As a control, the nuclear fluorescence caused by the spliceosomal SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B fused to GFP (Pro35S:SmB-GFP; Lorković et al., 2004) remained stable under the same conditions (Fig. 1A, inset), indicating that the decay observed for COP1 is not the result of fluorescence bleaching caused by the light treatment. We conclude that YFP-COP1 remobilization is much faster than that previously reported for COP1-GUS. One possible explanation might be the larger shift in size caused by GUS compared with GFP fusions, and the tendency of GUS to form tetramers that might interfere with mobility and translocation (von Arnim, 2007).
Figure 1.
Rapid decline in COP1 nuclear abundance in response to light. A, Time course of the fluorescence intensity (normalized to the maximum of each experiment) produced by YFP-COP1 in the nucleus of hypocotyl cells of dark-grown seedlings of Arabidopsis transferred to white light at time 0. Data are means ± se of eight to 11 seedlings. The inset shows the stable nuclear fluorescence produced by SmB-GFP under the same conditions, as a negative control for the fluorescence decay. B, Confocal microscopy images of representative hypocotyls of cop1-4 Pro35S:YFP-COP1 seedlings grown in darkness (0 h) or exposed to 6 h white light (6 h). Chlorophyll fluorescence is depicted in red, and the insets show the overlap between confocal and transmission images. [See online article for color version of this figure.]
THERE IS A LAG IN LIGHT-INDUCED STABILIZATION OF HY5
The light-induced decay of COP1 in the light is much faster than previously considered. However, the biological significance that could be attached to this decay depends on the actual kinetics of stabilization observed for COP1 targets.
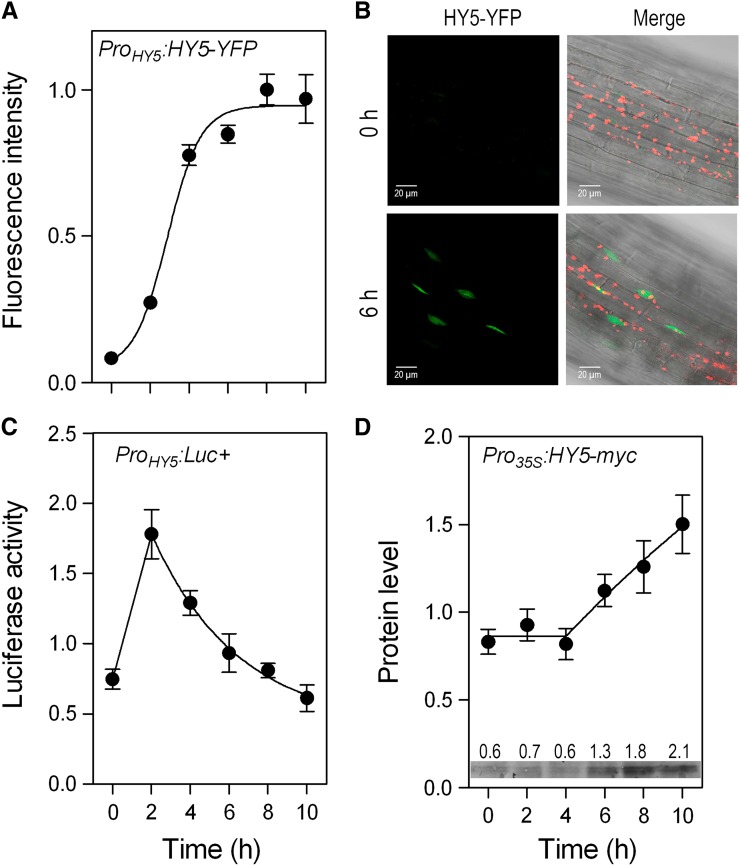
We investigated the kinetics of accumulation of the basic Leu zipper transcription factor HY5 (Oyama et al., 1997), one of the key targets of COP1 (Osterlund et al., 2000a), by using transgenic seedlings of the hy5-1 background bearing the ProHY5:HY5-YFP fusion (Oravecz et al., 2006), under the same conditions involved in the analysis of COP1. By using confocal microscopy (Karayekov et al., 2013), we observed that upon light exposure, the nuclear pool of HY5-YFP undergoes a rapid increase with a half-life of 2.9 ± 0.2 h (mean ± se; Fig. 2A), which leads to elevated levels that remained relatively stable between 6 and 10 h (Fig. 2, A and B). The results are consistent with a recent report showing the time course of HY5 protein blots (Pokhilko et al., 2011).
Figure 2.
Kinetics of HY5 nuclear accumulation, HY5 promoter activity, and HY5 protein stability in dark-grown seedlings transferred to white light. A, Time course of fluorescence intensity (normalized to the maximum of each experiment) produced by HY5-YFP in the nucleus of hypocotyl cells. Data are means ± se of eight to 14 seedlings. B, Confocal microscopy images of representative hypocotyls of hy5-1 ProHY5:HY5-YFP seedlings grown in darkness (0 h) or exposed to 6 h white light (6 h). Chlorophyll fluorescence is depicted in red, and the merge between confocal and transmission images are included. C, Time course of luciferase activity driven by the HY5 promoter (normalized to the median of each experiment). Data are means ± se of seven plates with seedlings. D, Time course of HY5 protein abundance (normalized to the median of each experiment) in hy5-211 Pro35S:HY5-MYC seedlings. HY5-MYC was detected by using the mouse anti-c-Myc monoclonal antibody (Life Technologies) as primary antibody and goat anti-mouse IgG-horseradish peroxidase as secondary antibody and Amersham ECL Prime Western Blotting Detection Reagent (Roche). Data are means ± se of five boxes with seedlings. The inset shows a representative protein blot and the quantification of its bands. [See online article for color version of this figure.]
One of the components of the light-induced accumulation of HY5 is the enhanced expression of the HY5 gene. The activity of the HY5 gene promoter was investigated by using a line bearing the ProHY5:LUC+ transgene (Ulm et al., 2004) and measuring luciferase activity as described (Karayekov et al., 2013). The activity of the HY5 promoter showed a peak at or before the 2 h of irradiation followed by a gradual decline (Fig. 2C). These data are consistent with the kinetics of HY5 mRNA shown previously (Tepperman et al., 2001; Pokhilko et al., 2011). The decay in HY5 promoter activity began when the rate of HY5 accumulation was increasing (Fig. 2, B and C), indicating the contribution of posttranscriptional regulation.
In darkness, COP1 ubiquitylates HY5, which becomes targeted to degradation (Osterlund et al., 2000a, 2000b). To investigate the kinetics of the HY5 protein stability without the interference from light-induced changes in HY5 transcription, we used transgenic seedlings of the hy5-211 background complemented by the Pro35S:HY5-MYC fusion (Shin et al., 2007). Protein extraction and protein blots were as described (Sellaro et al., 2009). HY5 protein abundance remained stable during the first 4 h of irradiation and only then increased at a constant rate (Fig. 2D). Therefore, by the time HY5 protein stabilization became obvious, the nuclear pool of COP1 had already decreased to less than one-half of the levels present in darkness.
MECHANISMS OF LIGHT-INDUCED INACTIVATION OF COP1
We propose that, against the currently established idea, the translocation of COP1 out of the nucleus is sufficiently rapid to regulate COP1 activity against its nuclear targets and contribute to the early stabilization of HY5. In fact, upon the dark-to-light transition, the nuclear fluorescence signal followed an exponential decline with a half-life of only 2.4 h, while the stabilization of its target HY5 showed a lag of 4 h. Interestingly, also mammalian COP1 dynamically migrates between the cytoplasm and the nucleus, and this process is important to regulation of its target proteins (Su et al., 2010). Noteworthy, the combination of light-induced HY5 gene expression and HY5 stabilization generated a switch from the weak background levels of nuclear HY5 observed in darkness to relatively stable, elevated levels beyond 6 h.
The occurrence of a rapid decay in nuclear levels does not exclude other mechanisms of inactivation. Compared with dark controls, the interaction between COP1 and SPA1 is reduced 15% and 33%, respectively, after 2 and 4 h of blue light (50 µmol m–2 s–1; Lian et al., 2011), indicating that this pathway of inactivation of COP1 would not be substantially faster or more intense than that involving the remobilization out of the nucleus. The comparable time frames suggest that dissociation from SPA1 could contribute to COP1 remobilization. Alternatively, if dissociation from SPA1 affects only the biochemical activity of COP1, COP1 remobilization would contribute simultaneously with the reductions in intrinsic activity to generate a sharper inactivation of COP1 toward its nuclear targets than any of the two mechanisms in isolation.
Acknowledgments
We thank the following researchers for their kind provision of transgenic seeds: Sabrina Sánchez and Marcelo J. Yanovsky (Fundación Instituto Leloir) for their unpublished Pro35S:SmB-GFP line, Roman Ulm (University of Geneva) for the cop1-4 Pro35S:YFP-COP1, hy5-1 ProHY5:HY5-YFP, and ProHY5:LUC+ lines, and Giltsu Choi (Korea Advanced Institute of Science and Technology) for the hy5-211 Pro35S:HY5-MYC line.
Footnotes
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (grant no. PICT 1819 to J.J.C.) and the University of Buenos Aires (grant no. 20020100100437 to J.J.C.).
These authors contributed equally to the article.
Some figures in this article are displayed in color online but in black and white in print.
References
- Chen M, Chory J. (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G β homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Ulm R. (2011) Light-regulated interactions with SPA proteins underlie cryptochrome-mediated gene expression. Genes Dev 25: 1004–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Quail PH. (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173–38178 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Karayekov E, Sellaro R, Legris M, Yanovsky MJ, Casal JJ. (2013) Heat shock-induced fluctuations in clock and light signaling enhance phytochrome B-mediated Arabidopsis deetiolation. Plant Cell 25: 2892–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X. (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148, doi/10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Hilscher J, Barta A. (2004) Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol Biol Cell 15: 3233–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Komeda Y. (2004) Flowering of Arabidopsis cop1 mutants in darkness. Plant Cell Physiol 45: 398–406 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Deng XW. (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000a) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. (2000b) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ. (2013) COP1 re-accumulates in the nucleus under shade. Plant J 75: 631–641 [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Ramos JA, Holtan H, Maszle DR, Khanna R, Millar AJ. (2011) Ubiquitin ligase switch in plant photomorphogenesis: a hypothesis. J Theor Biol 270: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Hoecker U, Yanovsky M, Chory J, Casal JJ. (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr Biol 19: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Stacey MG, Kopp OR, Kim TH, von Arnim AG. (2000) Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiol 124: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CH, Zhao R, Velazquez-Torres G, Chen J, Gully C, Yeung SC, Lee MH. (2010) Nuclear export regulation of COP1 by 14-3-3σ in response to DNA damage. Mol Cancer 9: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Kim BH, Lyssenko NN, Xu X, Johnson CH, von Arnim AG. (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci USA 101: 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG. (2007) Subcellular localization of GUS- and GFP-tagged proteins in onion epidermal cells. CSH Protoc 2007: t4689. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW. (1997) Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 114: 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C. (2010) The cryptochrome blue light receptors. Arabidopsis Book 8: e0135, doi/10.1199/tab.0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW. (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]