Histone methyltransferase promotes flowering time predominantly under long-day conditions in rice.
Abstract
Trithorax group proteins are chromatin-remodeling factors that activate target gene expression by antagonistically functioning against the Polycomb group. In Arabidopsis (Arabidopsis thaliana), Arabidopsis Trithorax protein1 (ATX1) regulates flowering time and floral organ identity. Here, we observed that suppression of Oryza sativa Trithorax1 (OsTrx1), an ortholog of ATX1, delayed flowering time in rice (Oryza sativa). Because the delay occurred only under long-day conditions, we evaluated the flowering signal pathways that specifically function under long-day conditions. Among them, the OsMADS50 and Heading date1 pathways were not affected by the mutation. However, the Grain number, plant height, and heading date7 (Ghd7) pathway was altered in ostrx1. Transcript levels of OsGI, phytochrome genes, and Early heading date3 (Ehd3), which function upstream of Ghd7, were unchanged in the mutant. Because Trx group proteins form a complex with other proteins to modify the chromatin structure of target genes, we investigated whether OsTrx1 interacts with a previously identified protein that functions upstream of Ghd7. We demonstrated that the plant homeodomain motif of OsTrx1 binds to native histone H3 from the calf thymus and that OsTrx1 binds to Ehd3 through the region between the plant homeodomain and SET domains. Finally, we showed that the SET domain at the C-terminal end of OsTrx1 has histone H3 methyltransferase activity when incubated with oligonucleosomes. Our results suggest that OsTrx1 plays an important role in regulating flowering time in rice by modulating chromatin structure.
Trithorax group (TrxG) proteins are chromatin-remodeling factors that activate transcription or maintain the active transcribed states of target genes. Trithorax (Trx), first discovered in the fruitfly (Drosophila melanogaster), is a key regulator of downstream homeobox genes during early developmental stages (Ingham, 1998). SET Domain-Containing1 (Set1) in yeast (Saccharomyces cerevisiae) and Myeloid/Lymphoid Mixed Lineage Leukemia1 (MLL1) in humans are homologs of the fruitfly Trx (Gu et al., 1992; Tkachuk et al., 1992; Nislow et al., 1997). In Arabidopsis (Arabidopsis thaliana), Arabidopsis Trx protein1 (ATX1) is a homolog of Trx and regulates flowering time and floral organ identity (Alvarez-Venegas et al., 2003). The TrxG proteins activate target gene expression by antagonistically functioning against Polycomb group (PcG) proteins that silence target genes.
The TrxG proteins are components of complexes that consist of three groups of proteins: histone-modifying, ATP-dependent chromatin-remodeling, and sequence-specific DNA-binding proteins. They associate with each other in modifying histone marks or by forming a proper transcriptional status (Schuettengruber et al., 2011). Set1 is a component of the complex proteins associated with Set1 (COMPASS) composed of Set1 and several other subunits that are required for methylation of Lys-4 on histone H3 (Miller et al., 2001; Krogan et al., 2002). MLL1 and Trx are also found in COMPASS-like complexes (Tenney and Shilatifard, 2005; Mohan et al., 2011). MLL1 binds to a subcomplex composed of Absent, Small and Homeotic Discs 2-Like (ASH2L), WD Repeat-Containing5 (WDR5), and Retinoblastoma-Binding Protein5 (RbBP5; Dou et al., 2006). In the fruitfly, Trx is part of trithorax acetylation complex1 (TAC1), which interacts with the histone acetyltransferase cAMP-response element-binding protein and the SET-binding factor1 (Petruk et al., 2001).
The plant life cycle is affected by this formation of complicated complexes because of the association of TrxG and PcG proteins (Alvarez-Venegas et al., 2010). Among the TrxG proteins in Arabidopsis, ATX1 binds to the Arabidopsis Ash2 Relative (ASH2R)-RbBP5-Like (RBL)-WDR5a complex, forming a COMPASS-like complex (Jiang et al., 2011). ATX1 interacts with ULTRAPETALA1, which controls histone methylation patterns of AGAMOUS in the shoot apical meristem (Carles and Fletcher, 2009). Other TrxG proteins, Chromatin Remodeling11 and Chromatin Remodeling17, homologs of the imitation switch of yeast, are associated with RINGLET1 and RINGLET2, and modulate the transition of the vegetative-to-reproductive phase (Li et al., 2012).
Plant homeodomain (PHD) fingers appear among TrxG and PcG proteins. These fingers carry the four Cys/one His/three Cys (C4HC3) signature and are generally known to have two distinct roles: association with histone and interaction with other proteins (Bienz, 2006). For example, Bromodomain and PHD Finger Transcription Factor (BPTF) and Inhibitor of Growth Protein2 (ING2) contain PHD fingers that are critical to the protein-trimethylated Lys-4 on histone H3 (H3K4me3) association (Shi et al., 2006; Wysocka et al., 2006.
In rice (Oryza sativa), diverse proteins containing putative PHD fingers are involved in various developmental processes such as flowering time and the development of anthers and inflorescences (Li et al., 2011; Matsubara et al., 2011; Hu et al., 2012; Yang et al., 2013a). The PHD fingers in CHR729, a rice homolog of Chromodomain, Helicase/ATPase, and DNA-Binding Domain3 (CHD3) protein, recognize H3K4me2 and trimethylated Lys-27 on histone H3 (H3K27me3) and have an overall effect on plant development (Hu et al., 2012). Early heading date3 (Ehd3), encoding a nuclear protein with PHD fingers, regulates flowering time (Matsubara et al., 2011).
Trx proteins also contain a conserved SET domain that has histone methyltransferase (HMTase) activity. They modify specific Lys or Arg residues on histone H3 or H4, making the chromatin structure flexible (Ng et al., 2007). For example, MLL1 in humans possesses H3K4me3 methyltransferases when combined with the ASH2L-RbBP5-WDR5 subcomplex (Dou et al., 2006). In Arabidopsis, ATX1, ATX2, ATXR3, and ATXR7 regulate the flowering repressor Flowering Locus C via H3K4me2/H3K4me3 (Alvarez-Venegas et al., 2003; Pien et al., 2008; Tamada et al., 2009; Yun et al., 2012). Meanwhile, ATXR5 and ATXR6 control the methylation of H3K27 for heterochromatin formation (Jacob et al., 2009). PICKLE, a CHD3 homolog, modulates the levels of H3K27me3 and enhances root meristem activity by acting antagonistically with CURLY LEAF (Aichinger et al., 2011).
Rice has at least 37 genes that encode SET domain group (SDG) proteins. For example, a knockdown of SDG714 causes H3K9 methylation levels to decline, resulting in a deficiency of macro trichomes (Ding et al., 2007). Ectopic expression of SDG714 in Arabidopsis leads to a growth defect due to a global increase in H3K9me2 (Ding et al., 2010). Mutations in SDG724/Long Vegetative Phase1 show late flowering and reduced levels of H3K36me2/H3K36me3 at the OsMADS50 and Rice Flowering Locus T1 (RFT1) loci (Sun et al., 2012). SDG725 targets Dwarf 11 in brassinosteroid signaling via depositions of H3K36me2/H3K36me3. In addition, SDG725 suppression causes late flowering by altering those depositions in several flowering-control genes (Sui et al., 2012, 2013).
Rice is a facultative short-day (SD; 10-h light/14-h dark) plant. Several regulatory genes that control flowering time have been identified in rice. Heading date3a (Hd3a) and RFT1 encode florigens (Kojima et al., 2002; Tamaki et al., 2007; Komiya et al., 2008). They are controlled by Ehd1, which encodes a B-type response regulator (Doi et al., 2004).
Four types of elements modulate expression of Ehd1. The first comprises day length-independent regulators that function regardless of day length. This type includes Rice Indeterminate1 (RID1)/Oryza sativa Indeterminate1 (OsId1)/Ehd2, which acts as a constitutive inducer, as well as Oryza sativa CONSTANS-like4 (OsCOL4), which is a constitutive suppressor of Ehd1 (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008; Lee et al., 2010). The second type of element includes SD-preferential regulators. A mutation in OsMADS51 shows late flowering only under SD conditions (Kim et al., 2007).
The third type contains long-day (LD; 14-h light/10-h dark) preferential regulators. One example is the mutation in OsMADS50, which causes a late-flowering phenotype only under LD conditions (Lee et al., 2004; Ryu et al., 2009). Grain number, plant height, and heading date7 (Ghd7) suppresses flowering preferentially under LD conditions (Xue et al., 2008). Ehd3, acting as a repressor upstream of Ghd7, functions predominantly under LD conditions (Matsubara et al., 2011). Early flowering7 (Ef7)/Heading date17 (Hd17)/Oryza sativa Early Flowering3-1 (OsELF3-1) negatively regulates circadian clock genes and Ghd7 (Matsubara et al., 2012; Saito et al., 2012; Zhao et al., 2012; Yang et al., 2013b). Finally, the fourth type comprises flowering regulators that have conflicting functions depending upon photoperiodic conditions. For example, Hd1 acts as an activator under SD but as a suppressor under LD conditions (Yano et al., 2000).
Here, we report the characterization of a rice homolog of the trithorax gene, Oryza sativa Trithorax1 (OsTrx1), which acts preferentially as an activator of flowering under LD conditions.
RESULTS
Identification of a Late-Flowering Mutant
We screened flowering-time mutants from insertional mutant populations that we had generated previously in Japonica rice by transfer DNA (T-DNA; Jeon et al., 2000; Jeong et al., 2002; An et al., 2003, 2005; Ryu et al., 2004). This enabled us to identify Line 2D-31249, which displayed a late-flowering phenotype in the paddy field (PF; Fig. 1C). In that line, T-DNA was inserted into the ninth intron of LOC_Os09g04890 (Fig. 1A). The locus encodes SDG723, a SET domain-containing protein (Ng et al., 2007). This protein is the most homologous to ATX1, with 58% identity and 71% similarity. We designated this gene OsTrx1. Because ATX1 also functions to control flowering time in Arabidopsis, we speculated that the Trx genes are functionally conserved in the plant kingdom.
Figure 1.
Schematic diagram of OsTrx1 and flowering phenotype of T-DNA insertional ostrx1 mutant. A, OsTrx1 has 25 exons (black boxes) and 24 introns (lines between boxes). Gray boxes indicate 5′ and 3′ untranslated regions. T-DNA shown as a triangle is located at the ninth intron (Line 2D-31249). Primers F1, R1, and GUS were used for genotyping ostrx1; F2 and R2 were used for semiquantitative RT-PCR analyses of OsTrx1 transcript levels (Supplemental Table S1). B, Semiquantitative RT-PCR analyses of OsTrx1 transcripts in ostrx1 and the segregating wild type (WT). Samples were harvested at Zeitgeber time 2 h from uppermost leaf blades of 28 DAG plants grown under LD (14-h light/10-h dark) conditions. Actin1 (LOC_Os03g50885) was used as a reference. C, Phenotypes at ripening stage for ostrx1 plants and the segregating wild type grown in the PF condition. Bar = 20 cm. D, Days to flowering of ostrx1 and the segregating wild type grown under SD (10-h light/14-h dark), LD, or PF conditions. Values are shown as means. Error bars indicate sds. n = 5 or more.
Mutations in OsTrx1 Caused Late Flowering Preferentially under LD Conditions
Reverse transcription PCR (RT-PCR) analyses of the OsTrx1 transcript showed that the gene was not expressed in ostrx1, indicating that the mutant is a null allele (Fig. 1B). In the PF condition, the ostrx1 mutants flowered at approximately 161 d after germination (DAG), 54 d later than the segregating wild-type plants (Fig. 1, C and D). Flowering time of the heterozygous plants did not differ from the wild type, indicating that ostrx1 is a recessive allele.
To determine whether day length influences the ostrx1 phenotype, we monitored flowering time under both SD and LD conditions. Under LD conditions, ostrx1 mutant plants flowered at approximately 145 DAG, whereas wild-type plants flowered at 77 DAG. However, there was no obvious difference in flowering time between the two under SD conditions (Fig. 1D). These results implied that a lack of gene expression resulted in delayed flowering and that OsTrx1 promoted flowering preferentially under LD conditions.
OsTrx1 Interference RNA Transgenic Plants Confirm the Late-Flowering Phenotype
To confirm this late-flowering phenotype of ostrx1, we generated seven OsTrx1 interference RNA (RNAi) transgenic rice plants (Fig. 2A; Supplemental Fig. S1). Among them, three lines (RNAi-1, RNAi-2, and RNAi-3) had high levels of RNAi transcripts, resulting in very low levels of OsTrx1 transcripts (Fig. 2D; Supplemental Fig. S1B). Under the PF conditions, the three RNAi lines flowered approximately 50 d later than the wild type (Fig. 2, B and C; Supplemental Fig. S1A). Those plants also displayed late flowering under LD conditions (Fig. 2C). Therefore, these results confirmed that OsTrx1 is a flowering-time regulator that functions preferentially under LD conditions.
Figure 2.
Phenotypes of OsTrx1 RNAi transgenic plants. A, Schematic diagram of OsTrx1 RNAi construct. pUbi, maize Ubiquitin1 promoter; OsTrx1-C, 720 bp of the carboxy terminus of OsTrx1 cDNA; Tnos indicates nopaline synthase terminator. B, Phenotypes of OsTrx1 RNAi-3 and wild-type plants at heading stage grown under LD conditions. Bar = 20 cm. C, Days to flowering of OsTrx1 RNAi-3 and the wild type grown under LD (left) or PF (right) conditions. D, Quantitative real-time RT-PCR analyses of expression levels of OsTrx1 in OsTrx1 RNAi-3 and wild-type plants. Samples were harvested at Zeitgeber time 4 h from uppermost leaf blades of 56 DAG plants grown under LD conditions. Ubiquitin (LOC_Os03g13170) was used as a reference. Values are shown as means. Error bars indicate sds. n = 5.
Four genes have sequence similarity with OsTrx1: SDG701, SDG705, SDG717, and SDG721 (Ng et al., 2007; Supplemental Fig. S2A). Therefore, there was a possibility that OsTrx1 RNAi could interfere with transcripts of the genes. However, our real-time PCR analyses demonstrated that their expression levels were not altered in the OsTrx1 RNAi lines, indicating that the phenotypes in the RNAi plants were a result of reduced expression of OsTrx1 (Supplemental Fig. S2, B–E).
To examine whether overexpression of OsTrx1 affects flowering time, we generated five independent transgenic rice plants that expressed the full-length OsTrx1 complementary DNA (cDNA) under the maize (Zea mays) Ubiquitin1 promoter (Supplemental Fig. S3A). Two lines were then selected, OsTrx1 OX-1 and OX-2, in which OsTrx1 transcript levels were elevated by more than 50-fold (Supplemental Fig. S3D). The overexpressing plants did not show any significant change in flowering time or other visible phenotypes under either SD or LD conditions (Supplemental Fig. S3, B and C).
OsTrx1 Suppresses Ghd7 Expression
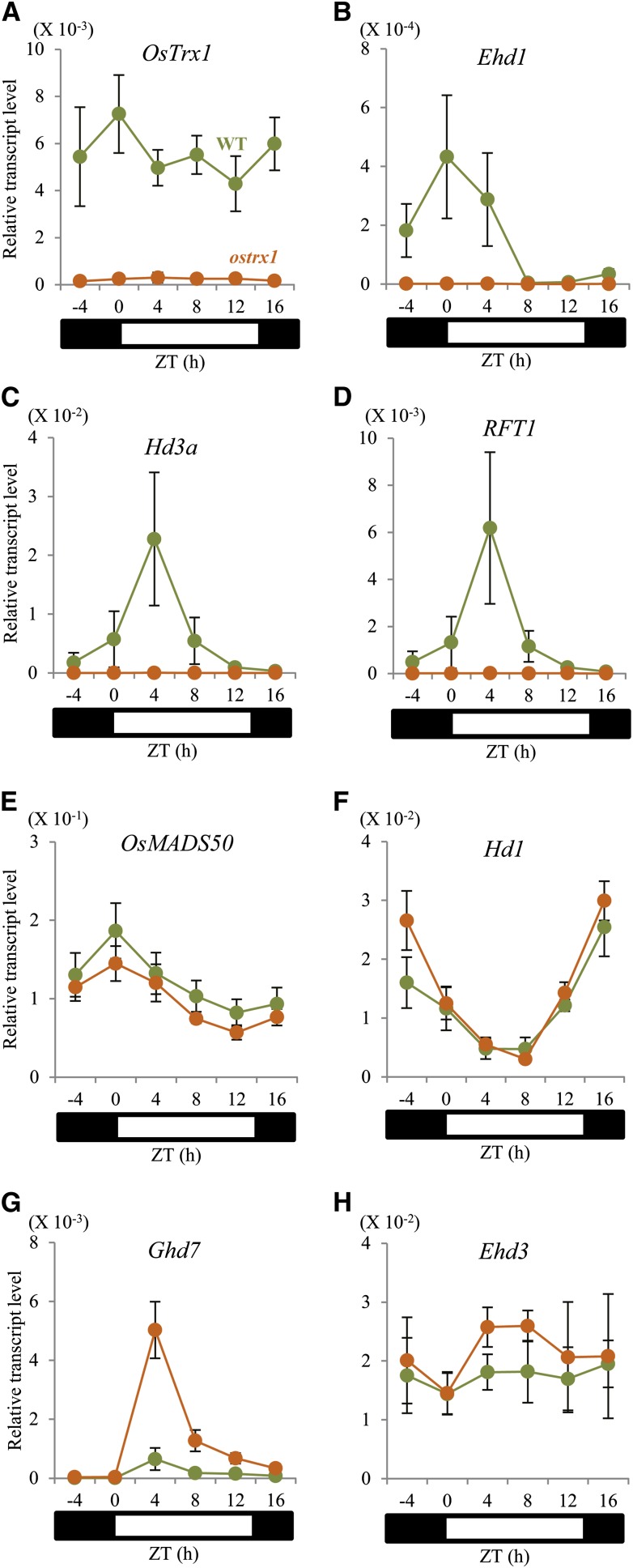
To understand the role of OsTrx1 within the flowering-time network, we studied the expression levels of genes known to control that phenomenon. Total RNA was extracted from leaf blades at 56 DAG, the time when flowering signals begin to induce. Transcripts for OsTrx1 in the wild type were at similar levels both day and night, not following any significant diurnal rhythm (Fig. 3A; Supplemental Fig. S4A). However, transcript levels of the florigen genes Hd3a and RFT1, as well as Ehd1, which is immediately upstream, showed diurnal rhythms. In the ostrx1 mutant, expression of Ehd1 was markedly low throughout the day (Fig. 3B). Similarly, transcripts of Hd3a and RFT1 were maintained at reduced levels in the mutant (Fig. 3, C and D). These results indicated that OsTrx1 functions upstream of Ehd1.
Figure 3.
Expression patterns of floral regulators in ostrx1 and segregating wild-type plants. Quantitative real-time RT-PCR analyses of OsTrx1 (A), Ehd1 (B), Hd3a (C), RFT1 (D), OsMADS50 (E), Hd1 (F), Ghd7 (G), and Ehd3 (H). Leaf samples were collected every 4 h from plants 56 DAG grown under LD conditions. Orange circle, ostrx1 plants; green circles, the segregating wild type. The y-axis shows the transcript level relative to rice Ubiquitin expression. Values are shown as means. Error bars indicate sds. n = 4 or more.
Because OsTrx1 functions preferentially under LD conditions, we studied transcript levels of OsMADS50, Ghd7, and Hd1, which are LD-preferential flowering-time regulators upstream of Ehd1. Transcripts of OsMADS50 were not altered in the ostrx1 mutant (Fig. 3E), and levels of Hd1 were almost identical between the mutant and the wild type (Fig. 3F). However, Ghd7 transcripts were significantly increased in the mutant compared with the wild type (Fig. 3G). Expression of Ghd7 peaked 1 to 2 h after the light was turned on (Supplemental Fig. S4C). The diurnal pattern for Ghd7 transcripts did not change in the ostrx1 plants, but expression of that gene increased by approximately 3-fold (Supplemental Fig. S4C). Transcript levels of Ghd7 were low during the seedling stage but were rapidly increased to the highest amount at 22 DAG before declining (Supplemental Fig. S5C). The overall pattern of expression by Ghd7 was similar between the ostrx1 mutant and the wild type, although transcript levels were two to three times higher in the former (Supplemental Fig. S5C).
Because Ehd3 and Hd17/Ef7/OsELF3-1 function upstream from Ghd7, we measured their transcript levels and found that neither of them was affected in the ostrx1 mutant (Fig. 3H; Supplemental Figs. S4B and S6A). In the wild type, Ehd3 transcripts were maintained at an almost constant level from the seedling to the mature stage. A similar, stage-related, expression pattern was observed from the ostrx1 mutant (Supplemental Fig. S5B). These results indicated that OsTrx1 functions as a negative regulator of Ghd7, independently from Ehd3 and Hd17/Ef7/OsELF3-1.
Several flowering-time regulators, such as OsGI, phytochrome genes, OsCOL4, and RID1/OsId1/Ehd2, control the expression of Ehd1 regardless of day length. Here, their transcript levels were not significantly different between the ostrx1 mutant and the wild type (Supplemental Fig. S6, B–G). These results were consistent with our observations that ostrx1 displayed an altered flowering time only under LD conditions.
Transcript Levels of OsTrx1 Are Not Affected in the Mutants of Other Flowering-Time Genes
To investigate whether other regulators function upstream of OsTrx1, we measured its transcript levels in mutants defective in flowering-control genes, using T-DNA knockout lines of OsMADS50 and OsCOL4 (Supplemental Fig. S7, B and D). We also used the activation tagging line of OsCOL4 (Supplemental Fig. S7E). Because we did not have knockout mutants for Ehd1 and OsId1, we used RNAi transgenic plants (Supplemental Fig. S7, A and C; Kim et al., 2007; Park et al., 2008). These analyses showed that OsTrx1 expression was not altered in the mutants (Fig. 4), suggesting that OsTrx1is not hypostatic to Ehd1, OsMADS50, OsId1, or OsCOL4.
Figure 4.
Expression analyses of OsTrx1 in mutants of genes regulating flowering time. Leaf samples of Ehd1 RNAi (A), osmads50 (B), OsId1 RNAi (C), oscol4 (D), and OsCOL4-d (E) plants grown under LD conditions were collected at 59 DAG, Zeitgeber time (ZT) 2 h (A); 56 DAG, ZT 0 h (B and C); and 54 DAG, ZT 0 h (D and E). The y-axis shows the transcript level relative to rice Ubiquitin expression. Values are shown as means. Error bars indicate sds. n = 4 or more. WT, wild type.
OsTrx1 Transcripts Are Abundant in the Leaf Blades
OsTrx1 transcript levels were higher in the leaf blades than in the roots. Expression was quite low in the leaf sheaths and stems compared with that in the leaf blades. This gene was also expressed at moderate levels in the flag leaf blades and panicles (Supplemental Fig. S4D). Such a leaf blade-preferential expression pattern is similar to that described for Ghd7 and Ehd3 (Xue et al., 2008; Matsubara et al., 2011). In the leaf blade, OsTrx1 transcripts were maintained at a rather constant level during the vegetative growth phase (Supplemental Fig. S5A).
The PHD Motif in OsTrx1 Is Associated with Histone H3
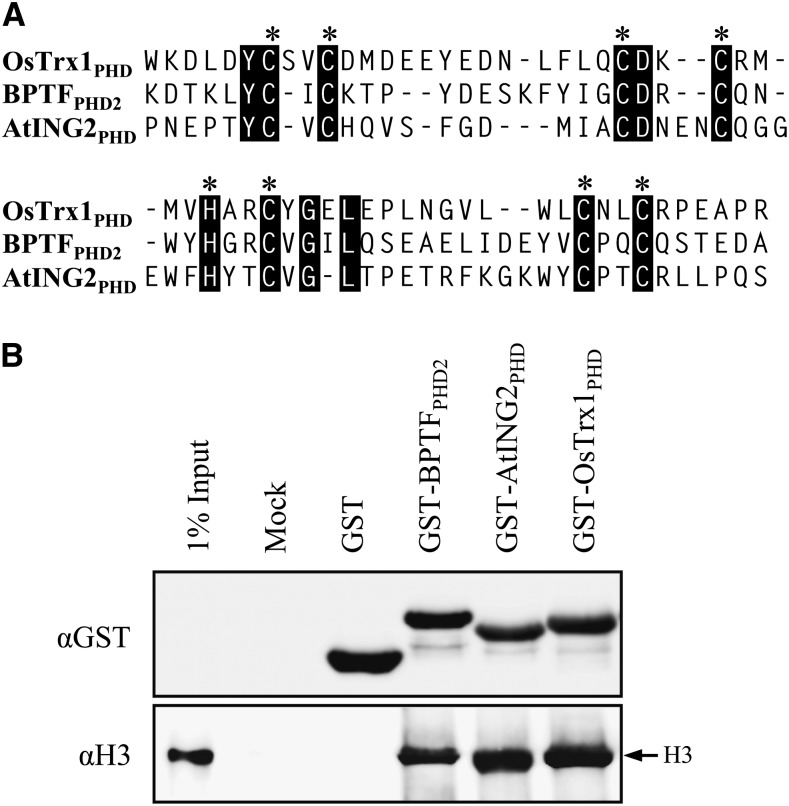
The PHD finger motif of OsTrx1 possessed the well-conserved C4HC3 signature (Fig. 5A). To examine whether that motif associates with histone, we generated a recombinant protein between the PHD motif and glutathione S-transferase (GST; i.e. GST-OsTrx1PHD). We used two positive controls that bind to H3: GST-BPTFPHD2 and GST-AtING2PHD (Wysocka et al., 2006; Lee et al., 2009). GST alone was used as a negative control. Our histone binding assay, using native histone H3 from calf thymus, showed that the PHD motif of OsTrx1 bound to histone H3, with similar affinity to the positive controls (Fig. 5B). Therefore, these results suggested that the OsTrx1 PHD finger motif is associated with histone H3.
Figure 5.
Histone binding assay using native histone H3. A, Comparison of PHD motifs present in OsTrx1, human BPTF, and Arabidopsis AtING2. Black boxes indicate the consensus sequence; asterisks indicate conserved C4HC3 residues in PHD fingers. B, Western-blot analyses of histone binding using native histone H3 from calf thymus. Detection is by anti-GST antibody (top) or antihistone H3 antibody (bottom).
OsTrx1 Binds to Ehd3 through the Region between the PHD and SET Domains
When the fusion gene was transiently expressed in mesophyll protoplasts from rice seedlings, the OsTrx1-synthetic green fluorescent protein (sGFP) fusion protein was localized in the nucleus, where it formed nuclear speckles (Supplemental Fig. S8, A–D). To determine whether OsTrx1 is colocalized with Ehd3, we introduced the OsTrx1-sGFP and Ehd3-modified red fluorescent protein (mRFP) fusion proteins together into protoplasts. As expected, Ehd3-mRFP was colocalized with OsTrx1-sGFP in the nucleus, but nuclear speckles were not colocalized with Ehd3 (Supplemental Fig. S8, E–H).
Because Trx protein is a subunit of a COMPASS-like complex, we proposed that the OsTrx1 protein might interact with another protein that functions as a flowering-time regulator. Among the proteins that function upstream of Ghd7, we speculated that Ehd3 was a potential interacting molecule because it contains PHD finger motifs (Matsubara et al., 2011).
To test whether OsTrx1 indeed interacts with Ehd3, we performed coimmunoprecipitation assays in which the former was tagged with Influenza Hemagglutinin (HA; OsTrx1-HA) and the latter with Myc (Ehd3-Myc). The fusion molecules were coexpressed in rice mesophyll protoplasts using the maize Ubiquitin1 promoter. Analysis with the anti-Myc antibody showed that OsTrx1-HA was coprecipitated with Ehd3-Myc (Fig. 6B). The results were similar with the anti-HA antibody (Fig. 6C), thereby suggesting that OsTrx1 binds to Ehd3.
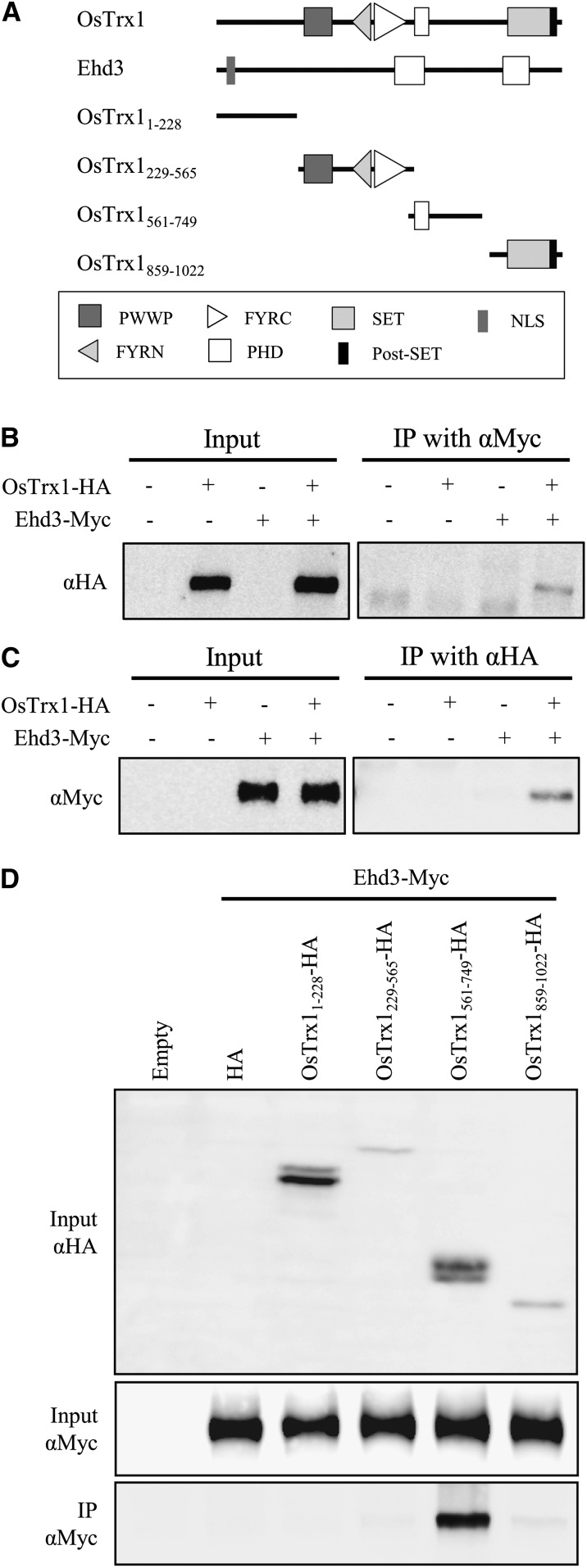
Figure 6.
Coimmunoprecipitation experiment between OsTrx1 and Ehd3. A, Schematic representation of their protein structures. OsTrx1 and Ehd3 indicate full-length proteins without a stop codon. Truncated forms of OsTrx1 contain indicated amino acid residues. B and C, Coimmunoprecipitation of OsTrx1 and Ehd3 proteins. Total proteins were extracted from mesophyll protoplasts of rice seedlings 7 to 14 DAG transiently expressing OsTrx1-HA and Ehd3-Myc. Extracts were immunoprecipitated with anti-Myc antibody (B) or anti-HA antibody (C), and signals were detected by SDS-PAGE using anti-HA antibody (B) or anti-Myc antibody (C). Inputs are extracts before immunoprecipitation, and IP indicates elutes from agarose beads after immunoprecipitation. Plus and minus signs indicate constructs introduced into protoplasts. D, Identification of the OsTrx1 motif that interacts with Ehd3. Expression of truncated OsTrx1-HA (top), expression of Ehd3-Myc (middle), and interaction between two proteins after coimmunoprecipitation (bottom).
To identify the motif that mediates this interaction with Ehd3, we generated four truncated constructs: OsTrx11-228, OsTrx1229-565, OsTrx1561-749, and OsTrx1859-1022. These peptides were fused to the HA tag and expressed in rice mesophyll protoplasts together with Ehd3-Myc (Fig. 6A). Western-blot analysis revealed that the truncated OsTrx1-HAs and Ehd3-Myc were expressed in the transient-expression system (Fig. 6D). Coimmunoprecipitation experiments also indicated the presence of an interaction signal from OsTrx1561-749-HA, which contains a PHD finger motif. Because the other three peptides did not bind to Ehd3 (Fig. 6D), we concluded that OsTrx1 interacted with Ehd3 via the region containing the PHD finger motif.
The OsTrx1561-749 region binds to native histone H3 as well as Ehd3. Therefore, we divided the region into two fragments: OsTrx1561-618 and OsTrx1673-749 (Supplemental Fig. S9A). The former fragment contains a typical PHD finger motif (C4HC3), whereas the latter fragment carries a C4HC2H zinc finger motif. Coimmunoprecipitation analysis showed that OsTrx1673-749-HA binds to Ehd3-Myc, whereas OsTrx1561-618 does not bind to Ehd3 (Supplemental Fig. S9B). These results indicated that the PHD motif region is not involved in interactions with Ehd3, whereas the region containing the C4HC2H motif is involved. It was previously reported that the C4HC2H signature participates in protein-protein interactions (Zhou et al., 2004; Sjøttem et al., 2007; Todd and Picketts, 2012).
OsTrx1 Methylates Histone H3 from Oligonucleosomes
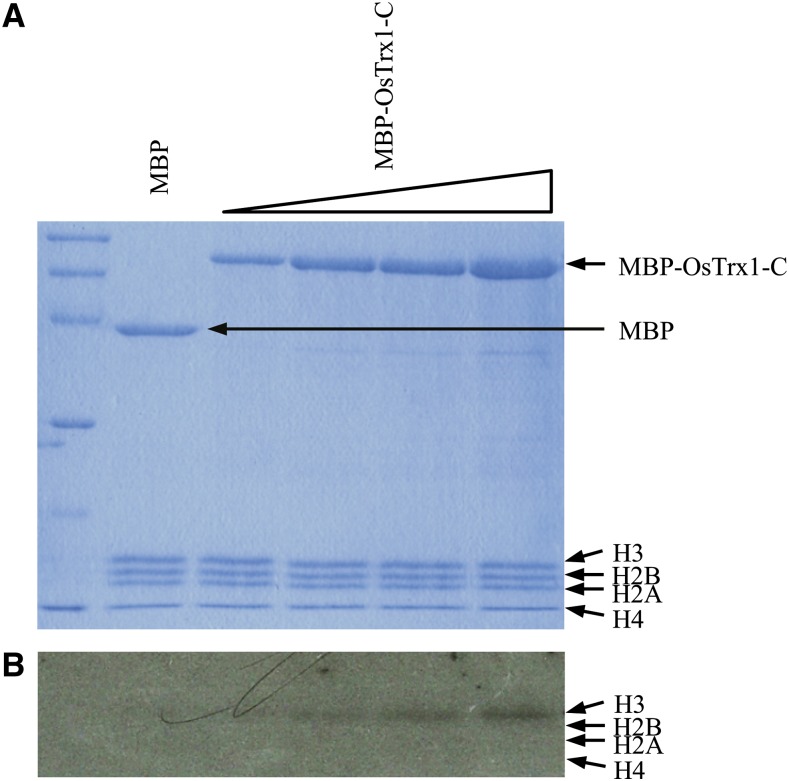
Because OsTrx1 has a SET domain, we examined whether it has HMTase activity. We generated a truncated recombinant protein of OsTrx1 that contained a carboxy terminus (OsTrx1-C), was composed of SET and post-SET domains, and was fused at the carboxy terminus of the maltose-binding protein (MBP). After the MBP-OsTrx1-C recombinant protein was expressed in Escherichia coli, it was purified. In the assays for enzymatic activity, the calf thymus oligonucleosome was used as a substrate and S-[methyl-3H]-adenosyl-l-Met was used as a methyl donor. This analysis showed that histone H3 from the oligonucleosomes was methylated by MBP-OsTrx1-C, but that the other histone molecules were not methylated (Fig. 7B). However, when free histones or myelin basic proteins were used as substrates, the recombinant protein did not show methyltransferase activity. These results suggested that the SET domain of OsTrx1 indeed has such activity toward histone H3 in the oligonucleosomes and that it is a bona fide methyltransferase in vivo.
Figure 7.
Methyltransferase assay of recombinant OsTrx1. Recombinant OsTrx1 C-terminal fragment (MBP-OsTrx1-C) containing SET and post-SET domains were incubated with oligonucleosomes in presence of methyl donor 3H-labeled SAM. MBP was used as negative control. After incubation, reaction mixtures were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (A). Autoradiograph was exposed to 3H-labeled SAM for 5 weeks (B). Positions of MBP-OsTrx1-C, histones, and MBP are indicated.
DISCUSSION
OsTrx1 May Form a Chromatin-Remodeling Complex with Other Proteins
Arabidopsis ATX1 forms a COMPASS-like complex with three structural core components: ASH2R, RBL, and WDR5a. The ASH2R-RBL-WDR5a subcomplex is also able to bind to SDG14 or SDG16 instead of ATX1 (Jiang et al., 2011). Likewise, the human H3K4 methyltransferases (hSet1, MLL1, and MLL2) are replaceable components in interactions with the ASH2L-RbBP5-WDR5 subcomplex (Dou et al., 2006; Ruthenburg et al., 2007). Because OsTrx1 has conserved motifs and is an ortholog of ATX1, it may function through an association with that same subcomplex. OsTrx1 is a member of the SDG family, which also includes SDG724 and SDG725. All are involved in modulating flowering time by controlling the methylation depositions of H3K36 in several floral regulators (Sun et al., 2012; Sui et al., 2013). Thus, ATX1 and these SDG family proteins may play important roles in that process because they belong to a complex with other chromatin-remodeling proteins.
We found that OsTrx1 binds to Ehd3. Because a single mutation in ostrx1 or ehd3 displayed a similar LD-preferential late-flowering phenotype, we concluded that both are needed for the activation of common downstream genes. The Ehd3 protein contains PHD finger motifs that occur in many chromatin-associated proteins (Shi et al., 2006; Wysocka et al., 2006; Lee et al., 2009). Highly homologous genes are present not only in monocots but also in eudicots such as Arabidopsis (Matsubara et al., 2011). However, it is unknown whether Ehd3 and related proteins are indeed involved in chromatin modification. Nevertheless, our observation that OsTrx1 binds to Ehd3 opens the possibility that the latter functions through the chromatin-modifying core complex with which TrxG is associated.
Because OsTrx1 interacts with Ehd3, it is possible that both are members of the COMPASS-like complex found from human MLL1 or Drosophila Trx (Tenney and Shilatifard, 2005; Mohan et al., 2011). However the ostrx1 phenotype differed from that of the ehd3 mutants, with the former flowering late but without any additional morphological alterations, whereas the latter displayed variations in both plant height and panicle length. The pleiotropic phenotype of ehd3 mutants may result from an interaction with other members of the SDG family. For example, it could function with SDG725 because mutations in the gene lead to several phenotypes, such as shorter internodes, panicles, and seeds, as well as an alteration in flowering time (Sui et al., 2012, 2013).
Overexpression of OsTrx1 did not affect flowering time. It is possible that OsTrx1 forms a functional complex with companion proteins, such as ATP-dependent chromatin-remodeling protein, sequence-specific DNA-binding protein, and other histone-modifying proteins. Therefore, this overexpression alone may not be sufficient to promote flowering time. Alternatively, the construct we used for transgenic expression may not have been functional. Although we obtained the full-length cDNA from the Knowledge-Based Oryza Molecular Biological Encyclopedia (KOME; http://cdna01.dna.affrc.go.jp/cDNA), it may not have been an active form if several mature mRNAs emerged via alternative splicing. Such splicing influences the amount and spatial distribution of transcripts, and also affects protein stability and translational efficiency (Ng et al., 2007). Further analysis will be needed to elucidate whether the KOME cDNA is functional in controlling flowering time.
The SET Domain in OsTrx1 Has HMTase Activity
In addition to the conserved PHD finger motif, OsTrx1 carries the SET domain at the C-terminal end. We showed that the truncated recombinant OsTrx1 fragment possesses the SET domain methylated histone H3 present in the oligonucleosomes. The human MLL1 has H3K4 methyltransferase activity; its catalytic function is required for associating with the ASH2L-RbBP5-DR5 subcomplex (Dou et al., 2006). Compared with human MLL1, our truncated OsTrx1 showed weak HMTase activity. Thus, an association with histones may enhance such activity. Furthermore, the PHD finger motif may act as an anchor in interactions with nucleosomes or other binding partners (Bienz, 2006). The PWWP domain present in OsTrx1 may also mediate protein-protein interactions (Stec et al., 2000), and Ehd3 might serve as a cofactor to increase HMTase activity by OsTrx1.
OsTrx1 Is an LD-Specific Flowering Activator That Suppresses Ghd7 Expression
Three independent pathways control flowering time preferentially under LD conditions. The first is the OsMADS50 pathway. Under LD conditions, a knockout mutation in OsMADS50 delays flowering preferentially by approximately 2 months, but does not cause any significant changes in the details of plant architecture, including plant height, tiller number, and grain number (Lee et al., 2004; Ryu et al., 2009). We noted the same phenotypic responses with our ostrx1 mutant, even though OsMADS50 transcript levels were not altered in that mutant. Conversely, OsTrx1 expression was not affected in the osmads50 mutant, indicating that OsTrx1 functions independently from that MADS box gene.
The second pathway is for Hd1, a gene that promotes flowering under SD conditions but inhibits it under LD conditions (Yano et al., 2000). Here, we observed no significant change in its level of transcripts, thus ruling out the possibility that OsTrx1 controls Hd1. The third pathway is for Ghd7. Mutations in that gene causes early flowering preferentially under LD conditions (Xue et al., 2008). We noted that its transcript levels were significantly reduced in the ostrx1 mutant, suggesting that OsTrx1 induces flowering by repressing Ghd7. Because expression of OsGI, Ehd3, and the phytochrome genes, which function upstream of Ghd7 (Hayama et al., 2002; Takano et al., 2005; Itoh et al., 2010; Matsubara et al., 2011), was not affected in the ostrx1 mutant, we deduced that OsTrx1 functions independently from these regulatory genes (Fig. 8).
Figure 8.
Flowering signal pathway under LD conditions in rice.
OsTrx1 protein is highly homologous to other TrxG proteins that activate target genes by modulating chromatin structure. Because OsTrx1 suppressed Ghd7 repression, Ghd7 is not likely a direct target of OsTrx1. Therefore, a yet unidentified gene might be present between the two genes.
OsTrx1 transcripts were more abundant in leaf blades than in the roots, and expression in the former remained relatively constant throughout the vegetative growth phase. A similar developmentally linked expression pattern has been observed with Ehd3 (Matsubara et al., 2011). However, we noted that the level of Ghd7 transcripts increased significantly at 22 DAG before slowly declining. This suggested that OsTrx1 does not control the transcription patterns of downstream genes during the development process. It is possible that the role of OsTrx1 is to open the chromatin structure of the target genes, and that it is other transcription factors that control the degree of expression. For example, other regulatory elements that control Ghd7 expression, such as OsGI and phytochromes, are likely responsible for developmental expression of the gene. It is also probable that OsTrx1 forms a complex with those other elements that are required for altering chromatin structure. Therefore, we do not rule out the possibility that expression levels of associated elements are altered during plant development.
MATERIALS AND METHODS
Plant Materials and Growing Conditions
T-DNA tagging lines were generated in rice (Oryza sativa, japonica cv Dongjin and Hwayoung; Jeon et al., 2000; Jeong et al., 2002; Yi and An, 2013). The flanking sequences were determined through inverse PCR (An et al., 2003; Ryu et al., 2004; Jeong et al., 2006). The production of T-DNA insertional mutants osmads50 and oscol4, activation tagging mutant OsCOL4-d, and RNAi knockdown plants of Ehd1 and OsId1 was previously described (Kim et al., 2007; Park et al., 2008; Ryu et al., 2009; Lee et al., 2010). All plants were grown either in the PF or in growth rooms under SD conditions (10 h of light at 28°C/14 h of darkness at 25°C) or LD conditions (14 h of light at 28°C/10 h of darkness at 25°C).
Plasmid Constructions and Rice Transformation
To construct the overexpressing binary vector of OsTrx1, the full-length cDNA of OsTrx1 was obtained from KOME using primer set T1-FL-F and T1-FL-R (Supplemental Table S1). The amplified PCR product was cloned into the pGA3426 binary vector, which contained the maize Ubiquitin1 promoter and the nopaline synthase terminator, through the SpeI site (Kim et al., 2009). To generate the knockdown binary vector of OsTrx1, the C-terminal fragment of the gene was amplified with primer set T1-RNAi-F and T1-RNAi-R (Supplemental Table S1). The amplified PCR fragments, with an ampicillin linker from the pGA3720, were cloned into the pGA3426 binary vector at the KpnI site located between the promoter and terminator (Kim et al., 2009). These constructs were transferred into Agrobacterium tumefaciens LBA4404 by the freeze-thaw method (An et al., 1988). Rice plants were transformed by the Agrobacterium tumefaciens-mediated cocultivation method, as previously described (Lee et al., 1999).
RNA Isolation and Quantitative RT-PCR Analyses
Total RNA was isolated using RNAiso Plus (Takara). First-strand cDNA was synthesized with 2 μg of total RNA, using Moloney murine leukemia virus reverse transcriptase (Promega), 10 ng oligo(dT) primer, and 2.5 mm deoxyribonucleotide triphosphate. Synthesized cDNA was used for quantitative real-time RT-PCR in a Rotor-Gene Q (Qiagen); the rice Ubiquitin gene served as a reference to normalize the cDNA quantity. The ΔΔCt method was followed for calculating alterations in expression. To ensure primer specificity, we analyzed data when the melting curve showed a single peak. Primers for analyzing gene expression are listed in Supplemental Table S1.
Protoplast Isolation and Transformation
Protoplast isolation from rice mesophyll or rice root-derived callus suspension (Oc) cells and polyethylene glycol (PEG)-mediated transformation were performed as described previously, with minor modifications (Bart et al., 2006; Hong et al., 2012). Briefly, leaves were excised from rice seedlings 7 to 14 DAG with a razor blade and incubated in enzyme solution A (1.5% [w/v] Cellulose RS, 0.3% [w/v] Macerozyme, 0.1% [w/v] pectolyase, 0.6 m mannitol, 10 mm MES, pH 5.7, 1 mM CaCl2, and 0.1% [w/v] bovine serum albumin) for 4 h in the dark with gentle shaking. Oc suspension solution was collected by centrifugation and the supernatant was removed. The cells were incubated in enzyme solution B (2% [w/v] Cellulose RS, 1% [w/v] Macerozyme, 0.4 m mannitol, 0.1% [w/v] MES, pH 5.7, 0.1% [w/v] CaCl2) for 4 h with gentle shaking. After adding an equal volume of W5 solution (154 mm sodium chloride, 125 mm CaC12, 5 mm KCl, and 2 mm MES, pH 5.7) and harvesting cells, protoplasts were resuspended in Mmg solution (0.6 m mannitol, 15 mm MgCl2, and 4 mm MES, pH 5.7) at 106 cells mL−1, as quantified with a hemocytometer. For transformation, 20 μg of each vector plus a 40% PEG solution (0.6 m mannitol, 100 mm CaCl2, 40% [w/v] PEG 3350) was added to the protoplasts for 10 min. The protoplasts were washed with 2 volumes of W5 solution and then resuspended in incubation solution (0.6 m mannitol, 4 mm MES, pH 5.7, and 4 mm KCl). They were then incubated overnight at 28°C in the dark.
Subcellular Localization of OsTrx1
The full-length cDNA of OsTrx1 and Ehd3 were amplified without its stop codon using gene-specific primer sets (Supplemental Table S1). Afterward, OsTrx1-sGFP was constructed using the SpeI site of the pGA3452 vector containing the maize Ubiquitin1 promoter and the sGFP coding region. Ehd3-mRFP was constructed using the SmaI and SpeI site of the pGA3574 vector containing the maize Ubiquitin1 promoter and the mRFP coding region (Kim et al., 2009). A nuclear protein marker, NLS-mRFP driven by the 35S promoter, was obtained from Dr. Inhwan Hwang (POSTECH). OsTrx1-sGFP, Ehd3-mRFP, and NLS-mRFP were transformed into protoplasts isolated from rice mesophyll or Oc cells, and were examined on an Axioplan 2 fluorescence microscope (Carl Zeiss) equipped with filter sets for GFP and RFP.
In Vitro Binding Assays
The OsTrx1 PHD finger was searched from domain predictions at the SMART Web site (http://smart.embl-heidelberg.de). The OsTrx1PHD corresponded to 561 to 618 residues. PHD fingers of BPTF and AtING2 were produced based on previous studies (Wysocka et al., 2006; Lee et al., 2009). All constructs were amplified with each primer set (Supplemental Table S1), and were cloned into pGEX-5X-1 using the SmaI and XhoI sites (GE Healthcare). The recombinant proteins were expressed in Escherichia coli strain BL21 (DE3) and were purified using Glutathione Excellose beads (Bioprogen). Histone binding assays were performed at 4°C, following previously described protocols (Matthews et al., 2007). Briefly, 10 μg of the target proteins was incubated with 50 μL of the Glutathione Excellose bead slurry for 1 h with continuous gentle shaking. After the supernatants were removed, the beads were blocked with buffer A (50 mm Tris-HCl, pH 7.5, 0.3 m sodium chloride, and 0.1% [v/v] NP-40) that contained 5% bovine serum albumin. The mixtures were incubated for 2 h to prevent nonspecific binding. After the beads were washed three times with buffer A (containing 1 mm phenylmethylsulfonyl fluoride), they were combined with 10 μg of a native histone H3 from calf thymus (Roche). After overnight incubation with continuous gentle shaking, the beads were washed five times with buffer B (50 mm Tris-HCl, pH 7.5, 1 m sodium chloride, and 0.1% [v/v] NP-40). Proteins that were bound to the beads were eluted with SDS sample buffer and electrophoresed on a 15% SDS-polyacrylamide gel. They were then transferred to a polyvinylidene difluoride membrane. GST-fused PHD fingers bound on that membrane were incubated with a horseradish peroxidase (HRP)-conjugated anti-GST monoclonal antibody (Millipore) or an antihistone H3 polyclonal antibody (Abcam). Protein signals were detected using the enhanced chemiluminescence prime western-blotting detection reagent in LAS-4000.
Coimmunoprecipitation Assays
Full-length OsTrx1, six OsTrx1 truncates, and full-length Ehd3 were amplified without the stop codon, using the primer sets listed in Supplemental Table S1. HA-tagged vectors were produced with the HpaI and KpnI sites of the pGA3698 vector, which contained the maize Ubiquitin1 promoter and the 3x HA coding region. An Myc-tagged full-length Ehd3 vector was constructed using the HpaI site of the pGA3697 vector that carried the maize Ubiquitin1 promoter and the 4x Myc coding region. Each DNA combination of all vectors was transformed into the protoplasts isolated from rice mesophyll or Oc cells. After incubation, the protoplasts were harvested and used for coimmunoprecipitation assays as previously reported, but with minor modifications (Yang et al., 2013a). All experiments were performed at 4°C. Briefly, the protoplasts were resuspended in Immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm sodium chloride, 1% [v/v] Triton X-100, 1 mm dithiothreitol, 2 mm NaF, 50 μm MG132, and an adequate amount of Protease inhibitor cocktail [Roche]. After brief vortexing, the samples were centrifuged at 12,000 rpm for 10 min. We added 10 μL each of protein A and G conjugated to agarose beads (Millipore) to the supernatant for 1 h of preclearing to prevent nonspecific binding. We used 10% (v/v) of the precleared extracts as input controls, whereas the rest were precipitated with an anti-HA monoclonal antibody (Roche) and incubated overnight with gentle shaking. The extracts were then combined with 10 μL each of protein A and G conjugated to agarose beads and incubated for 2 h. After washing five times with Immunoprecipitation buffer, the proteins bound to the beads were eluted with 20 μL of SDS sample buffer. The elutes were electrophoresed on a 10% (v/v) SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore). Proteins that bound to that membrane were incubated with an HRP-conjugated anti-Myc monoclonal antibody (Cell Signaling) and were detected using an enhanced chemiluminescence prime western-blotting detection reagent (GE Healthcare) in LAS-4000 (GE Healthcare). The experiment for monitoring the interaction between full-length OsTrx1 and Ehd3 was also performed reciprocally using the anti-Myc monoclonal antibody for precipitation and an HRP-conjugated anti-HA monoclonal antibody (Cell Signaling) for detection.
HMTase Assays
OsTrx1-C, containing the SET and post-SET domains, was amplified with primer set MBP-T1-HMT-F and MBP-T1-HMT-R (Supplemental Table S1). The amplified PCR fragments were ligated into MBP ligation-independent cloning vectors. The recombinant protein MBP-OsTrx1-C was expressed in E. coli strain BL21 (RIL) and purified. Methyltransferase assays were performed as previously described (Pei et al., 2007). Briefly, recombinant MBP-OsTrx1-C was incubated with substrates and S-[methyl-3H]-adenosyl-l-Met in Histone methyltransferase buffer (20 mm Tris-HCl, pH 8.0, 4 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 0.5 mm dithiothreitol) for 3 h at 30°C. The reaction mixture was separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Afterward, the gel was treated with Amplifier (GE Healthcare), and then dried and exposed to Kodak Biomax MS film (Sigma-Aldrich) at −80°C.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OsTrx1/SDG723 (Os09g0134500), SDG701 (Os08g0180100), SDG705 (Os01g0655250), SDG717 (Os12g0613200), and SDG721 (Os01g0218800).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of OsTrx1 RNAi transgenic plants.
Supplemental Figure S2. Expression analyses of OsTrx1 homologs in wild-type and OsTrx1 RNAi-3 plants.
Supplemental Figure S3. Phenotypes of OsTrx1-overexpressing transgenic plants.
Supplemental Figure S4. Spatio-temporal expression patterns of floral regulators.
Supplemental Figure S5. Expression patterns of flowering-time genes by developmental stage.
Supplemental Figure S6. Expression patterns of floral regulators in segregating wild-type and ostrx1 plants.
Supplemental Figure S7. Expression analyses of each floral regulator in mutant backgrounds compared to wild-type.
Supplemental Figure S8. Sub-cellular localization of OsTrx1\x{2013}sGFP and Ehd3\x{2013}mRFP revealed by transient expression using protoplasts.
Supplemental Figure S9. Coimmunoprecipitation experiment between Ehd3 and the region containing PHD motif of OsTrx1.
Supplemental Table S1. Primers used in study.
Glossary
- TrxG
trithorax group
- PcG
Polycomb group
- COMPASS
complex proteins associated with Set1
- HMTase
histone methyltransferase
- SD
short-day
- LD
long-day
- T-DNA
transfer DNA
- PF
paddy field
- RT-PCR
reverse transcription PCR
- DAG
days after germination
- RNAi
interference RNA
- cDNA
complementary DNA
- GST
glutathione S-transferase
- sGFP
synthetic green fluorescent protein
- mRFP
modified red fluorescent protein
- KOME
Knowledge-Based Oryza Molecular Biological Encyclopedia
- PEG
polyethylene glycol
- HRP
horseradish peroxidase
Footnotes
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aichinger E, Villar CB, Di Mambro R, Sabatini S, Köhler C. (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637 [DOI] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In Plant Molecular Biology Manual, SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Vol A3. Martinus Nijhoff Publishers, pp 1–18 [Google Scholar]
- An G, Lee S, Kim SH, Kim SR. (2005) Molecular genetics using T-DNA in rice. Plant Cell Physiol 46: 14–22 [DOI] [PubMed] [Google Scholar]
- An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, et al. (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133: 2040–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. (2006) The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40 [DOI] [PubMed] [Google Scholar]
- Carles CC, Fletcher JC. (2009) The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev 23: 2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Zhu Y, Bu ZY, Shen WH, Yu Y, Dong AW. (2010) SDG714 regulates specific gene expression and consequently affects plant growth via H3K9 dimethylation. J Integr Plant Biol 52: 420–430 [DOI] [PubMed] [Google Scholar]
- Ding Y, Wang X, Su L, Zhai J, Cao S, Zhang D, Liu C, Bi Y, Qian Q, Cheng Z, et al. (2007) SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. (1992) The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71: 701–708 [DOI] [PubMed] [Google Scholar]
- Hayama R, Izawa T, Shimamoto K. (2002) Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol 43: 494–504 [DOI] [PubMed] [Google Scholar]
- Hong SY, Seo PJ, Cho SH, Park CM. (2012) Preparation of leaf mesophyll protoplasts for transient gene expression in Brachypodium distachyon. J Plant Biol 55: 390–397 [Google Scholar]
- Hu Y, Liu D, Zhong X, Zhang C, Zhang Q, Zhou DX. (2012) CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc Natl Acad Sci USA 109: 5773–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. (1998) trithorax and the regulation of homeotic gene expression in Drosophila: a historical perspective. Int J Dev Biol 42: 423–429 [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. (2009) ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G. (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y. (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G. (2007) OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol 145: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Lee DY, Yang JI, Moon S, An G. (2009) Cloning vectors for rice. J Plant Biol 52: 73–78 [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. (2002) COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G. (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42: 310–316 [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G. (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Lee WY, Lee D, Chung WI, Kwon CS. (2009) Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J 58: 511–524 [DOI] [PubMed] [Google Scholar]
- Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, et al. (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63: 18–30 [DOI] [PubMed] [Google Scholar]
- Li G, Zhang J, Li J, Yang Z, Huang H, Xu L. (2012) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J 72: 261–270 [DOI] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D. (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M. (2012) Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol 53: 709–716 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M. (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66: 603–612 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M. (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. (2011) The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769: 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell 8: 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, et al. (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 56: 1018–1029 [DOI] [PubMed] [Google Scholar]
- Pei Y, Niu L, Lu F, Liu C, Zhai J, Kong X, Cao X. (2007) Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol 144: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. (2001) Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334 [DOI] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U. (2008) ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32: 1412–1427 [DOI] [PubMed] [Google Scholar]
- Ryu CH, You JH, Kang HG, Hur J, Kim YH, Han MJ, An K, Chung BC, Lee CH, An G. (2004) Generation of T-DNA tagging lines with a bidirectional gene trap vector and the establishment of an insertion-site database. Plant Mol Biol 54: 489–502 [DOI] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, et al. (2012) Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53: 717–728 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. (2011) Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 12: 799–814 [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjøttem E, Rekdal C, Svineng G, Johnsen SS, Klenow H, Uglehus RD, Johansen T. (2007) The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res 35: 6648–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. (2000) The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473: 1–5 [DOI] [PubMed] [Google Scholar]
- Sui P, Jin J, Ye S, Mu C, Gao J, Feng H, Shen WH, Yu Y, Dong A. (2012) H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J 70: 340–347 [DOI] [PubMed] [Google Scholar]
- Sui P, Shi J, Gao X, Shen WH, Dong A. (2013) H3K36 methylation is involved in promoting rice flowering. Mol Plant 6: 975–977 [DOI] [PubMed] [Google Scholar]
- Sun C, Fang J, Zhao T, Xu B, Zhang F, Liu L, Tang J, Zhang G, Deng X, Chen F, et al. (2012) The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24: 3235–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, et al. (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM. (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tenney K, Shilatifard A. (2005) A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem 95: 429–436 [DOI] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML. (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71: 691–700 [DOI] [PubMed] [Google Scholar]
- Todd MAM, Picketts DJ. (2012) PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res 11: 4326–4337 [DOI] [PubMed] [Google Scholar]
- Wu C, You C, Li C, Long T, Chen G, Byrne ME, Zhang Q. (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105: 12915–12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G. (2013a) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J 73: 566–578 [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng Q, Chen GX, Li XH, Wu CY. (2013b) OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol Plant 6: 202–215 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, An G. (2013) Utilization of T-DNA tagging lines in rice. J Plant Biol 56: 85–90 [Google Scholar]
- Yun JY, Tamada Y, Kang YE, Amasino RM. (2012) Arabidopsis trithorax-related3/SET domain GROUP2 is required for the winter-annual habit of Arabidopsis thaliana. Plant Cell Physiol 53: 834–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, Wang S, Deng XW, Li S. (2012) OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE 7: e43705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MI, Wang H, Foy RL, Ross JJ, Cohen HT. (2004) Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res 64: 1278–1286 [DOI] [PubMed] [Google Scholar]