An ethylene response transcription factor helps in the maintenance of phosphate homeostasis by altering root and shoot growth in Arabidopsis.
Abstract
Inorganic phosphate (Pi) availability is a major factor determining growth and consequently the productivity of crops. However, it is one of the least available macronutrients due to its high fixation in the rhizospheres. To overcome this constraint, plants have developed adaptive responses to better acquire, utilize, and recycle Pi. Molecular determinants of these adaptive mechanisms include transcription factors (TFs) that play a major role in transcriptional control, thereby regulating genome-scale networks. In this study, we have characterized the biological role of Arabidopsis thaliana Ethylene Response Factor070 (AtERF070), a Pi starvation-induced TF belonging to the APETALA2/ETHYLENE RESPONSE FACTOR family of TFs in Arabidopsis (Arabidopsis thaliana). It is localized to the nucleus and induced specifically in Pi-deprived roots and shoots. RNA interference-mediated suppression of AtERF070 led to augmented lateral root development resulting in higher Pi accumulation, whereas there were reductions in both primary root length and lateral root number in 12-d-old transgenic seedlings overexpressing AtERF070. When the overexpressing lines were grown to maturity under greenhouse conditions, they revealed a stunted bushy appearance that could be rescued by gibberellic acid application. Furthermore, a number of Pi starvation-responsive genes were modulated in AtERF070-overexpressing and RNA interference lines, thereby suggesting a potential role for this TF in maintaining Pi homeostasis.
Inorganic phosphate (Pi) is one of the essential macronutrients that plays a pivotal role in metabolic processes (Marschner, 1995). Despite high Pi content in natural and agroclimatic environments, its availability is limited by its high fixation and slow diffusion rates in rhizospheres (Raghothama, 1999; Plaxton and Tran, 2011). Plants adapt to Pi deficiency by evolving an array of developmental, biochemical, and molecular responses facilitating optimal acquisition, mobilization, and distribution of Pi (Franco-Zorrilla et al., 2004; Lynch, 2011; Smith et al., 2011; Jain et al., 2012). In Arabidopsis (Arabidopsis thaliana), Pi deficiency triggers modulation of root system architecture (RSA), and increases in the number and length of root hairs, thereby enabling plants to acquire Pi from soils (Bates and Lynch, 1996; Ma et al., 2001; Williamson et al., 2001; López-Bucio et al., 2002; Jain et al., 2007b, 2012). In addition, increased production of phosphatases and RNases and activation of alternative nonphosphorylating metabolic pathways further facilitate the adaptation to limited Pi availability (López-Bucio et al., 2002; Rausch and Bucher, 2002; Vance et al., 2003; Hammond et al., 2004; Plaxton and Tran, 2011). Global gene expression analyses using microarrays have resulted in the identification of several Pi starvation-responsive (PSR) genes that are implicated in Pi homeostasis (Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007; Calderon-Vazquez et al., 2008; Thibaud et al., 2010). In addition, developmental and molecular changes during Pi starvation are mediated by phytohormones like auxin, cytokinin, ethylene, and GA3 (Franco-Zorrilla et al., 2004; Jain et al., 2007a; Jiang et al., 2007; O’Rourke et al., 2013).
In Arabidopsis, transcriptional regulation of Pi-deficiency responses involves an array of transcription factors (TFs; PHOSPHATE STARVATION RESPONSE1 [PHR1, At4g28610], PHOSPHATE STARVATION RESPONSE1-LIKE1 [PHR1-like1, At5g29000], PHOSPHATE ROOT DEVELOPMENT [PRD, At1g79700], BASIC HELIX-LOOP-HELIX32 [bHLH32, At3g25710], WRKY6 [At1g62300], HYPERSENSITIVITY TO LOW PHOSPHATE-ELICITED PRIMARY ROOT SHORTENING1 [HRS1, At1g13300], WRKY75 [At5g13080], ZINC FINGER OF ARABIDOPSIS6 [ZAT6, At5g04340], MYB62 and MYB DOMAIN PROTEIN62 [MYB62, At1g68320]) that have been functionally characterized (Rubio et al., 2001; Chen et al., 2007, 2009; Devaiah et al., 2007a, 2007b, 2009; Liu et al., 2009; Bustos et al., 2010). Among these TFs, extensively studied PHR1 has been implicated in regulating a subset of Pi-starvation responses, in spite of not being induced during Pi deficiency. To identify PSR genes that are implicated in transcriptional regulation, a microarray analysis using Affymetrix was carried out by subjecting Arabidopsis to short-term (3, 6, and 12 h pooled), medium-term (1 and 2 d pooled), and long-term (10 d) Pi starvation (Misson et al., 2005). A total of 866 genes were found to be spatiotemporally and differentially regulated, of which 80 were involved in transcriptional regulation and some of them belonged to APETALA2, WRKY, BASIC REGION/LEUCINE ZIPPER MOTIF, MYB, and Zinc Finger superfamilies (Misson et al., 2005). MYB62 and ZAT6, which showed induction only during long-term Pi deprivation in the leaf, have been functionally characterized (Misson et al., 2005; Devaiah et al., 2007b, 2009). The overexpression of ZAT6 resulted in the suppression of PSR genes and alteration of RSA (Devaiah et al., 2007b), whereas overexpression of MYB62 resulted in modulated RSA, elevated Pi uptake and acid phosphatase activity, attenuation in the expression of PSR genes, and an interaction with GA signaling pathway (Devaiah et al., 2009). These studies suggested that MYB62 and ZAT6 play critical roles in the maintenance of Pi homeostasis. Interestingly, among the 80 TFs identified from this microarray analysis, only AtERF070 (At1g71130), belonging to the B5 subfamily of ETHYLENE RESPONSE FACTOR (ERF) TFs, showed early induction during short-term Pi deprivation and also remained induced when the Pi deficiency treatment was extended for a long term (Misson et al., 2005). Although the microarray analysis suggested this TF to be highly responsive to Pi deficiency, it has not yet been functionally characterized.
In this study, we have employed RNA interference (RNAi)-mediated suppression and constitutive overexpression of AtERF070 to determine the role of this TF in maintaining Pi homeostasis. Although RNAi-mediated suppression of AtERF070 resulted in a pronounced increase in root growth and Pi content, there was no significant effect on the expression of PSR genes. On the contrary, the overexpression of this gene resulted in a retarded growth phenotype and modulation in the expression of several PSR and GA biosynthetic genes. Thus, this study indicates the role of this TF in maintaining Pi homeostasis and potential cross talk with the phytohormone signaling pathway.
RESULTS
Pi Deficiency-Mediated Differential Spatial Expression of AtERF070 and Subcellular Localization of Its Fusion Protein
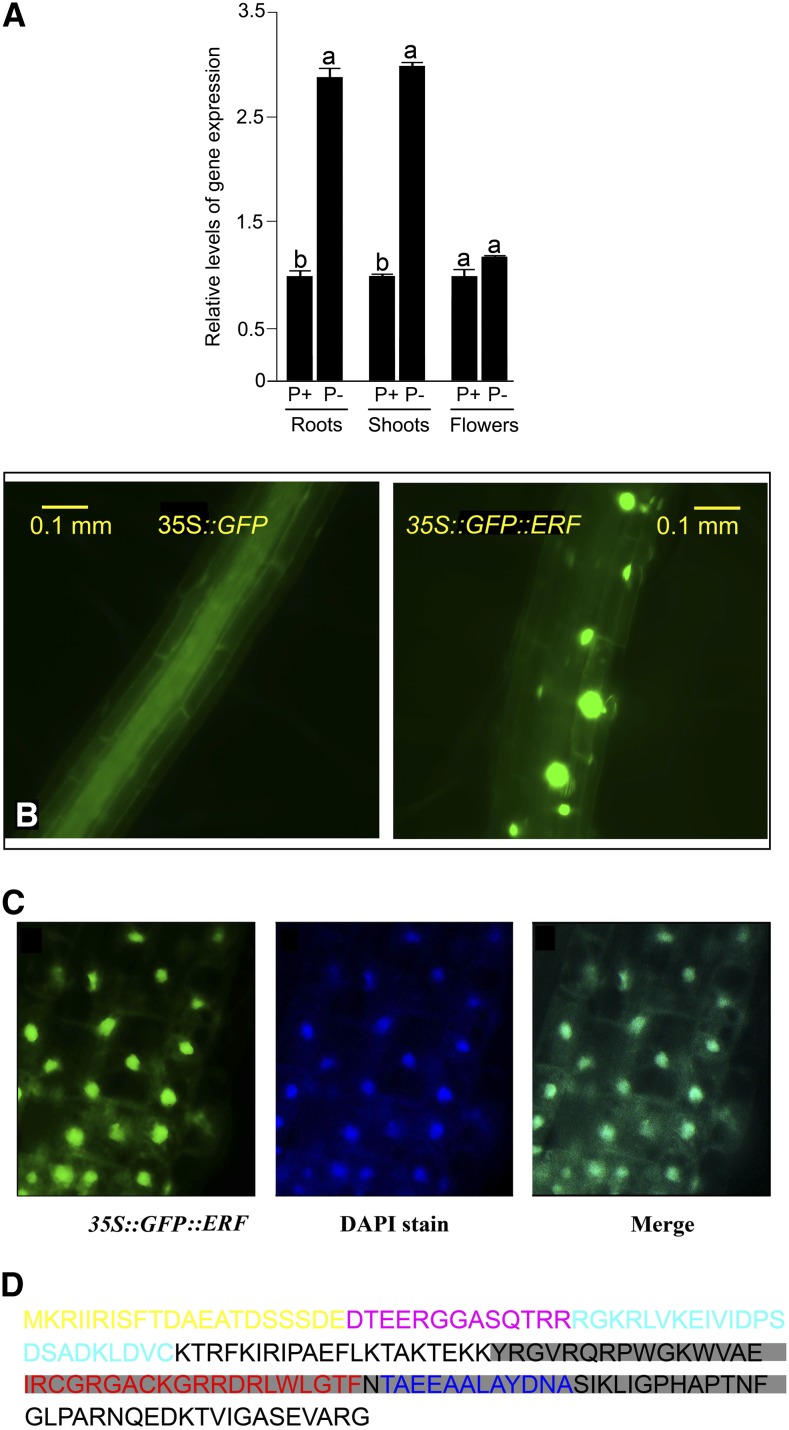
Pi deficiency triggers differential regulation of an array of PSR genes. To obtain a more comprehensive insight into the effects of Pi deprivation on the spatial expression of AtERF070, real-time PCR analyses were carried out for both vegetative (roots and shoots) and reproductive (flowers) tissues of the wild-type seedlings grown hydroponically in the greenhouse under P+ (250 µm Pi) and P− (0 µm Pi) conditions for 7 d. Pi deficiency triggered a significant increase in the relative expression of AtERF070 in both roots and shoots, but its relative expression was comparable in the flowers irrespective of the Pi regime (Fig. 1A). The data not only confirmed earlier microarray analysis (Misson et al., 2005) but also highlighted the differential spatial regulation of this gene, signifying its potential role in the maintenance of Pi homeostasis. To further determine the effect of other nutrient deficiencies on the relative expression of AtERF070, wild-type Arabidopsis seedlings were also subjected to potassium, nitrogen, and iron deficiencies for 7 d. None of these nutrient deficiencies illicit any significant induction of this gene in the seedlings (data not shown), thereby demonstrating the specific responsiveness of this gene to Pi deprivation.
Figure 1.
Pi deficiency-induced differential spatial expression of AtERF070 and subcellular localization of its fusion protein. A, Four-week-old wild-type seedlings were grown hydroponically and then subjected to P+ (250 µm Pi) and P− (0 µm Pi) treatments for 7 d. Real-time PCR analyses of AtERF070 in different tissues (roots, shoots, and flowers) of hydroponically grown seedlings under P+ and P− conditions was performed. AtACTIN2 was used as an internal control, and P+ values were normalized to 1. Data presented are means of six technical replicates ± se. Different letters indicate significant differences in the relative level of gene expression. B, Subcellular localization of a GFP::AtERF070 fusion protein. C, Colocalization of GFP::AtERF070 and the nucleus-specific dye DAPI. Shown from left to right are GFP fluorescence, staining of the nucleus by DAPI, and a merged image. D, Amino acid sequence of AtERF070 with the shaded region depicting the ERF domain. The β-sheet and α-helix are depicted in red and blue, respectively, in the shaded region of the sequence. Letters in yellow and cyan represent the two conserved domains CMVI-1 and CMVI-2, respectively, while the letters in pink indicate an overlapping region between them. [See online article for color version of this figure.]
Subcellular localization is an important characteristic of TFs, and it plays a critical role in regulating the nutrient starvation responses (Beck and Hall, 1999). Therefore, for ascertaining the subcellular localization of AtERF070, its coding region was fused with the 3′ end of an EGFP (for enhanced GFP) reporter gene and expressed constitutively under the control of the cauliflower mosaic virus (CaMV) 35S promoter. EGFP under the control of the CaMV 35S promoter was used as a control. Stable transgenic Arabidopsis plants expressing reporter 35S::EGFP alone (control) and chimeric 35S::EGFP::AtERF070 were analyzed for their subcellular localization under P+ and P− conditions (Fig. 1B). The GFP fluorescence was localized to the nucleus in the 35S:GFP::AtERF070 transgenic lines irrespective of the Pi regime, whereas the distribution of GFP fluorescence was uniform throughout the cell of the control plant. Furthermore, the nuclear localization of GFP::AtERF070 protein was confirmed by staining the nucleus with 4′,6-diamidino-2-phenylindole (DAPI; Fig. 1C). Bioinformatic analysis of the amino acid sequence of AtERF070 showed an ERF domain toward the C-terminal end of the protein (Fig. 1D).
AtERF070 RNAi Lines Show Altered Morphophysiological Traits
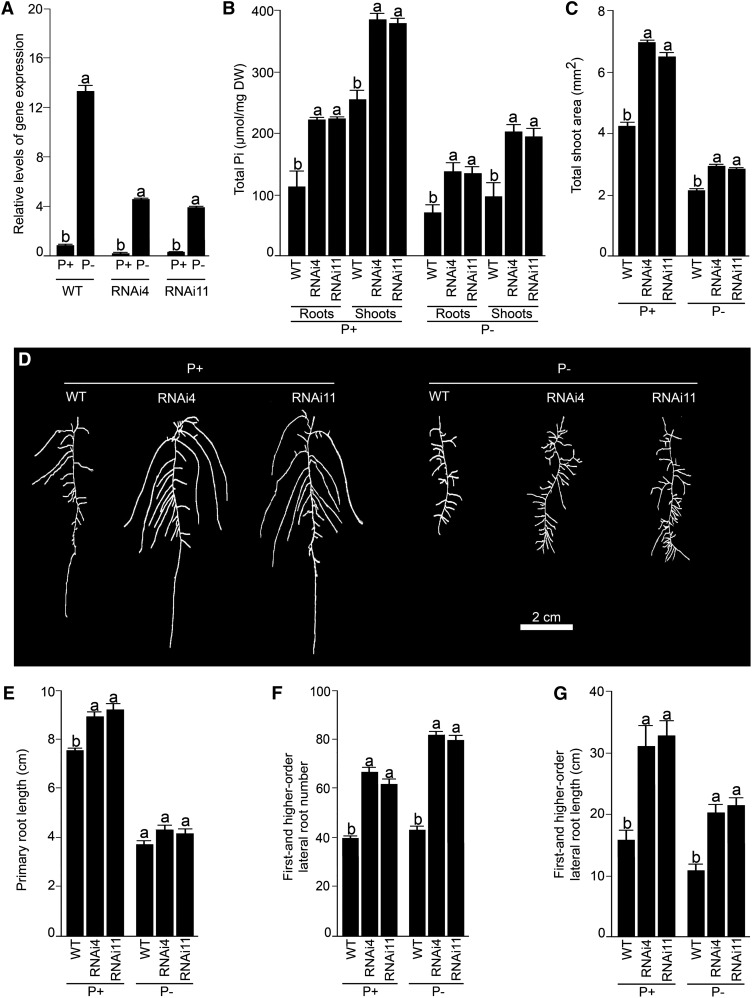
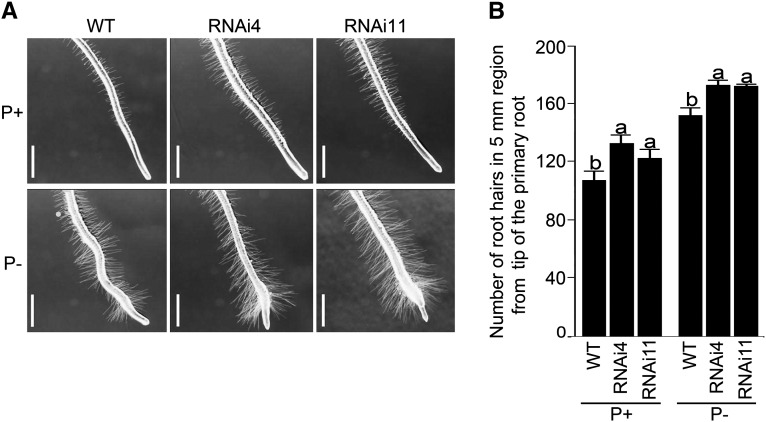
RNAi-mediated gene silencing has been successfully used for the functional characterization of different TFs in Arabidopsis (WRKY75; Devaiah et al., 2007a) and rice (Oryza sativa; OsPHR1 and OsPHR2 [Zhou et al., 2008] and OsMYB2P-1 [Dai et al., 2012]) that play a pivotal role in the maintenance of Pi homeostasis. Therefore, a similar strategy was used to determine the role of AtERF070, if any, in the acquisition and/or mobilization of Pi. Two independent RNAi mutant lines (RNAi4 and RNAi11) were used for comprehensive evaluation. Real-time PCR analysis was employed for determining the extent of attenuation in the expression of AtERF070 in RNAi seedlings grown under P+ and P− conditions on agar petri plates for 7 d (Fig. 2A). Under both P+ and P− conditions, RNAi-mediated silencing triggered about 65% to 70% reductions in the expression of AtERF070 in RNAi4 and RNAi11 seedlings compared with their respective wild-type seedlings. These two RNAi lines were subsequently used for detailed morphophysiological characterization (Fig. 2, B–G). Under both P+ and P− conditions, there were significant (P ≤ 0.05) increases in total Pi content of roots and shoots, and total shoot area, of both the RNAi lines compared with their respective wild-type controls (Fig. 2, B and C). Since RNAi4 and RNAi11 exhibited elevated Pi content and shoot area, it was assumed that these morphophysiological changes could possibly be due to modulation of the root system that plays a pivotal role in Pi acquisition and mobilization (Williamson et al., 2001; López-Bucio et al., 2002, 2005; Jain et al., 2007b, 2009; Lynch, 2011). Also, mutations in the Pi-responsive TFs WRKY75 and PRD have been shown to affect different root traits under different Pi regimes (Devaiah et al., 2007a; Camacho-Cristóbal et al., 2008; Jain et al., 2012). Therefore, different traits of the RSA of AtERF070 RNAi lines grown under P+ and P− conditions for 7 d on agar petri plates were compared with their respective wild types (Fig. 2, D–G). There was a noticeable exaggerated root growth of both P+ and P− RNAi4 and RNAi11 lines compared with their respective wild-type seedlings (Fig. 2D) due to significant (P ≤ 0.05) increases in both primary root growth (Fig. 2E) and the number and length (Fig. 2, F and G) of first- and higher-order lateral roots. Mutant analyses of some of the TFs (PHR1, WRKY75, and BHLH32) demonstrated their differential regulatory influence on root hair development under different Pi regimes (Chen et al., 2007; Devaiah et al., 2007a; Bustos et al., 2010). Therefore, we compared the developmental responses of root hairs in RNAi4 and RNAi11 lines with the wild type during growth under P+ and P− conditions (Fig. 3). Under both P+ and P− conditions, there was a significant (P ≤ 0.05) increase in the number of root hairs in the 5-mm region of the primary root tip of RNAi4 and RNAi11 compared with their corresponding wild types (Fig. 3).
Figure 2.
RNAi-mediated silencing of AtERF070 modulates morphophysiological traits. Arabidopsis (wild-type [WT] and RNAi lines) seedlings were raised initially on vertically oriented agar petri plates containing one-half-strength MS for 5 d and then transferred to P+ and P− media for 7 d. A, The whole seedling was used for real-time PCR analyses of AtERF070 as described in the legend to Figure 1. The wild-type P+ value was normalized to 1. Different letters indicate significant differences in the relative level of gene expression. B to G, Roots and shoots of the wild-type and AtERF070 RNAi lines were separated at the hypocotyl-root junction. Data are presented for total Pi content (B), total shoot area (C), root architectural details (D), primary root length (E), first- and higher-order lateral root number (F), and their length (G). Values are means ± se, and n = 6 replicates of 10 seedlings each (B) or n = 10 (C and E–G). Different letters on the histograms indicate that the means differ significantly (P ≤ 0.05). DW, Dry weight.
Figure 3.
Root hair development is enhanced in AtERF070 RNAi lines. Arabidopsis (wild-type [WT] and RNAi lines) seedlings were raised as described in the legend to Figure 2 under P+ and P− conditions for 2 d. A, Development of root hairs in a 5-mm region of the primary root tip of wild-type and RNAi seedlings grown under P+ and P− conditions. Bars = 1 mm. B, Total number of root hairs in a 5-mm region of the primary root tip. Values are means ± se; n = 10. Different letters on the histograms indicate that the means differ significantly (P ≤ 0.05).
Overexpression of AtERF070 Attenuates Pi Content and Different Traits of RSA
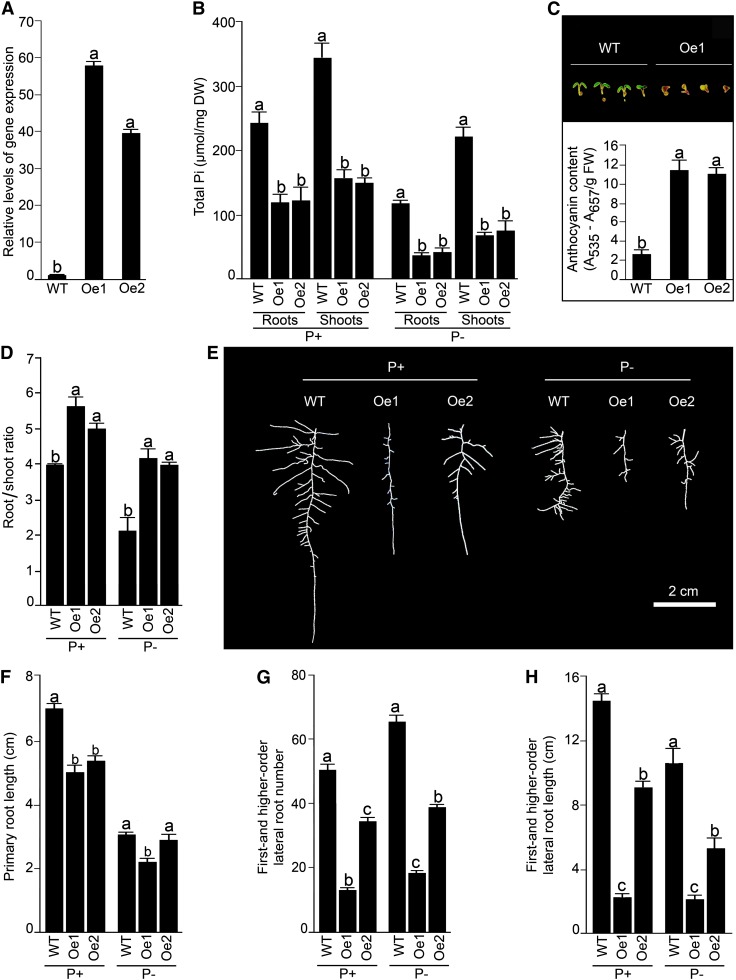
Overexpression lines, harboring the full-length complementary DNA (cDNA) of AtERF070 under the control of a CaMV 35S promoter, were generated (hereafter referred to as Oe1 and Oe2) and evaluated for changes in Pi content and different traits of RSA under P+ and P− conditions (Fig. 4). Two independently generated overexpressors revealed 40- to 60-fold increases in the relative expression of AtERF070 in 12-d-old seedlings grown under nutrient-rich conditions compared with the wild type (Fig. 4A). These overexpressors were subsequently used for morphophysiological characterization. Under both P+ and P− conditions, the overexpression lines (Oe1 and Oe2) showed significant (P ≤ 0.05) reductions in Pi contents in both roots and shoots (Fig. 4B). The overexpression lines germinated on one-half-strength Murashige and Skoog (MS) medium showed a distinct accumulation of anthocyanin that intensified on day 4 post germination (Fig. 4C, top), which was 6-fold higher compared with the wild-type seedlings (Fig. 4C, bottom). Also, under P+ and P− conditions, there were significant (P ≤ 0.05) increases in root-shoot ratio of both the overexpression lines compared with the wild type (Fig. 4D). Furthermore, the RSA of overexpression lines grown on P+ and P− media on vertically oriented agar plates for 7 d was documented (Fig. 4E). Irrespective of Pi regime, both the overexpression lines overall displayed a relatively smaller root system compared with the wild type. The effect of overexpression on the growth of primary root was more pronounced in Oe1 under both P+ and P− conditions (Fig. 4F). Interestingly though, the inhibitory effects of AtERF070 overexpression were evident on both the number and the length of first- and higher-order lateral roots of Oe1 and Oe2 (Fig. 4, G and H). Relatively, the effect of the overexpression of AtERF070 was more pronounced on lateral root development than on primary root growth. The differential effect of AtERF070 on the primary and lateral roots could be attributed to their distinct ontogeny. These results thus suggested a potential role of AtERF070 in regulating the growth and development of RSA that, in turn, affects Pi homeostasis.
Figure 4.
Overexpression of AtERF070 affects morphophysiological responses. Arabidopsis (wild-type [WT] and Oe1 and Oe2) seedlings were raised as described in the legend to Figure 2. A, Whole seedlings (the wild type and Oe1 and Oe2) raised on one-half-strength MS medium were used for real-time PCR analyses of AtERF070 as described in the legend to Figure 1. The wild-type P+ value was normalized to 1. Different letters indicate significant differences in the relative level of gene expression. B to H, Roots and shoots of the wild type and Oe1 and Oe2 were dissected at the hypocotyl-root junction. Data are presented for total Pi contents in roots and shoots; n = 6 replicates of 25 seedlings each (B), accumulation (top) and quantification (bottom) of anthocyanin content in the shoots; n = 6 replicates of 100 seedlings each (C), root-shoot ratio; n = 6 replicates of 25 seedlings each (D), root architectural details (E), primary root length (F), first- and higher-order lateral root number (G), and their length (H); n = 10 for F to H. Values are means ± se, and different letters on the histograms indicate that the means differ significantly (P ≤ 0.05). DW, Dry weight; FW, fresh weight. [See online article for color version of this figure.]
AtERF070 Overexpression Lines Display Retarded Growth Responses
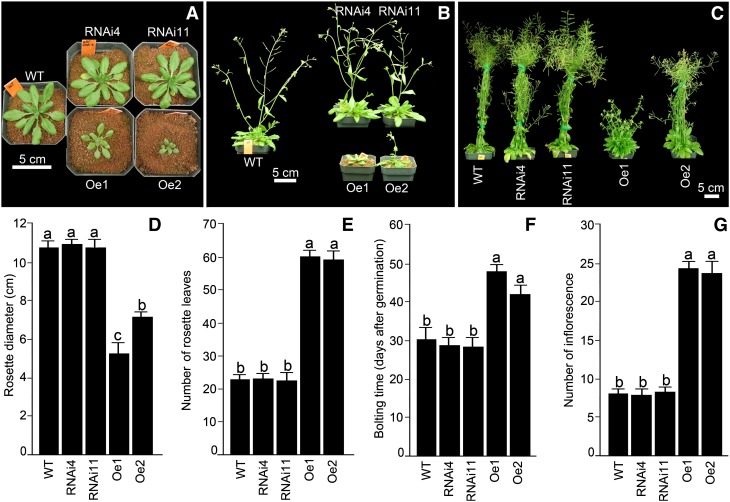
Wild-type, AtERF070 RNAi, and overexpression lines were germinated and grown to maturity on a Premier ProMix PGX peat mix under greenhouse condition for documenting different morphometric traits (Fig. 5). The overexpression lines exhibited retarded growth until 45 d after germination, resulting in a smaller rosette diameter compared with wild-type and RNAi lines (Fig. 5). Subsequently, overexpression lines developed a large number (around 60) of rosette leaves, resulting in a dwarf bushy appearance (Fig. 5, C–E). Furthermore, the bolting was delayed by 2 weeks in both the overexpression lines compared with the wild-type and RNAi lines (Fig. 5F). Interestingly, overexpression lines also produced around 20 to 24 inflorescence branches, which were about 3-fold higher than that produced by either wild-type or RNAi lines (Fig. 5G). The overexpression lines exhibited a delay in the onset of senescence and eventually completed their life cycle 2 weeks later than the wild-type and RNAi lines (data not shown).
Figure 5.
Differential temporal growth responses of AtERF070 RNAi and overexpression lines. A to C, Wild-type (WT), AtERF070 RNAi, and overexpression lines were germinated in potting mix and grown under greenhouse conditions, and their temporal growth responses were documented 30 d (A), 45 d (B), and 70 d (C) after germination. D to G, Data are presented for rosette diameter (D), number of rosette leaves (E), bolting time (F), and number of inflorescences (G). Values are means ± se, and n = 10. Different letters on the histograms indicate that the means differ significantly (P ≤ 0.05). [See online article for color version of this figure.]
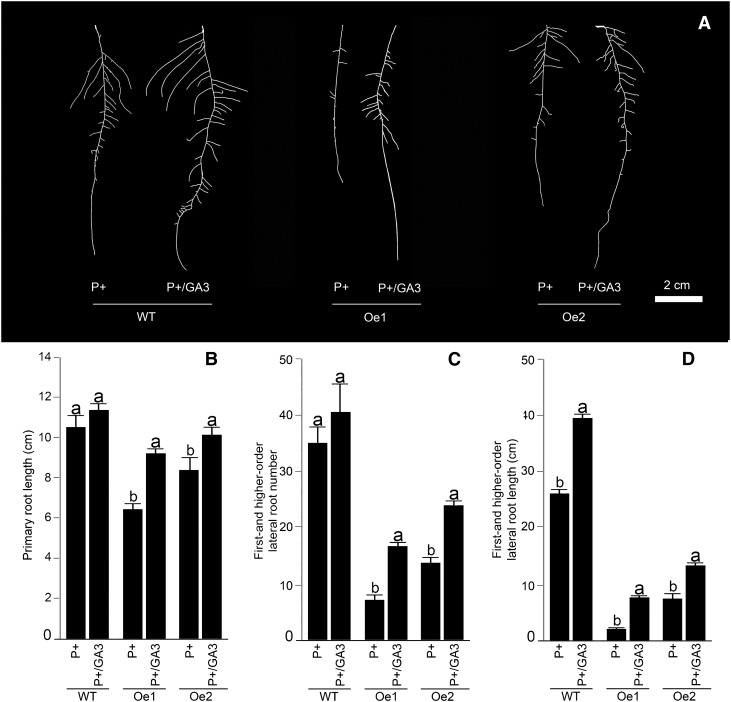
GA Supplementation-Mediated Partial Recovery of the Growth Responses of the Overexpression Lines
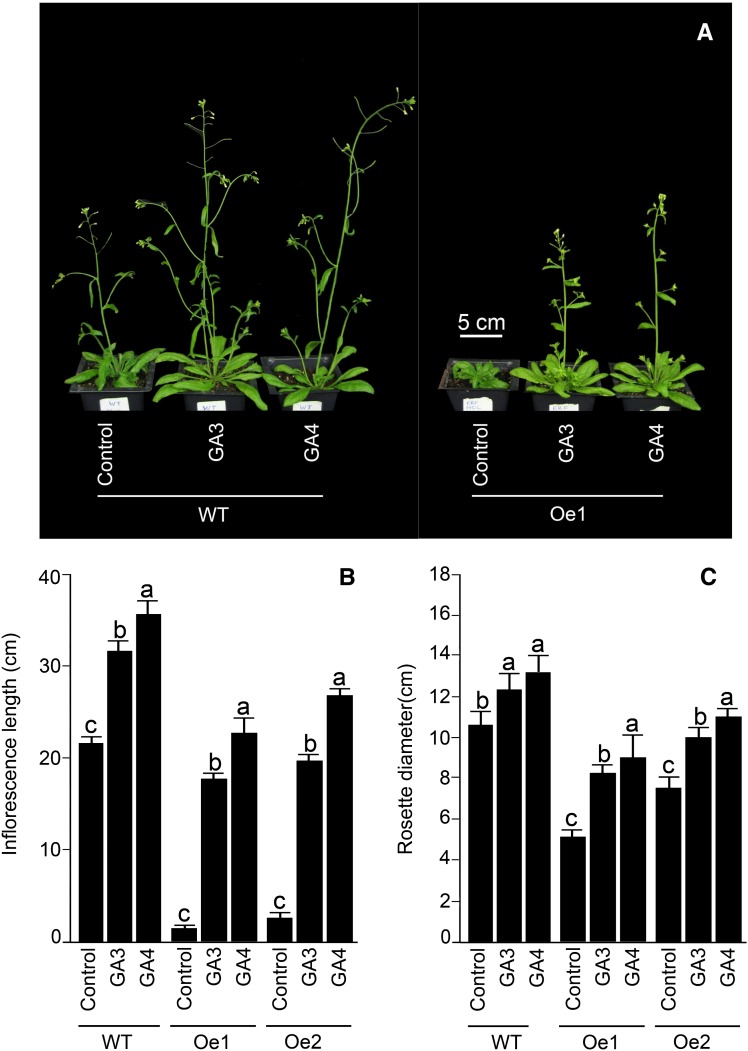
Since AtERF070 overexpression lines displayed a phenotype similar to GA-deficient mutant (Fu and Harberd., 2003) and MYB62 overexpression (Devaiah et al., 2009) lines, we presumed that the AtERF070 overexpression phenotype could possibly be rescued by supplementing the growth medium with GA. Therefore, wild-type and overexpression lines were grown under Pi-replete condition supplemented with and without GA3 (1 µm) on vertically oriented petri plates for 7 d (Fig. 6). The primary root length and number of first- and higher-order lateral roots of the wild-type seedlings were comparable during growth on P+ and GA3-supplemented P+ medium (Fig. 6, A–C). On the contrary, there was a significant (P ≤ 0.05) increase in both the length of the primary root and the number of first- and higher-order lateral roots of the overexpression lines when grown on GA3-supplemented P+ medium (Fig. 6, A–C), whereas the effect of GA3 supplementation in P+ medium on the augmented growth of the first- and higher-order lateral roots was relatively comparable in wild-type and overexpression lines (Fig. 6D). Although GA3 supplementation in the P+ medium triggered an increase in the growth responses of different root traits in overexpression lines, the root phenotype was still not comparable to that of the wild-type seedling grown on P+ medium. This suggested a rather partial recovery of the root phenotype of overexpression lines by GA3 treatment. Furthermore, the role of GA in recovering the phenotype of the aerial parts of the overexpression lines grown under greenhouse conditions was tested. The overexpression lines and the wild type were grown on Premier ProMix PGX peat mix for 2 weeks and sprayed eight times on alternate days with GA3 (100 µm) or GA4 (50 µm). As a control, both overexpression lines and wild-type plants were sprayed with water for the same duration. The apical dominance and time of bolting in both overexpression lines could be restored by spraying with GA3 or GA4 (Fig. 7A). In addition, spraying with either GA3 or GA4 partially recovered the phenotype of lateral branches, length of primary inflorescence, and rosette diameter of the overexpression lines (Fig. 7, B and C). However, spraying with either GA3 or GA4 did not have any effect on the number of rosette leaves produced by overexpression lines, which remained significantly (P ≤ 0.05) higher than those of the wild type (data not shown).
Figure 6.
Growth medium supplemented with GA modulates the RSA of AtERF070 overexpression lines. A, Wild-type (WT) and AtERF070 overexpression lines were germinated on one-half-strength MS medium for 7 d and then transferred to P+ medium with or without 1 µm GA3. Lateral roots were spread to reveal details of RSA and are representative of 10 seedlings each for the wild-type and overexpression lines. B to D, Data are presented for the effects of GA treatment on primary root length (B), first- and higher-order lateral root number (C), and first- and higher-order lateral root length (D) of wild type and transgenics. Values are means ± se, and n = 10. Different letters on the histograms indicate that the means differ significantly (P ≤ 0.05).
Figure 7.
Exogenous GA application partially restores the shoot phenotype of AtERF070 overexpression lines. A, Effects of GA application on wild-type (WT) and AtERF070 overexpression lines. Wild-type and AtERF070 overexpression lines were germinated on soil, and 20-d-old plants were treated with 50 µm GA4 or 100 µm GA3 on alternate days. A total of eight sprays were carried out, and control plants were treated with water. B and C, Data are presented for the effects of exogenous GA application on inflorescence length (B) and rosette diameter (C) of wild type and transgenics. Values are means ± se (n = 10). Different letters on the histograms indicate that the means differ significantly (P ≤ 0.05). [See online article for color version of this figure.]
Overexpression of AtERF070 Alters the Expression of a Subset of PSR and GA Biosynthetic Genes
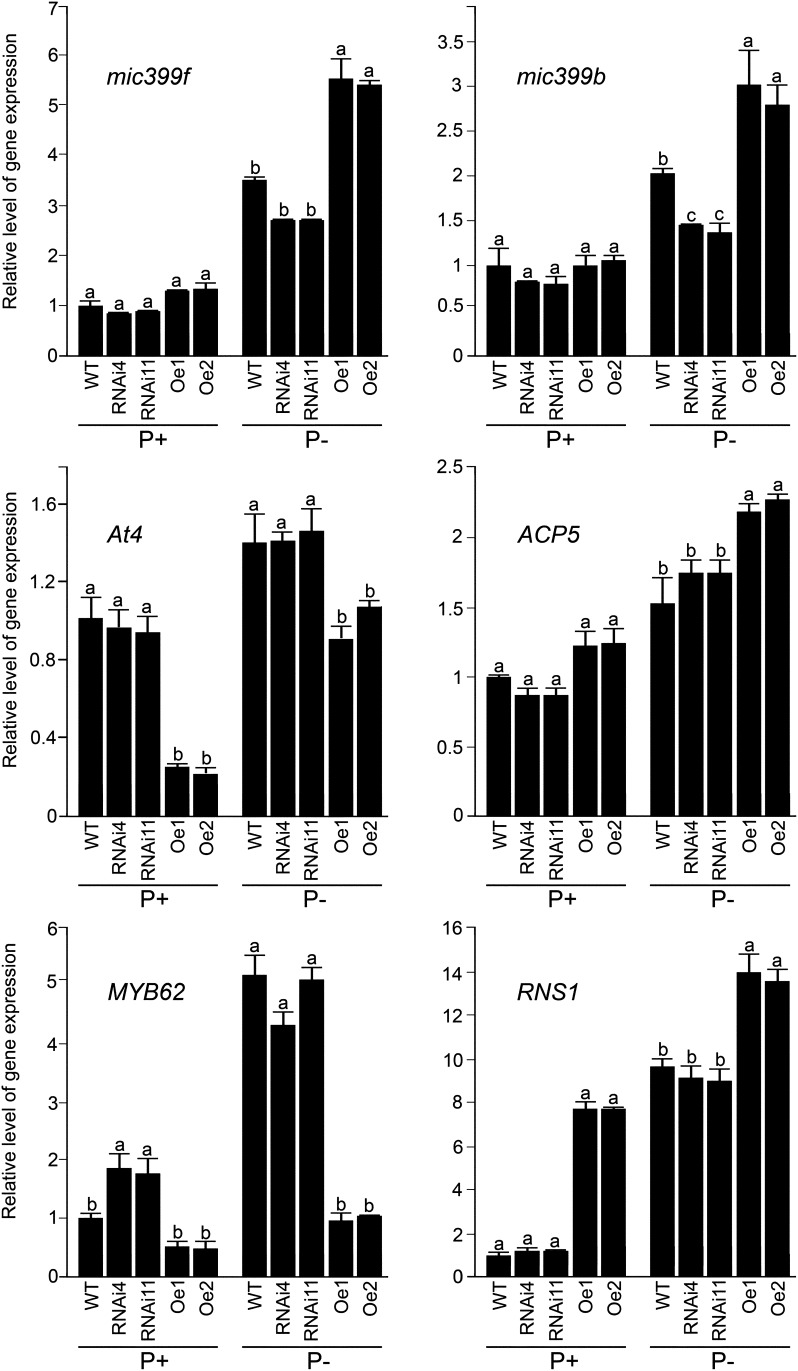
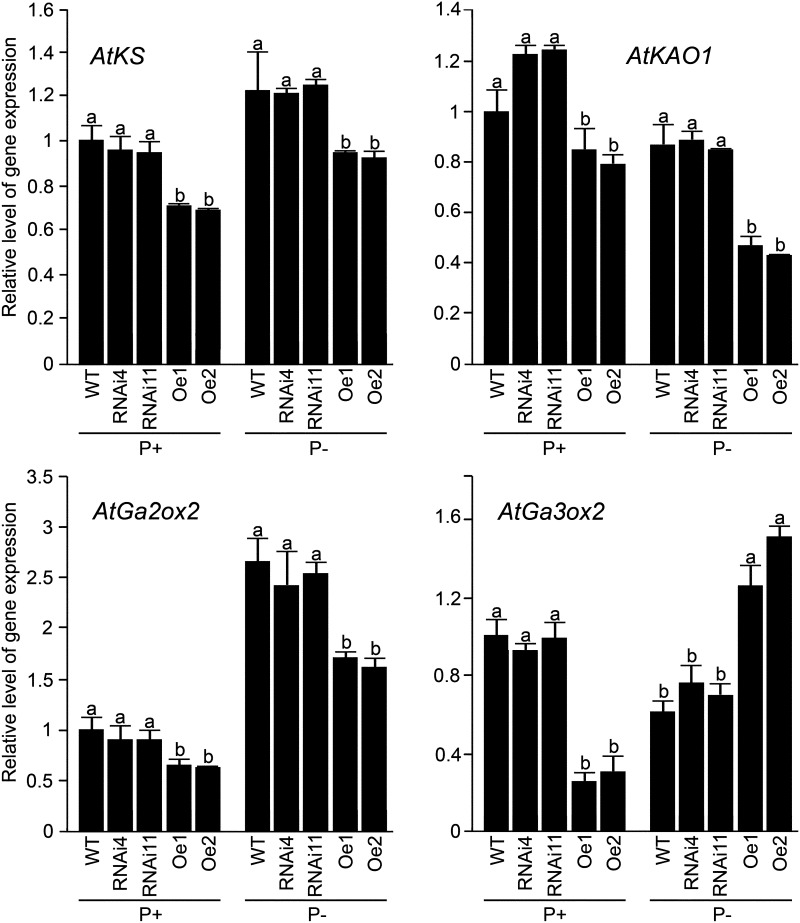
To determine the relative expression of some of the functionally characterized PSR genes (i.e. HIGH-AFFINITY PHOSPHATE TRANSPORTER1;1 [Pht1;1], Pht1;4, INDUCED BY PHOSPHATE STARVATION1 [IPS1], ACID PHOSPHATASE5 [ACP5], RIBONUCLEASE1 [RNS1], At4, microRNA399b, and microRNA399f; Muchhal et al., 1996; Bariola et al., 1999; del Pozo et al., 1999; Martín et al., 2000; Karthikeyan et al., 2002; Shin et al., 2004, 2006; Fujii et al., 2005) in the seedlings by real-time PCR, the wild-type, RNAi, and overexpression lines were grown under P+ and P− conditions for 7 d in vertically oriented agar plates. The relative expression levels of these PSR genes were comparable in both wild-type and RNAi seedlings under both P+ and P− conditions (Fig. 8), whereas there was a significant increase in the expression of RNS1 in overexpression seedlings compared with wild-type seedlings. On the contrary, the relative expression of At4 was suppressed in both P+ and P− overexpression seedlings compared with wild-type seedlings. There was an increase in the relative expression of microRNA399b, microRNA399f, and ACP5 only in P− overexpression seedlings compared with wild-type seedlings. However, the relative expression of AtIPS1, Pht1;1, and Pht1;4 in overexpression lines was comparable irrespective of Pi regime. These results clearly suggested differential effects of overexpressing AtERF070 on a subset of PSR genes. Since the root and shoot phenotypes of overexpression lines could be recovered partially by exogenous GA application (Figs. 6 and 7), this suggested a likely effect of overexpressing AtERF070 on the relative expression of some of the genes involved in GA biosynthesis. Interestingly, the relative expression levels of Arabidopsis thaliana ent-Kaurene synthase (AtKS), mediating the first committed step in GA biosynthesis, along with Arabidopsis thaliana ent-Kaurenoic acid oxidase1 and Arabidopsis thaliana GA2oxidase2 (AtGa2ox2) were down-regulated in overexpression lines under both P+ and P− conditions. Although the relative expression of AtGa3ox2 was down-regulated in P+ overexpression lines, there was a significant increase in the relative expression of this gene in P− overexpression lines. However, the relative expression levels of several other genes that are involved in the GA biosynthetic pathway (AtKO, AtGa3ox1, AtGa2ox1, AtGa20oxs) were comparable in overexpression and wild-type seedlings under both P+ and P− conditions. These results further suggest the effect of overexpressing AtERF070 on a subset of genes that are involved in the GA biosynthetic pathway (Fig. 9).
Figure 8.
Real-time PCR analysis of PSR genes in AtERF070 RNAi and overexpression lines. Wild-type (WT), RNAi, and overexpression seedlings were germinated on one-half-strength MS medium for 7 d and then transferred to vertically oriented agar petri plates containing P+ and P− media for 1 week. Whole seedlings were harvested and used for real-time PCR analysis revealing the relative expression levels of PSR genes in the wild type and transgenics. The Arabidopsis housekeeping gene AtACTIN2 (At3g18780) was used as an internal control. Expression values for the PSR genes in wild-type P+ were set at 1. The values presented are means of three technical replicates ± se. Different letters indicate significant differences.
Figure 9.
Real-time PCR analysis of GA biosynthetic genes in AtERF070 RNAi and overexpression lines. Wild-type (WT), RNAi, and overexpression seedlings were germinated on one-half-strength MS medium for 7 d and then transferred to vertically oriented agar petri plates containing P+ and P− media for 1 week. Whole seedlings were harvested and used for real-time PCR analysis revealing the relative expression levels of GA biosynthetic genes in the wild type and transgenics. The Arabidopsis housekeeping gene AtACTIN2 (At3g18780) was used as an internal control. Expression values for the GA biosynthetic genes in wild-type P+ were normalized to 1. Different letters indicate significant differences in the relative level of gene expression. The values presented are means of three technical replicates ± se.
DISCUSSION
TFs regulate a diverse group of genes during stress responses and are important components of gene regulatory networks (Yamaguchi-Shinozaki and Shinozaki, 2006). TFs also play an important role in the maintenance of Pi homeostasis (Jain et al., 2012). In Arabidopsis, the AP2/ERF TF superfamily comprises 147 members, including the AP2 and ERF families (Riechmann et al., 2000; Feng et al., 2005). AP2/ERF proteins are known to regulate the responses of plants to various biotic and abiotic stresses and developmental processes (Elliott et al., 1996; Stockinger et al., 1997; Gu et al., 2000; Boutilier et al., 2002; Dubouzet et al., 2003). In this study, we report the functional characterization of AtERF070, a member of the AP2/ERF family.
AtERF070 Is a PSR Nucleus-Localized TF
The expression of AtERF070 is induced significantly and specifically in Pi-starved roots and leaves (Fig. 1A), and the results were consistent with an earlier study (Misson et al., 2005). Several other functionally characterized TFs (i.e. WRKY6-1, WRKY75, ZAT6, MYB62, PRD, HRS1-1, bHLH32, BnPHR1 OsPTF1, and OsMYB2P-1) have been shown to be differentially regulated under different Pi regimes and to play a role in the maintenance of Pi homeostasis (Yi et al., 2005; Chen et al., 2007; Devaiah et al., 2007a, 2007b, 2009; Camacho-Cristóbal et al., 2008; Liu et al., 2009; Dai et al., 2012; Ren et al., 2012). Although PHR1, OsPHR1, and OsPHR2 are not responsive to Pi deficiency, they have also been implicated in regulating a subset of PSR genes (Rubio et al., 2001; Bari et al., 2006; Zhou et al., 2008; Bustos et al., 2010).
The localization of TFs could play a major role in determining their function. For example, the target of Rapamycin signaling pathway is implicated in the cytoplasmic retention of several TFs, thereby preventing the transcription of downstream genes induced by nitrogen limitation (Beck and Hall, 1999). In the yeast (Saccharomyces cerevisiae) phosphatase (PHO) regulon, Pi-starvation-induced nuclear localization of Pho4 has been shown to be a prerequisite for its activity in transcriptional regulation (Ogawa et al., 1995; Lenburg and O’Shea, 1996). Interestingly, AtERF070 is localized to the nucleus irrespective of Pi regime. This pattern of Pi-independent localization is also a characteristic feature of some of the Pi-starvation-induced TFs in Arabidopsis (WRKY75, Devaiah et al., 2007a; ZAT6, Devaiah et al., 2007b; MYB62, Devaiah et al., 2009) and rice (OsPTF1, Yi et al., 2005). Regulation of these TFs by posttranslational modifications, protein-protein interaction, DNA-protein interaction, and/or chromatin modifications has been postulated (Schwechheimer and Bevan, 1998; Miura et al., 2005).
AtERF070 RNAi Lines Have Higher Pi Accumulation and Increased Root Growth
To decipher the in planta role of AtERF070 in regulating Pi homoeostasis, two independent RNAi lines were analyzed. As anticipated, there was a significant decrease in the expression of AtERF070 in both RNAi lines. The AtERF070 RNAi lines displayed increases in Pi content and shoot area irrespective of Pi regime. A similar increase in Pi content was also observed in the WRKY75RNAi lines under both P+ and P− conditions (Devaiah et al., 2007a), whereas wrky6-1 showed an increased accumulation of Pi only in P+ shoots (Chen et al., 2009). In contrast, the Pi contents of prd, hrs1-1, Osphr1RNAi, and Osphr2RNAi lines were comparable to their respective wild types irrespective of the Pi regime. On the contrary, phr1 showed a significant reduction in the Pi content of the P+ seedlings compared with the wild type (Rubio et al., 2001; Bustos et al., 2010). Therefore, higher Pi accumulation as a result of the reduced expression of AtERF070 in the RNAi lines suggests that it is a negative regulator of Pi homeostasis.
RSA is a critical component of plant adaptation to the heterogenous distribution of Pi in soils. RSA comprises ontogenetically distinct embryonic and postembryonically developed primary and lateral roots, respectively. Primary root growth was augmented in the AtERF070 RNAi lines under Pi-sufficient conditions, suggesting that it is a negative regulator of this root trait. This is in contrast to an inhibitory effect on the primary root growth in P− prd seedlings (Camacho-Cristóbal et al., 2008), which suggested a positive regulatory effect of PRD on Pi-deficiency-mediated primary root growth. Relatively, there were no noticeable variations in primary root growth of OsPHR2RNAi, WRKY75RNAi, bhlh32, and hrs1-1 compared with their respective wild type (Zhou et al., 1997; Chen et al., 2007; Devaiah et al., 2007a; Liu et al., 2009). In addition, AtERF070 RNAi lines also showed a significant increase in root hair number (Fig. 3). Similar increases in root hair numbers have also been reported for bhlh32 and WRKY75 RNAi lines (Chen et al., 2007; Devaiah et al., 2007a). These results indicate that AtERF070, in coordination with other TFs, regulates root hair development. An earlier report implicated ethylene in both root hair initiation and elongation (Dolan, 2001). Under nutrient deficiency, including Pi starvation, auxin and ethylene are known to play major roles in root hair development (Masucci and Schiefelbein, 1994). Our results show that an ERF that is the downstream target of the ethylene signaling pathway is involved in root hair development.
In addition, the AtERF070 RNAi lines displayed significant increases in the number and length of the lateral roots, which was similar to the augmented lateral root growth of the WRKY75RNAi lines (Devaiah et al., 2007a). Interestingly, the PSR TFs ZAT6, MYB62, PRD (Devaiah et al., 2007b, 2009; Camacho-Cristóba et al., 2008), and AtERF070 modulate the growth of both primary and lateral roots, while WRKY75 regulates the growth of lateral roots only (Devaiah et al., 2007a). These results suggest that different TFs have differential effects on the developmental responses of the root system.
Overexpression of AtERF070 Displays Stunted Root Growth
Overexpression of AtERF070 resulted in reductions of primary root length, lateral root number, and lateral root length irrespective of Pi regime (Fig. 4). This decrease in the primary root growth was similar to the stunted primary root exhibited by ZAT6 and MYB62 overexpression lines (Devaiah et al., 2007b, 2009). While the ZAT6 overexpressors compensated this by augmenting the lateral root growth, the AtERF070 and MYB62 overexpression lines (Devaiah et al., 2009) displayed a decreased lateral root growth relative to their respective wild types. Interestingly, ZAT6 and MYB62 overexpression lines also showed altered root parameters under both P+ and P− conditions (Devaiah et al., 2007b, 2009).
In the case of the AtERF070 overexpression lines, the reduced root growth could have led to the lower Pi content relative to the wild type (Fig. 4B). Comparatively, the ZAT6 overexpression lines exhibited decreased Pi content (Devaiah et al., 2007b), while the MYB62 overexpressors had a higher Pi content in P+ shoots and a lower Pi content in the P− seedlings, relative to the wild type (Devaiah et al., 2009). The decrease in the Pi content of the AtERF070 overexpression lines could have triggered higher anthocyanin accumulation (Fig. 4C), which is similar to that observed in the ZAT6 and MYB62 overexpressors (Devaiah et al., 2007b, 2009). Taken together, these studies suggest that different TFs induced during Pi deficiency coordinately regulate multiple Pi-starvation responses.
Overexpression of AtERF070 Modulates Plant Growth
Overexpression of AtERF070 resulted in stunted plants lacking apical dominance with delayed bolting and senescence, which are reminiscent of a GA-deficient plant (Fig. 5). Interestingly, the AtERF070 overexpression lines had a phenotype similar to the MYB62 overexpression line (Devaiah et al., 2009), suggesting that both TFs may be involved in the cross talk between GA and Pi starvation. This assumption was confirmed by treating AtERF070 overexpression lines with GA.
GA3-treated 7-d-old seedlings of AtERF070 overexpression lines showed increased primary root growth, higher lateral root number, modest increase in lateral root length, and restoration of apical dominance, pointing toward a partial recovery of the phenotype. This response is similar to the recovery of ga1-3, a GA-deficient mutant, by an exogenous supplementation of GA (Fu and Harberd, 2003). Partial reversal of the phenotype by GA application further substantiated the reasoning that the overexpression lines are GA deficient. To further confirm this, we analyzed the relative expression of GA biosynthetic genes in the AtERF070 overexpression and RNAi lines relative to the wild-type plants. In Arabidopsis, AtKS, Arabidopsis thaliana ent-Kaurenoic acid oxidase1, and AtGA2oxs are implicated in the GA biosynthetic pathway. Overexpression of AtERF070 resulted in significant attenuation in the relative expression of these genes. Even a marginal attenuation in the expression of AtKS could have an impact on GA content, since it is encoded by a single gene and is involved in the first committed step in GA biosynthesis (Olszewski et al., 2002). This suggested that the AtERF070 overexpression lines had altered expression of a subset of GA biosynthetic genes, which could possibly result in reduced GA in these lines. Interestingly, the subset of GA biosynthetic genes attenuated in the MYB62 overexpression lines was different (Devaiah et al., 2009) from those altered in AtERF070 overexpression lines. Although these two TFs affect different aspects of the GA biosynthetic pathway, they both seem to modulate GA levels, causing a similar plant phenotype in their overexpression lines. An earlier study demonstrated a reduction in bioactive GA levels during Pi starvation (Jiang et al., 2007). A number of the Pi-deficiency-mediated morphological changes are associated with a lack of GA (O’Rourke et al., 2013). Therefore, potential roles of AtERF070 and MYB62 in mediating a cross talk between phytohormone and Pi-sensing and -signaling cascades could be assumed.
PSR Genes Are Modulated in AtERF070 Overexpression Lines
Among the PSR genes, microRNA399f and microRNA399b were up-regulated in the Pi-deprived AtERF070 overexpression lines. However, an elevated relative expression of microRNA399b in the AtERF070 overexpression lines neither altered Pho2 level nor increased Pi content as reported in an earlier study (Lin et al., 2008). A similar increase in the expression of microRNA399f was reported in response to an increased expression of the transcriptional activator AtMYB2 (Baek et al., 2013). In contrast, the AtERF070 RNAi lines showed a modest suppression of microRNA399b only under Pi-deficient conditions. The up-regulation of microRNAs in the AtERF070 overexpression lines draws focus to the fact that both TFs and microRNAs share their role as regulatory elements. Both of them are trans-acting and bind to cis-elements present in DNA/RNA. In addition, they can regulate a single gene or a number of genes. The point of interest is that they can act together in a specific cell type and establish a particular gene expression pattern in that cell (Hobert, 2004, 2008). This would be of great relevance in Pi starvation, where spatiotemporal regulation of gene expression is a well-established phenomenon. Determining whether AtERF070 and microRNA399 work together or in a hierarchical manner could help in understanding regulatory relationships during Pi starvation.
Furthermore, relative expression levels of Pht1:1; Pht1:4, and AtIPS1 remained unaltered in the AtERF070 transgenic lines. This is in contrast to the modulated expression levels of all these genes in the MYB62 and ZAT6 overexpression lines (Devaiah et al., 2007b, 2009). Interestingly, MYB62 is suppressed in the AtERF070 overexpression lines. This points to the fact that although they might be acting in the same Pi-starvation signaling pathway, they have different and nonoverlapping downstream targets. On the other hand, At4 was suppressed in the AtERF070 and ZAT6 overexpression lines (Devaiah et al., 2007b), although the expression of ZAT6 remained unaltered in the AtERF070 overexpression lines. This is indicative of complex interactive pathways of TFs during Pi starvation. A similar sequential gene regulation has been reported in rice, where the overexpression of Iron Deficiency-Responsive cis-Acting Element Binding Factor1 caused an increase in the expression of IRON-RELATED TRANSCRIPTION FACTOR2, both being TFs involved in iron deficiency responses (Kobayashi et al., 2007).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for generation of the transgenic lines and other experimentation in this study. Wild-type and transgenic seeds were surface sterilized and stratified at 4°C for 2 d. Seeds (20–30) were germinated per petri plate (150 mm × 15 mm) containing one-half-strength MS medium supplemented with 1.5% (w/v) Suc and 1.2% (w/v) agar (Sigma A1296, lot no. 110K0195 with an impurity of 15 μM Pi in 1.2% [w/v] agar [Jain et al., 2009]). The tissue culture and greenhouse growth conditions used were as described (Ramaiah et al., 2011).
Plant Transformation
Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998) to generate stable transgenic lines. AtERF070 RNAi, overexpression, and GFP fusion constructs were used for generating the transgenic lines, and in each case, an empty vector was used for transformation for generating control plants.
The binary vector pGSA1131 was utilized to generate the RNAi construct for AtERF070. A 250-bp fragment, unique to the AtERF070 coding sequence, was amplified using the primers 5′-TCTAGACCATGGTAAAGAGATCGTAAT-3′ and 5′-CCTAGGGGCGCGCCATAAGCTAGAGC-3′. The PCR-amplified fragment was digested with NcoI and AscI to facilitate the cloning of the fragment in the sense orientation in the binary vector. The same fragment was then digested with XbaI and AvrII and cloned in the antisense orientation in the binary vector next to a GUS intron spacer separating the two multiple cloning sites. The presence of the two inserts in the desired orientation was confirmed by sequencing.
The binary vector pEGAD with EGFP as an N-terminal translational fusion was used to generate the GFP::AtERF070 construct. Full-length cDNA of AtERF070 was amplified using the primers 5′-CCCGGGATGAAGCGTATTATCAGAAT-3′ and 5′-AAGCTTTCACGAGCCTCTAGCA-3′. The amplified fragment was cloned in frame with the EGFP after digestion with SmaI and HindIII to generate an N-terminal fusion.
A modified pEGAD vector (Devaiah et al., 2009) was used to generate the overexpression construct for AtERF070. Full-length cDNA of AtERF070 was amplified with the primers 5′-CCCGGGATGAAGCGTATTATCAGAA-3′ and 5′-AAGCTTTCACGCGCCTCTAGCA-3′. The full-length cDNA fragment so amplified was digested with SmaI and HindIII enzymes and cloned next to the CaMV 35S promoter to generate the overexpression construct.
The binary vectors pGSA1131 and pEGAD confer Basta resistance, and the transformed seedlings were recovered by spraying with 50 µL L–1 Basta.
GA Treatment
The stocks for GA4 and GA3 (Sigma-Aldrich) were prepared by dissolving in alcohol, and the required dilutions were made with water. For greenhouse experiments, GA was sprayed on the aerial parts of the plant. For in vitro experiments, filter-sterilized GA3 was added to Pi-sufficient medium after autoclaving, and plates were prepared. The spray routine was based on an earlier study involving the rescue of MYB62-overexpressing plants by GA application (Devaiah et al., 2009).
Documentation of GFP Fluorescence
A Nikon E800 compound microscope (Diagnostic Instruments) was used for GFP imaging. The documentation was facilitated by a SPOT RT-slider digital camera connected to the microscope. Fluorescein isothiocyanate filters on the microscope were used to provide the GFP excitation. To confirm the nuclear localization, the transgenic and control plants were stained with DAPI as described (Devaiah et al., 2009).
RSA Analysis
The seedlings grown on P+ and P− media were spread out on petri plates and scanned at 200 dots per inch using a desktop scanner (UMAX Powelook 2100 XL). The lengths of the primary and lateral roots were measured using the ImageJ program (http://rsb.info.nih.gov/ij).
Determination of Shoot Area
Leaf area was measured by dissecting the leaves from the shoot individually and spreading them on agar plates. These plates were then scanned at 200 dots per inch, and ImageJ was used to compute the leaf area.
Root Hair Measurements
Seven-day-old seedlings germinated on one-half-strength MS petri plates containing 1.5% (w/v) Suc and 1.2% (w/v) agar were transferred to P+ or P− media. Two days post transfer, root hairs within the 5-mm region from the root tip were captured using a stereomicroscope (Nikon SMZ-U) equipped with a digital camera. The number and length of the root hairs were determined using ImageJ (http://rsb.info.nih.gov/ij/).
Anthocyanin Estimation
Seedlings germinated on one-half-strength MS plates were harvested 4 d after germination and ground into fine powder with liquid nitrogen. About 100 mg of tissue was used for anthocyanin quantification as described (Lange et al., 1971).
Quantification of Total Pi
A modified U.S. Environmental Protection Agency method 365.2 was used to determine the total Pi content in the wild-type and transgenic lines. The seedlings raised in petri plates were harvested, and about 50 mg of fresh sample was transferred into a preweighed vial and oven dried. The dry weight of the samples was recorded before flaming them to ash and dissolving them in 100 µL of concentrated HCl. Sample dilutions were prepared by adding 10 µL of the sample from the previous step to 790 µL of water. A total of 200 µL of mixed reagent (4.8 mm NH4MoO4, 2.5 n H2SO4, and 35 mm ascorbic acid) was added to this diluted sample (800 µL), and the reaction was incubated at 45°C for 20 min. Appropriate standards were used to measure the total Pi content at A650, and the values were expressed as total Pi mg−1 tissue dry weight.
Real-Time PCR
RNA extraction and subsequent real-time PCR were performed as described (Ramaiah et al., 2011). The primers used for the amplification of AtERF070 and the GA biosynthetic genes are listed in Supplemental Table S1.
Statistical Analysis
The data generated were subjected to Student’s t test, and different letters on the histograms indicate means that were statistically different at P ≤ 0.05.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number BT025618.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of primers used for real-time PCR.
Glossary
- Pi
inorganic phosphate
- RSA
root system architecture
- PSR
Pi starvation-responsive
- TF
transcription factor
- RNAi
RNA interference
- CaMV
cauliflower mosaic virus
- DAPI
4′,6-diamidino-2-phenylindole
- cDNA
complementary DNA
- MS
Murashige and Skoog
Footnotes
This work was supported by the McKnight Foundation (grants to K.G.R.) and by the Ministry of Science and Technology, Department of Biotechnology, Government of India (Ramalingaswamy Fellowship [BT/HRD/35/02/26/2009] to A.J.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Baek D, Park HC, Kim MC, Yun DJ. (2013) The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal Behav 8: e23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ. (1999) Regulation of S-like ribonuclease levels in Arabidopsis: antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19: 529–538 [Google Scholar]
- Beck T, Hall MN. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Camacho-Cristóbal JJ, Rexach J, Conéjéro G, Al-Ghazi Y, Nacry P, Doumas P. (2008) PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta 228: 511–522 [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J 405: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai X, Wang Y, Yang A, Zhang WH. (2012) OsMYB2P-1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. (2007a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. (2001) The role of ethylene in root hair growth in Arabidopsis. J Plant Nutr Soil Sci 164: 141–145 [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JX, Liu D, Pan Y, Gong W, Ma LG, Luo JC, Deng XW, Zhu YX. (2005) An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol 59: 853–868 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. (2000) Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ. (2004) Genetic responses to phosphorus deficiency. Ann Bot (Lond) 94: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2004) Common logic of transcription factor and microRNA action. Trends Biochem Sci 29: 462–468 [DOI] [PubMed] [Google Scholar]
- Hobert O. (2008) Gene regulation by transcription factors and microRNAs. Science 319: 1785–1786 [DOI] [PubMed] [Google Scholar]
- Jain A, Nagarajan VK, Raghothama KG. (2012) Transcriptional regulation of phosphate acquisition by higher plants. Cell Mol Life Sci 69: 3207–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. (2007a) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144: 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Smith AP, Nagarajan VK, Lahner B, Meagher RB, Raghothama KG. (2009) Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiol 150: 1033–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vasconcelos MJ, Sahi SV, Raghothama KG (2007b) Molecular mechanisms of plant adaptation to phosphate deficiency. In J Janick, ed, Plant Breeding Reviews, Vol 29. John Wiley & Sons, Hoboken, NJ, pp 359–419 [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. (2007) Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, Nishizawa NK. (2007) The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc Natl Acad Sci USA 104: 19150–19155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Shropshire W, Jr, Mohr H. (1971) An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol 47: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O’Shea EK. (1996) Signaling phosphate starvation. Trends Biochem Sci 21: 383–387 [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ. (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang H, Wu C, Feng J, Liu X, Qin H, Wang D. (2009) Overexpressing HRS1 confers hypersensitivity to low phosphate-elicited inhibition of primary root growth in Arabidopsis thaliana. J Integr Plant Biol 51: 382–392 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L. (2005) An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis: identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137: 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 29: 459–467 [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, Saitoh H, Miura K, Magbanua JP, Bun-ya M, Harashima S, Oshima Y. (1995) Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol Gen Genet 249: 406–416 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu J, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D, et al. (2013) An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161: 705–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ramaiah M, Jain A, Baldwin JC, Karthikeyan AS, Raghothama KG. (2011) Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol 157: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C, Bucher M. (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB. (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7: e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Bevan M. (1998) Transcriptional regulation of plant gene expression. Trends Plant Sci 3: 378–383 [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ. (2006) Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smith SE, Jakobsen I, Grønlund M, Smith FA. (2011) Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol 156: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]