Abstract
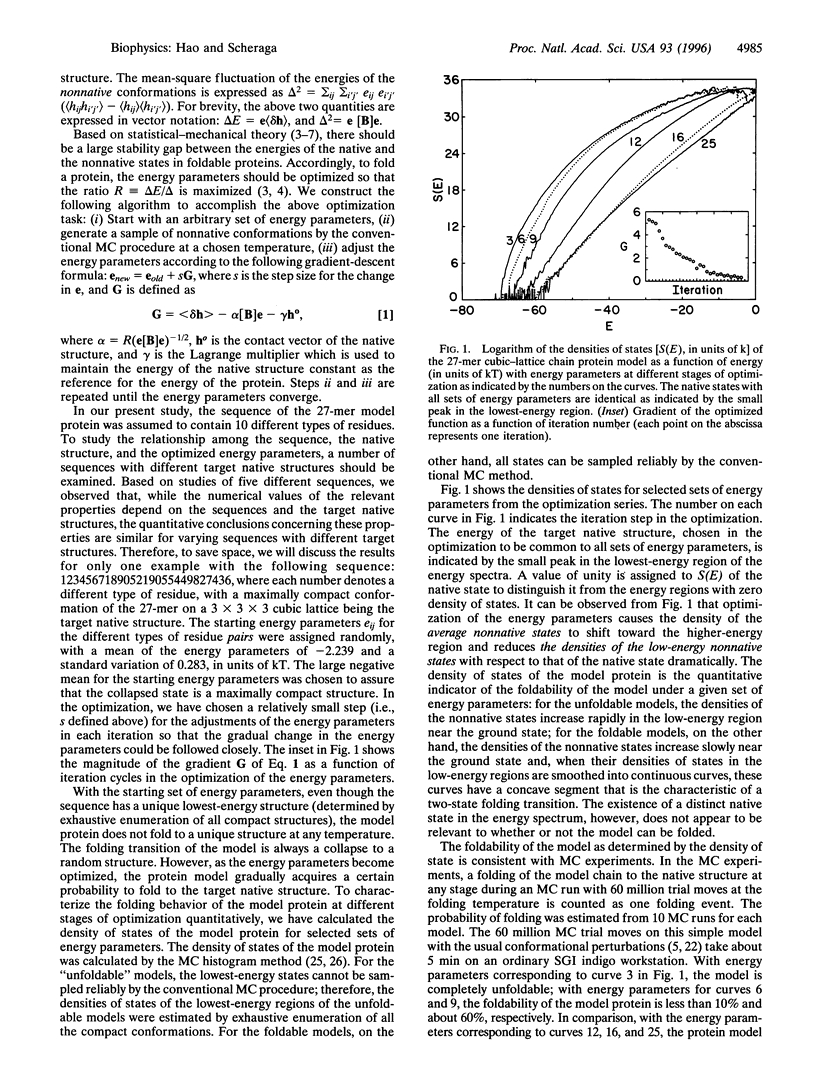
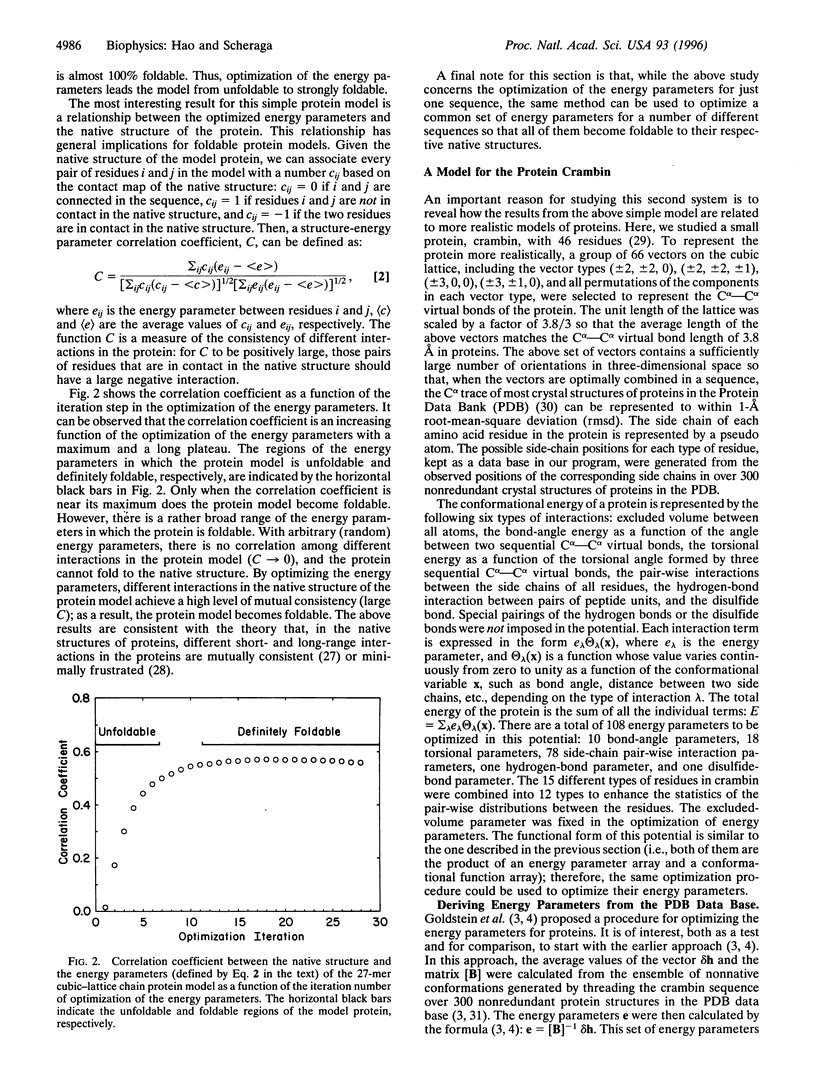
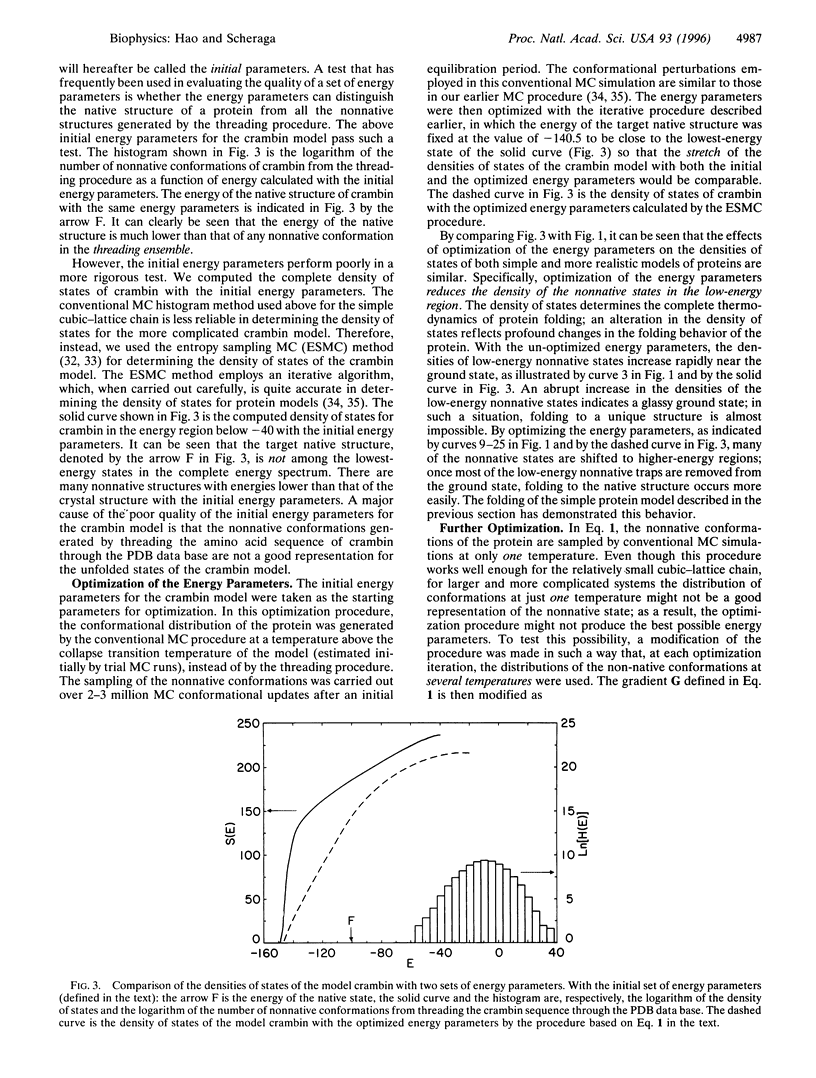
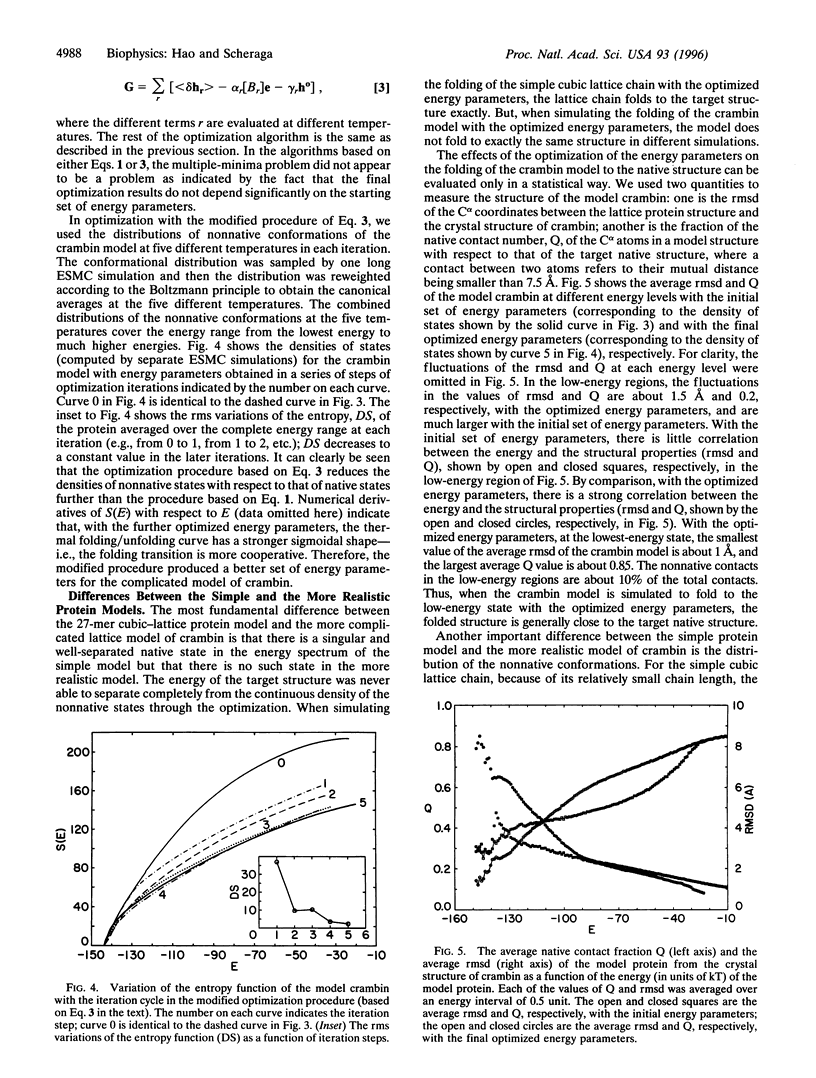
The relationship between the optimization of the potential function and the foldability of theoretical protein models is studied based on investigations of a 27-mer cubic-lattice protein model and a more realistic lattice model for the protein crambin. In both the simple and the more complicated systems, optimization of the energy parameters achieves significant improvements in the statistical-mechanical characteristics of the systems and leads to foldable protein models in simulation experiments. The foldability of the protein models is characterized by their statistical-mechanical properties--e.g., by the density of states and by Monte Carlo folding simulations of the models. With optimized energy parameters, a high level of consistency exists among different interactions in the native structures of the protein models, as revealed by a correlation function between the optimized energy parameters and the native structure of the model proteins. The results of this work are relevant to the design of a general potential function for folding proteins by theoretical simulations.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg BA, Celik T. New approach to spin-glass simulations. Phys Rev Lett. 1992 Oct 12;69(15):2292–2295. doi: 10.1103/PhysRevLett.69.2292. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Wolynes P. G. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrenberg AM, Swendsen RH. New Monte Carlo technique for studying phase transitions. Phys Rev Lett. 1988 Dec 5;61(23):2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- Ferrenberg AM, Swendsen RH. Optimized Monte Carlo data analysis. Phys Rev Lett. 1989 Sep 18;63(12):1195–1198. doi: 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- Go N. Theoretical studies of protein folding. Annu Rev Biophys Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- Goldstein R. A., Luthey-Schulten Z. A., Wolynes P. G. Optimal protein-folding codes from spin-glass theory. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4918–4922. doi: 10.1073/pnas.89.11.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. A., Luthey-Schulten Z. A., Wolynes P. G. Protein tertiary structure recognition using optimized Hamiltonians with local interactions. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9029–9033. doi: 10.1073/pnas.89.19.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler A. T., Huler E., Lifson S. Energy functions for peptides and proteins. I. Derivation of a consistent force field including the hydrogen bond from amide crystals. J Am Chem Soc. 1974 Aug 21;96(17):5319–5327. doi: 10.1021/ja00824a004. [DOI] [PubMed] [Google Scholar]
- Hendlich M., Lackner P., Weitckus S., Floeckner H., Froschauer R., Gottsbacher K., Casari G., Sippl M. J. Identification of native protein folds amongst a large number of incorrect models. The calculation of low energy conformations from potentials of mean force. J Mol Biol. 1990 Nov 5;216(1):167–180. doi: 10.1016/S0022-2836(05)80068-3. [DOI] [PubMed] [Google Scholar]
- Lee J. New Monte Carlo algorithm: Entropic sampling. Phys Rev Lett. 1993 Jul 12;71(2):211–214. doi: 10.1103/PhysRevLett.71.211. [DOI] [PubMed] [Google Scholar]
- Leopold P. E., Montal M., Onuchic J. N. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorov V. N., Crippen G. M. Contact potential that recognizes the correct folding of globular proteins. J Mol Biol. 1992 Oct 5;227(3):876–888. doi: 10.1016/0022-2836(92)90228-c. [DOI] [PubMed] [Google Scholar]
- Nishimura M., Obayashi H., Ohta M., Uchiyama T., Hao Q., Saida T. No association of the 11778 mitochondrial DNA mutation and multiple sclerosis in Japan. Neurology. 1995 Jul;45(7):1333–1334. doi: 10.1212/wnl.45.7.1333. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994 May 19;369(6477):248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. A new approach to the design of stable proteins. Protein Eng. 1993 Nov;6(8):793–800. doi: 10.1093/protein/6.8.793. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Engineering of stable and fast-folding sequences of model proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7195–7199. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich EI. Proteins with selected sequences fold into unique native conformation. Phys Rev Lett. 1994 Jun 13;72(24):3907–3910. doi: 10.1103/PhysRevLett.72.3907. [DOI] [PubMed] [Google Scholar]
- Shrivastava I., Vishveshwara S., Cieplak M., Maritan A., Banavar J. R. Lattice model for rapidly folding protein-like heteropolymers. Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9206–9209. doi: 10.1073/pnas.92.20.9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M. J. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990 Jun 20;213(4):859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Scheraga H. A. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules. 1976 Nov-Dec;9(6):945–950. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- Teeter M. M., Hendrickson W. A. Highly ordered crystals of the plant seed protein crambin. J Mol Biol. 1979 Jan 15;127(2):219–223. doi: 10.1016/0022-2836(79)90242-0. [DOI] [PubMed] [Google Scholar]
- Yue K., Fiebig K. M., Thomas P. D., Chan H. S., Shakhnovich E. I., Dill K. A. A test of lattice protein folding algorithms. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):325–329. doi: 10.1073/pnas.92.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]



