Abstract
The timing and localization of events during mitosis is controlled by the regulated phosphorylation of proteins by the mitotic kinases, which include Aurora A, Aurora B, Nek2, Plk1, and the cyclin-dependent kinase complex Cdk1/cyclin B. Although mitotic kinases can have overlapping subcellular localizations, each kinase appears to phosphorylate its substrates on distinct sites. To gain insight into the relative importance of local sequence context in kinase selectivity, identify previously unknown substrates of these five mitotic kinases, and explore potential mechanisms for substrate discrimination, we determined the optimal substrate motifs of these major mitotic kinases by Positional Scanning Oriented Peptide Library Screening (PS-OPLS). We verified individual motifs with in vitro peptide kinetic studies and used structural modeling to rationalize the kinase-specific selection of key motif-determining residues at the molecular level. Cross comparisons among the phosphorylation site selectivity motifs of these kinases revealed an evolutionarily conserved mutual exclusion mechanism in which the positively and negatively selected portions of the phosphorylation motifs of mitotic kinases, together with their subcellular localizations, result in proper substrate targeting in a coordinated manner during mitosis.
Introduction
Reversible protein phosphorylation is a major mechanism for regulating signal transduction events in mammalian cells (1, 2). For many kinases, substrate selection depends not only on the amino acid sequence motif surrounding the phosphorylation site, but also on additional contextual information provided by kinase-substrate subcellular colocalization or formation of multicomponent complexes through binding proteins, adaptor molecules, or additional protein-domain-mediated interactions, or combinations thereof. Contextual information is critical for correctly identifying substrates of specific DNA damage-activated protein kinases, protein kinase C family members, and growth factor receptor tyrosine kinases (3). During mitosis, however, when the genomic content of the nucleus is being equally partitioned into two daughter cells through cell division, much of this contextual information may be lost because many subcellular boundaries are dissolved or disrupted.
The mitotic process includes centrosome separation and maturation, chromatin condensation, nuclear envelope breakdown, Golgi disassembly, spindle formation, attachment of chromosomes to the mitotic spindle, and chromosome segregation (4-7). The timing and mechanics of each of these events are carefully regulated, because uncorrected errors committed during these processes often result in aneuploidy and genetic instability, and can lead to cancer (8, 9). Regulation of these dynamic processes involves the precise phosphorylation of mitotic effector proteins at specific sites through the actions of mitotic kinases, as well as other posttranslational events, such as ubiquitin-mediated proteolysis (6, 10). Major mitotic kinases include the complex of cyclin-dependent kinase 1 and cyclin B (Cdk1/cyclin B), the kinases Aurora A and Aurora B, Nek2 (never in mitosis kinase 2), and Plk1 (Polo-like kinase 1), all of which play key roles during various mitotic substages (6). Cdk1/cyclin B is a master regulator of mitosis with roles in chromatin condensation, nuclear envelope breakdown, and spindle formation (11-16), and its activation marks entry into mitosis. Aurora A is involved in centrosome separation and maturation; Aurora B is involved in chromosome condensation and assuring chromosome attachment to the spindle (amphitelic chromosome attachment) prior to the metaphase to anaphase transition (17-23). Nek2 is required for centrosome separation and plays a role in centrosome maturation (24, 25). Plk1 facilitates Cdk1/cyclin B activation, centrosome maturation, spindle formation, amphitelic chromosome attachment, and cytokinesis, and thus is involved in each of the different substages of mitosis (26-34).
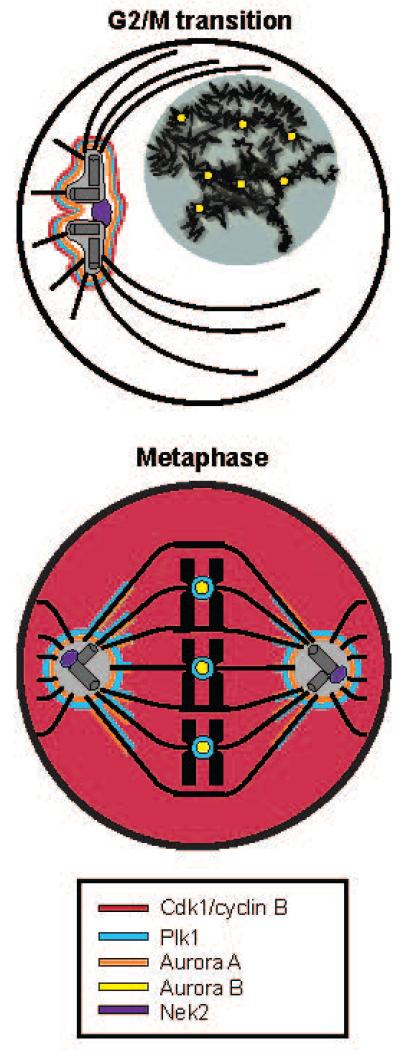
Because these kinases cooperate to regulate processes within mitosis, they have variously overlapping localizations (Fig. 1). At and just prior to the G2/M transition, Aurora A and Plk1 are localized to the pericentriolar material, while Nek2 localizes to the proximal centriole at the core of the centrosome and Aurora B localizes to the centromeres of the decondensed, but duplicated, chromosomes (24, 35-37). Cdk1/Cyclin B activity is low at this time, but increases rapidly as cells progress into prophase and prometaphase (28, 38). By metaphase, Cdk1/cyclin B is maximally active and diffusely localized throughout the cell. Intense activity of Cdk1/cyclin B, which first appeared on centrosomes during prophase, remains at the centrosomes, and also extends to spindle microtubules and kinetochores (11, 28, 38-40). Aurora A and Plk1 colocalize with Cdk1/Cyclin B to the pericentriolar material and the spindle microtubules early in mitosis (37, 41), whereas Plk1 later colocalizes with Aurora B and Cdk1/Cyclin B at the kinetochores during and after prometaphase (11, 35, 37).
Fig. 1. Major mitotic kinases have both discrete and overlapping subcellular localizations in mitotic cells.
Schematic representation showing the localizations of Cdk1/Cyclin B, Aurora A, Aurora B, Plk1, and Nek2 are indicated at the G2/M transition or early prophase (top) and in metaphase (bottom). In the top panel, a magnified representation of the centrosome with nucleated microtubules is shown, illustrating Nek2 localization near the proximal centrioles in the centrosomal core. The nucleus is shaded grey, containing duplicated but uncondensed chromosomes (black). In the bottom panel, condensed chromosomes are shown aligned on the metaphase plate, with their kinetochores indicated by central co-axial circles, attached to spindle microtubules.
Various substrates for these kinases are known, but more remain to be identified because the known substrates are not sufficient to explain all of the phenotypic events regulated by these kinases. In addition, despite overlapping subcellular localizations, and therefore access to overlapping sets of potential phosphorylation sites on substrates, each of these mitotic kinases appears to have distinct and apparently mutually exclusive substrate phosphoryation sites in vivo. How the activities of these kinases, along with their substrate targeting, are coordinated so that each kinase maintains a distinct set of phosphorylation sites is unclear. With Positional Scanning Oriented Peptide Library Screening (PS-OPLS) (42), we identified optimal motifs for Cdk1/cyclin B, Aurora A and B, Nek2, and Plk1 in order to shed light on the issue of substrate selection, as well as to identify previously unknown substrates of these kinases. Our analysis revealed both positively selected motifs and “anti-motifs”, which represented specific residues that were strictly selected against. Integration of the motif data with localization data suggested that these mitotic kinases exist in two functionally orthogonal spaces, a localization space and a motif space, such that the major mitotic kinases with overlapping localizations do not have overlapping motifs and major mitotic kinases with overlapping motifs do not have overlapping localizations.
Results
Comparison of the optimal consensus phosphorylation motifs of Cdk1/cyclin B and Plk1 identified with PS-OPLS
To investigate the role of sequence specificity in substrate selection by mitotic kinases, and to facilitate further substrate identification, we determined and compared the optimal consensus phosphorylation motifs of Cdk1/cyclin B and Plk1 (Fig. 2) using PS-OPLS (42), in which the kinase of interest is incubated individually in solution with each of 180 different peptide libraries. Each library contains a C-terminal biotin tag, a central Ser or Thr that acts as the phosphoacceptor, and a second fixed amino acid located at any of the residues from five before the phosphoacceptor site (Ser/Thr-5) to the residue four after the phosphoacceptor site (Ser/Thr+4) (fig. S1). At all other positions within this 10 amino acid window, a degenerate mixture containing all 20 naturally occurring amino acids except Cys, Ser, or Thr is present. The position and identity of the second fixed residue acts as the primary determinant for substrate phosphorylation.
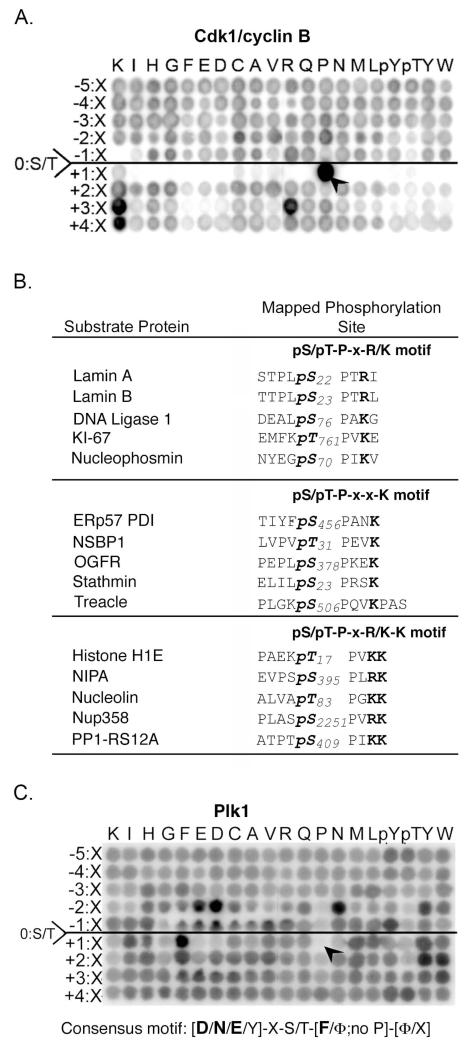
Fig. 2. Phosphorylation motifs for Cdk1/cyclin B and Plk1.
(A) The substrate motif for Cdk1/cyclin B reveals near exclusive selection for Pro in the Ser/Thr+1 position, strong selectivity for Arg and Lys in the Ser/Thr+3 position, and Lys in the Ser/Thr+4 position. (B) Comparison of the expanded Cdk1/cyclin B motif with mapped substrate sites (49). Bold represents residues that distinguish the submotifs. (C) The substrate motif for Plk1 reveals selection for Asp, Asn, or Glu in the Ser/Thr-2 position and hydrophobic amino acids (Φ) in the Ser/Thr+1 position, and discrimination against Pro in the Ser/Thr+1 position.
We performed experiments with Cdk1/cyclin B as a control, because the optimal phosphorylation motif for this kinase is generally accepted as Ser/Thr-Pro-X-Arg/Lys, where X is any amino acid, and the slashes represent “or” (43-47), and a large number of in vivo substrates are known (12, 48-54). Peptide library phosphorylation by Cdk1/cyclin B showed a near absolute requirement for Pro in the +1 position, as well as strong selection for Arg or Lys in the +3 position, as expected (Fig. 2A). We also found a strong positive selection for Lys in the +4 position and modest selection for Pro or Cys in the −2 position, along with weaker selection for other amino acids in this and other positions. On the basis of these results, we expanded the optimal Cdk1/cyclin B peptide phosphorylation motif to [P/C/X]-X-[S/T]-P-X-[R/K]-K, where bold indicates the most strongly selected residues.
To examine whether the amino acid preferences revealed by this peptide library screen are physiologically relevant, we compared our results with the reported collection of Cdk1/cyclin B substrates and phosphorylation sites determined by with an analog-sensitive human Cdk1 combined with thiophosphate tagging and covalent capture (49). Approximately 25% of Cdk1 substrates contained a Lys in the Ser/Thr+4 position, either alone (pS/pT-P-X-X-K motif) or together with an Arg/Lys in the Ser/Thr+3 position (pS/pT-P-X-R/K-K motif) (Fig. 2B). Many of the substrates also contained a Pro in the Ser/Thr-2 position, consistent our peptide library results. Thus, the PS-OPLS approach can reveal new motif information even for well-studied kinases like Cdk1.
The results of PS-OPLS for Plk1 (using the active phosphomimicking T210D form of the kinase) revealed a strong positive selection for Asp, Asn, or Glu in the Ser/Thr-2 position along with modest selection for aromatic residues, particularly Tyr (Fig. 2C). There was less amino acid selectivity in the Ser/Thr-1 position, although peptides containing either Gly or Pro in this position were poorly phosphorylated. In addition, Plk1 showed strong positive selection for hydrophobic amino acids in the Ser/Thr+1 position, particularly Phe, Tyr, Ile, and Met, as well as some additional hydrophobic selection in the Ser/Thr+2 position. The presence of a Pro residue in the Ser/Thr+1 position was very strongly discriminated against, as revealed by an almost complete absence of phosphorylation of this sub-library of peptides (Fig. 2C, arrowhead). These data suggest an optimal substrate phosphorylation motif for Plk1 of [D/N/E/Y]-X-[S/T]-[F/Φ; no P]-[Φ/X], where Φ is any hydrophobic amino acid.
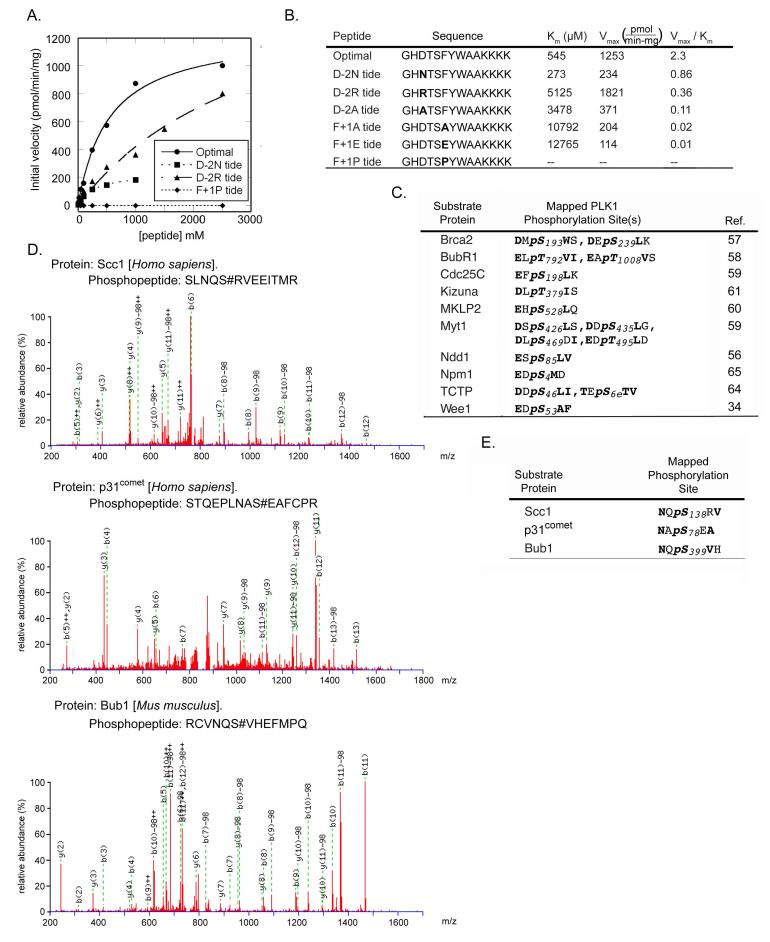
To validate the PS-OPLS results, we measured kinetic parameters for Plk1-dependent phosphorylation of an optimal peptide substrate based on the PS-OPLS consensus motif (GHDTSFYWAAYKKKK) and for several peptide variants containing single amino acid substitutions (Fig. 3A, B). In general, the Vmax/Km ratios of the optimal and variant peptides followed the same trends as those displayed in the PS-OPLS data. The optimal peptide had neither the lowest Km nor the highest Vmax, but had the highest Vmax/Km ratio, which is consistent with the peptide library screening approach identifying optimal kinase motif sequences on the basis of the maximal substrate turnover rate rather than on Km or Vmax alone (55). All variations from the optimal peptide displayed decreased Vmax/Km ratios indicating that the peptide chosen as the optimal substrate by individually selecting the best amino acid in each position was correct, and verifying that optimal phosphorylation motifs can be determined from PS-OPLS data by choosing the most highly selected amino acid in each position independently. Changing the Asp to an Ala in the Ser/Thr −2 position resulted in a greater than 20-fold drop in the Vmax/Km ratio (due to a 6.3-fold rise in Km, and a 3.3-fold drop in Vmax relative to those of the optimal peptide), and the ability of Plk1 to phosphorylate the peptide with a Pro in the Ser/Thr+1 was so minimal that it was not possible to fit the data to a Michaelis-Menten curve, recapitulating the lack of activity observed in the PS-OPLS screen for the sublibrary of peptides that all contained Pro in the Ser/Thr+1 position (Fig. 2C, arrowhead).
Fig. 3. Kinetics of Plk1 phosphorylation and identification of an expanded consensus motif.
(A) Kinetics of peptide phosphorylation by the Plk1 T210D kinase domain using an optimal substrate peptide and peptides with single amino acid substitutions at the indicated positions. Peptide sequences are shown in panel B. (B) Kinetic parameters (Km, Vmax, and Vmax/Km ratio) for the phosphorylation of the indicated peptides by Plk1 were determined by fitting the data to the Michaelis-Menten equation. Bold residues in the sequences represent the ones substituted. (C) The substrate specificity motif of Plk1 matches previously mapped Plk1 phosphorylation sites, which contain Asp or Glu in the Ser/Thr-2 and hydrophobic amino acids in the Ser/Thr+1 position. Bold represents motif-distinguishing residues. (D) Annotated MS/MS spectra for the phosphopeptides containing three Plk1 inhibitor-sensitive phosphosites containing Asn in the Ser/Thr-2 position. S# in the phosphopeptide sequence represents phosphoserine. (E) Sequence motifs of the sites mapped from the data shown in panel D.
The motif that we determined is consistent with previously mapped Plk1 phosphorylation sites on known substrates (Fig. 3C) (34, 56-65). Most of these Plk1 phosphorylation sites contain an Asp or Glu in the Ser/Thr-2 position and a hydrophobic amino acid in the Ser/Thr+1 position, in agreement with the consensus motif revealed by the PS-OPLS method. Hydrophobic amino acids in the Ser/Thr+2 position were also occasionally observed, but did not appear to be a strong discriminator for known in vivo Plk1 sites. The PS-OPLS-derived motif is similar to a previously published motif for Plk1, D/E-X-S/T-Φ-X-D/E, which was determined by mutagenic analysis of a single peptide sequence surrounding Ser198 in Cdc25C, which is phosphorylated by Plk1 (59). The PS-OPLS-derived motif differs from the other motif by having a strong selection for Asn in the Ser/Thr-2 position and no general selectivity for Asp or Glu in the Ser/Thr+3 position.
This Plk1 phosphorylation motif revealed by PS-OPLS screening suggested the possible existence of previously unrecognized Plk1 substrates with phosphorylation sites containing Asn in the Ser/Thr-2 position. Although no such Plk1 substrates matching this motif have been identified in higher eukaryotes, an in vivo study of yeast mitotic cohesins revealed eleven Cdc5 (the yeast Plk1 ortholog) sites on Rec8, of which 5 contained Asn in the Ser/Thr-2 position (66). To directly explore whether a subset of mitotic Plk1 substrates in mammalian cells contained a similar N-X-S/T motif, we performed a mass-spectrometry based screen in nocodazole-arrested HeLa cells for phosphorylated mitotic proteins containing Asn in the pSer/pThr-2 position that were not present in the nocodazole-treated cells if they were pre-incubated with the Plk1 inhibitor BI 4834 (fig. S2).
We identified three Plk1-dependent phosphorylation sites containing an Asn in the Ser/Thr-2 position (Fig. 3D, E). Only the nonphosphorylated forms of these same peptides, but not the phosphorylated forms, were observed following Plk1 inhibition. Two of the proteins containing these Plk1 sites, Bub1 and p31comet, are involved in spindle assembly checkpoint and the third, Scc1 (also known as Rad21), is involved in sister chromatid cohesion in mitosis (23, 67, 68), processes that are controlled by Plk1 (69, 70).
Determination of the optimal consensus motif of Aurora A and Aurora B
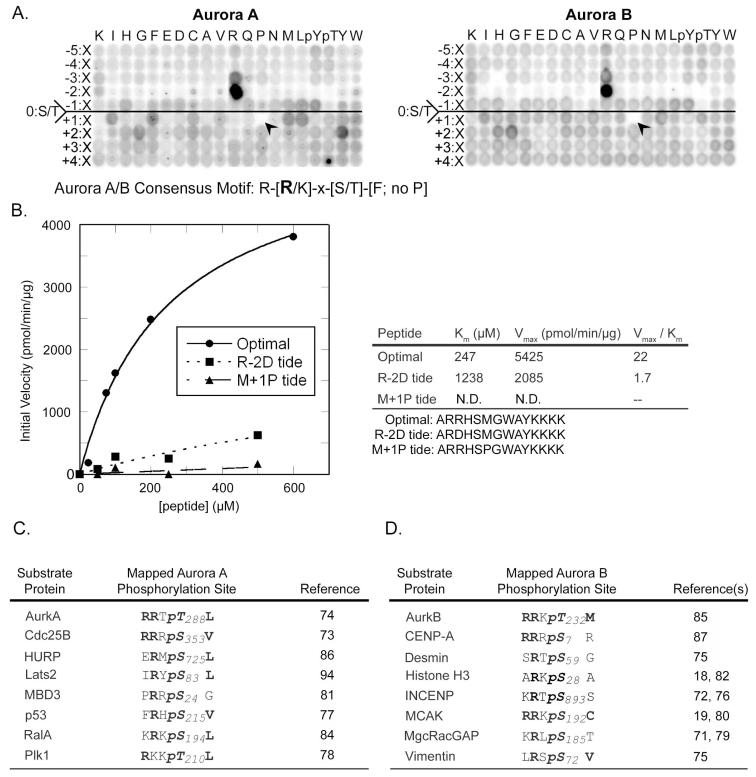
We used the PS-OPLS method to determine the optimal sequence motifs phosphorylated by Aurora A and Aurora B, and compared the results with those obtained for Cdk1/cyclin B and Plk1. Both Aurora A and Aurora B showed similar motifs, with extremely strong selection for Arg in the Ser/Thr-2 position and strong discrimination against all other amino acids at this position (Fig. 4A). Even Lys in the Ser/Thr-2 position was a poor substitute for Arg. There is a smaller selectivity for Arg in the Ser/Thr-3 position. In the Ser/Thr+1 position, although both Aurora A and B selected hydrophobic amino acids, the specific set of preferred amino acids differed slightly between them. Aurora B showed some preference for Ile and Met in this position, whereas residues preferred by Aurora A also included Phe and Leu. Like Plk1, both Aurora A and Aurora B showed very strong discrimination against libraries containing Pro in the Ser/Thr+1 position (Fig. 4A, arrowheads). Moderate selection for certain aromatic or hydrophobic amino acids, along with Gly and Pro, was noted in the Ser/Thr+2 position for both kinases, with little selectivity for particular amino acids beyond the Ser/Thr-2 to Ser/Thr+2 positions for either kinase.
Fig. 4. Phosphorylation motif selectivity of Aurora A and B.
(A) PS-OPLS blots of Aurora A (left) and Aurora B:INCENP complex (right) reveal positively and negatively selected residues for motifs recognized by these kinases. The arrowheads identify the complete selectivity against Pro at position Ser/Thr+1. (B) Peptide phosphorylation and determination of kinetic parameters (Km, Vmax, and Vmax/Km ratio) for reactions of Aurora B:INCENP with the optimal substrate peptide and peptides with single amino acid substitutions. A graph of reaction data fitted to the Michaelis-Menten equation for peptides is indicated on the left, and a table of kinetic parameters is shown on the right. R-2D tide and M+1P tide sequences are indicated below the table with the substituted positions shown in bold. (C) Previously published phosphorylation sites on mapped substrates of Aurora A (left) and Aurora B (right) conform to the optimal phosphorylation motif determined by PS-OPLS. Bold represents motif selected residues.
The position of maximal substrate selectivity for both of the Aurora kinases revealed by PS-OPLS screening -- a strongly basic amino acid in the Ser/Thr-2 position -- is the opposite of the requirement for an acidic (or Asn) residue in the Ser/Thr-2 position that we observed for Plk1. We verified the PS-OPLS-predicted kinase selectivity by measuring the kinetics of Aurora B-dependent phosphorylation of an optimal PS-OPLS-determined peptide (ARRHSMGWAYKKK), along with two peptide variants in which the Arg in the Ser/Thr-2 position was changed to an Asp (rendering it a more Plk1-like substrate), or the Met in the Ser/Thr+1 position was changed to a Pro (rendering it a more Cdk1-like substrate) (Fig. 4B). Substitution of Asp for Arg in the Ser/Thr-2 position both increased the Km and decreased the Vmax, resulting in a 13-fold drop in the Vmax/Km ratio relative to the Aurora B optimal peptide. As with Plk1, the ability of Aurora B to phosphorylate an otherwise consensus peptide that contained Pro in the Ser/Thr+1 position was so low that it was not possible to fit the data to a Michaelis-Menten equation. From these data, the minimal optimal consensus phosphorylation sequence shared by both Aurora A and Aurora B is R-X-S/T-Φ, where X is any amino acid and Φ is any hydrophobic amino acid except Pro. These motifs are consistent with mapped Aurora A and Aurora B phosphorylation sites (Fig. 4C), nearly all of which contain an Arg in the Ser/Thr −2 position. Many of these previously mapped sites also contain a hydrophobic amino acid in the Ser/Thr+1 position, particularly those that are subtrates of Aurora A (18, 19, 71-87).
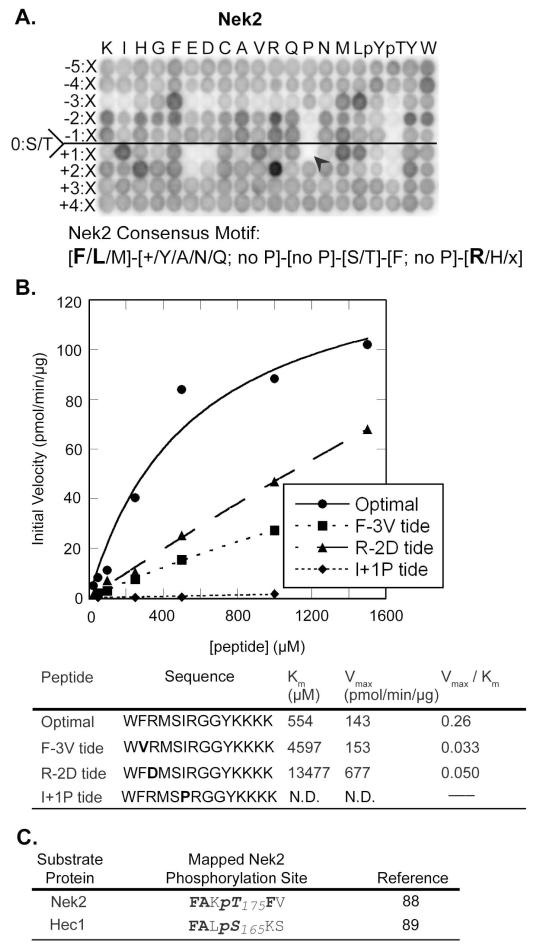
Determination of the optimal consensus motif of Nek2
In contrast to Plk1 and Aurora A and B, which discriminated against both Cdk1 sites by selection against Pro in the Ser/Thr+1 position, and against each other by selection for or against acidic or basic residues in the Ser/Thr-2 position, the Nek2 motif [determined for the active stabilized T175A form (88)] exhibited strong amino acid selectivity in the Ser/Thr-3 and Ser/Thr+2 positions (Fig. 5A). In the Ser/Thr-3 position, Nek2 selected for specific hydrophobic amino acids, particularly Phe, Met, and Leu, and strongly discriminated against hydrophilic amino acids. Both basic (Arg or Lys) and acidic (Asp or Glu) residues were disfavored in the Ser/Thr-3 position. Unlike Plk1 and Aurora A and B kinases, Nek2 did not show a strong and narrow preference for specific amino acids in the Ser/Thr-2 position, although Arg, Lys, Phe, Tyr, and Trp were somewhat favored. Pro was strongly disfavored in both the Ser/Thr-2 and Ser/Thr-1 positions. In the Ser/Thr+1 position, Nek2 also showed moderately strong selection for hydrophobic amino acids, along with strong discrimination against Pro, similar to the strong Pro deselection observed for Plk1 and Aurora A and B. Nek2 also showed strong discrimination against Asp and Glu in the Ser/Thr+1 position, as well as against Glu in the Ser/Thr+2 position, where instead we found pronounced selection for Arg, along with modest selection for His.
Fig. 5. Phosphorylation motif selectivity for Nek2.
(A) PS-OPLS screening reveals that Nek2 strongly selects a subset of hydrophobic amino acids in the Ser/Thr-3 and the Ser/Thr+1 positions and Arg in the Ser/Thr+2 position, but strongly discriminates against Pro in the Ser/Thr+1 position. Additional selection against acidic residues in the Ser/Thr-3, Ser/Thr+1 and +2 positions is also observed, as is positive selection for certain residues in the Ser/Thr-2 position. Within the indicated motif, + denotes basic residues, Φ denotes hydrophobic residues, and Ψ denotes aromatic residues. (B) Peptide phosphorylation and determination of kinetic parameters (Km, Vmax, and Vmax/Km ratio) for reactions of Nek2 with the optimal substrate peptide and peptides with single amino acid substitutions. A graph of reaction data fitted to the Michaelis-Menten equation for peptides is indicated on the top, and a table of kinetic parameters is shown on the bottom. F-3V tide, R-2D tide, and I+1P tide, sequences are indicated in the table with the substituted positions relative to the optimal peptide shown in bold. (C) Previously published mapped Nek2 phosphorylation sites are in reasonable agreement with the Nek2 PS-OPLS phosphorylation motif. Bold represents motif selected residues.
To verify the PS-OPLS data, we measured the kinetic parameters for the Nek2-dependent phosphorylation of its optimal PS-OPLS-determined peptide (WFRMSIRGGYKKK) along with those of three peptide variants containing single amino acid substitutions (Fig. 5B). Replacement of the strongly selected Phe in the Ser/Thr-3 position with Val, a similarly hydrophobic residue but one that was somewhat disfavored in the PS-OPLS screen, resulted in an approximately 8-fold decrease in the Vmax/Km ratio due primarily to an increase in Km relative to that of the optimal peptide. In contrast, complete charge reversal in the Ser/Thr-2 position of the Nek2 optimal peptide by substitution of Asp for Arg resulted in only a 5-fold decrease in the Vmax/Km ratio due to compensating effects on Km and Vmax. As with Plk1 and Aurora B, the kinase activity of Nek2 against an otherwise optimal peptide containing a Pro substitution in the Ser/Thr+1 position was minimal, precluding any determination of kinetic parameters.
These data reveal an optimal consensus phosphorylation motif for Nek2 as [F/L/M]-X′-X′′-S/T-Φ-[R/H/X], where both X′ and X′′ denote any amino acid except Pro, and X′ also includes some positive selection for basic and hydrophobic residues, Φ denotes any hydrophobic amino with Pro, Asp, and Glu excluded from this position, and X denotes any amino acid including Pro. The motif that we determined for Nek2 agrees reasonably well with the known phosphorylation sites that are present in the few well-verified Nek2 substrates, including Nek2 itself and the centrosomal and centromeric protein Hec1 (88, 89) (Fig. 5C).
An evolutionarily conserved structural basis for mitotic kinase motif exclusivity
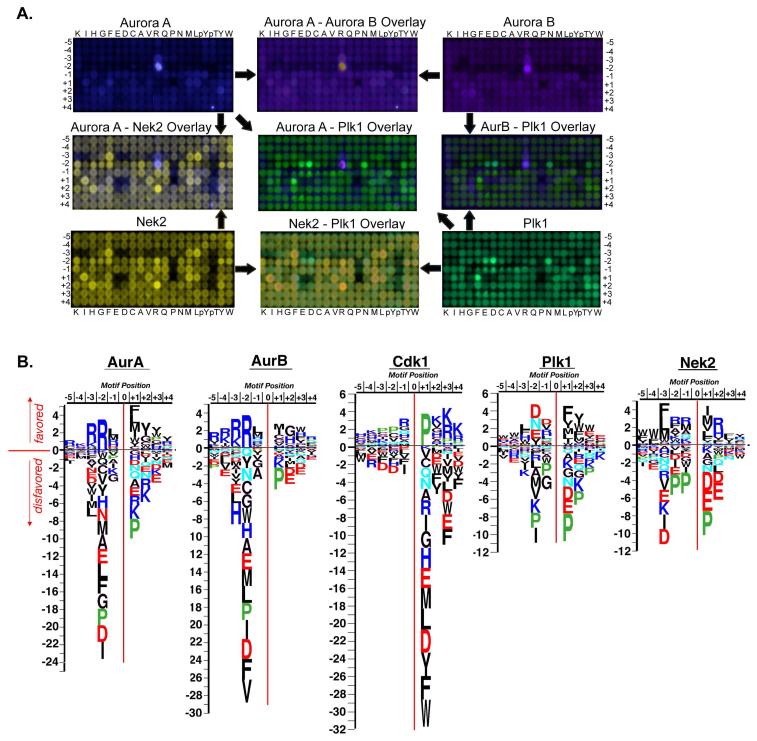
The mitotic kinase phosphorylation motifs identified with PS-OPLS suggested a mechanism for dictating mitotic protein kinase specificity. If the identity of the Ser/Thr+1 residue of the substrate site is a Pro residue, then the substrate is recognized by Cdk1/cyclin B, and not by Plk1, Aurora A, Aurora B, or Nek2. The presence of amino acids other than Pro, particularly hydrophobic ones, in the Ser/Thr +1 position determines whether phosphorylation is performed by Plk1, Aurora A or B, or Nek2. This critical Ser/Thr+1 specificity determinant prevents Cdk1 from phosphorylating putative Plk1, Aurora A, Aurora B, or Nek2 sites and likewise prevents Plk1, Aurora A, Aurora B and Nek2 from phosphorylating Cdk1 sites. However, additional criteria, must dictate whether the non-Cdk1 sites are preferentially phosphorylated by Plk1, one of the Aurora kinases, or Nek2. A comparison of the PS-OPLS blots and motif logos for Plk1, Aurora A, Aurora B, and Nek2 suggested that additional substrate-kinase specificity for mitotic kinases appears to arise from mutual exclusivity of the different mitotic kinase motifs themselves (Fig. 6A, B). This mutual exclusivity means that the strongest specificity-determining residues for one mitotic kinase were strongly deselected by the others, particularly in the case of Plk1 and Aurora A or Aurora B.
Fig. 6. Exclusivity of phosphorylation motifs for major mitotic kinases.
(A) PS-OPLS blots for Plk1, Aurora A, Aurora B, and Nek2 are pseudocolored and superimposed. Note the near identity of the Aurora A and B motifs in the merged view, and the distinct lack of superposition of the Aurora A/B and Plk1 motifs, as revealed by the largely distinct green and purple spots. The Nek2 motif shows substantial overlap with that of Plk1 and Aurora at multiple positions. (B) Spot intensities from the PS-OPLS blots were quantified and used to generate motif logos for each of the major mitotic kinases. Basic residues are colored blue, acidic residues red, amide side chain-containing residues cyan, Pro green, and the remaining residues in black. AurA, Aurora A; AurB, Aurora B.
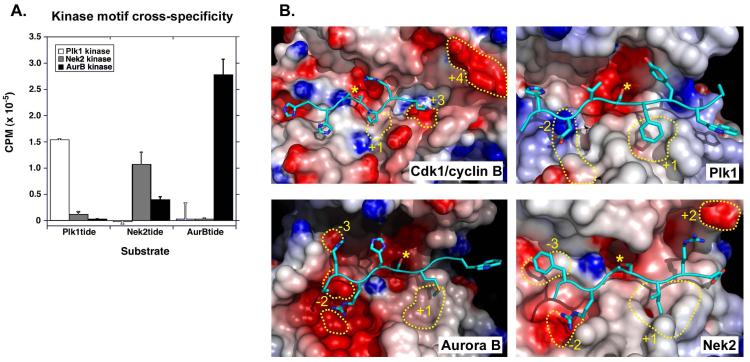
We compared the ability of Aurora B, Nek2, and Plk1 to phosphorylate a common set of peptides, each of which had been optimized for a particular family member (Fig. 7A). Each kinase phosphorylated its optimal peptide the best. Neither Aurora B nor Nek2 displayed much activity against the optimal Plk1 peptide, producing <1% and ~8%, respectively, of the phosphorylation displayed by Plk1 in this assay. Similarly, neither Plk1 nor Nek2 had much activity against the optimal Aurora B peptide, resulting in ~1% or less of the phosphorylation displayed by Aurora B. In contrast, although Plk1 had essentially no activity for the optimal Nek2 peptide (<1%), Aurora B did phosphorylate the Nek2-optimized peptide (~23% of the amount observed for Nek2 kinase itself).
Fig. 7. Structural basis for kinase motif selectivity and exclusivity.
(A) Optimal peptides for Plk1 (Plk1tide), Nek2 (Nek2tide), and Aurora B (AurBtide) were used as substrates in phosphorylation reactions containing the indicated kinases. The amount of radioactivity incorporated into each peptide after a one-hour incubation is indicated, showing mean values and standard deviations from triplicate measurements. (B) Modeled structures of kinases bound to optimal substrates (Cdk1/cyclin B:HHASPRK; Plk1:HDTSFYWA; Aurora B:RRHSMGW; Nek2:FRASIR). The molecular surfaces corresponding to the active site regions for each kinase are shown with red and blue regions indicating negative and positive electrostatic potentials, respectively. Active site features important for substrate selectivity are circled with yellow dotted lines and numbered relative to the position of the phospho-acceptor. Peptide substrates are shown with cartoon rendering for the backbone and stick rendering for relevant side chains. The phosphoacceptor Ser residues are indicated with yellow asterisks. For Cdk1, the +1 region corresponds to Val164 of Cdk1, the +3 region corresponds to pThr160 of Cdk1, and the +4 region corresponds to Glu265 and Asp268 of cyclin B. For Plk1, the −2 regions correspond to Lys178 and Asn216, and the +1 region corresponds to Leu211, Pro215, Ile218 and Val222. For Aurora B, the −3 region corresponds to Glu177, the −2 regions correspond to Glu220 and Glu281, and the +1 region corresponds to Trp237, Met249 and Leu256. For Nek2, the −3 region corresponds to Ala95 and Ala145, the −2 region corresponds to Glu208, and the +1 region corresponds to Phe176, Pro180 and Met183.
To examine the molecular basis for this motif exclusivity, we performed molecular modeling studies of kinase-substrate complexes (Fig. 7B) based on published X-ray crystal structures (see Materials and Methods for details). Despite the near absolute requirement for a Ser/Thr-Pro motif for substrate phosphorylation by Cdk1, we did not find any structural evidence in the Cdk1/cyclin B model for a direct interaction between the cyclin or Cdk subunits and the Ser/Thr+1 Pro residue in the substrate peptide (Fig. 7B), which is consistent with the reported Cdk2/cyclin A:peptide crystal structure (90). Instead, the strong selection for Pro observed in this position likely results from a combination of relatively weak effects. The backbone conformation of Val164 in the activation loop prevents hydrogen bonding between its carbonyl oxygen and the NH group of any amino acid (but would allow Pro to interact because it lacks an NH) in the Ser/Thr+1 position as described for the Cdk2/cyclin A structure (90). This conformation of Val164, combined with the need to correctly position the phosphoacceptor Ser/Thr residue near the end of the peptide-binding cleft, while simultaneously avoiding a steric clash of the remaining C-terminal residues of the peptide substrate with the cyclin subunit, means that the kink in the substrate main chain backbone resulting from the Ser/Thr+1 Pro would facilitate favorable interactions of basic side chains in the Ser/Thr+3 and Ser/Thr+4 positions with the cyclin and Cdk subunits. The peptide Ser/Thr+3 Lys in the Cdk2/cyclin A:peptide structure made a salt bridge to the regulatory phosphothreonine (pThr160) in the activation loop of the kinase domain, as well as a hydrogen bond to the main chain carbonyl of Ile270 on cyclin A; we expect that equivalent residues in the Cdk1/cyclin model (pThr161 in Cdk1 and the main chain carbonyl of Met261 in cyclin B1) are likely responsible for the observed Ser/Thr +3 Arg or Lys selectivity. The selection that we observed for Lys in the Ser/Thr+4 position of the substrate peptides appears to arise from interactions with acidic side chains of residues Glu265 and Asp268 in cyclin B, rather than from any direct contact with residues in the substrate-binding cleft of the kinase itself.
For Plk1, selection for Asp, Asn, and Glu in the Ser/Thr-2 position of the optimal substrate motif likely results from electrostatic or hydrogen-bonding interactions with the side chains of Lys178 and Asn216, or a combination of these kinds of interactions (Fig. 7B). This interaction model is supported by the observation that mutation of the Ser/Thr-2 residue to one incapable of forming either type of interaction (for example, Ala) increased the Km by 6-fold, whereas replacement with a positively charged residue (for example, Arg) increased the Km nearly 10-fold relative to the optimal Plk1 peptide. Hydrogen-bonding interactions may dominate over electrostatic interactions for favorable substrate selection, because substitution of Asn for Asp at position Ser/Thr-2 lowered the peptide Km by half, but decreased the Vmax by greater than a factor of 5 relative to the optimal Plk1 peptide, suggesting that the high degree of negative charge on the Asp-containing peptide following phosphorylation contributes to its dissociation from the catalytic cleft. Both Lys178 and Asn216 are conserved in the Plk1 homologs of vertebrates, worms, flies, and yeast, implying evolutionary conservation of motif selection for Asp, Asn, and Glu, and strong deselection for Arg or Lys, in the Ser/Thr-2 position of potential substrates (fig. S3). Phe, along with other aromatic or bulky hydrophobic amino acids that were selected in the Ser/Thr+1 position in the PS-OPLS screening can be accommodated in a hydrophobic pocket formed by Leu211, Pro215, Ile218, and Val222 in the activation loop. Substitution of small or negatively charged amino acids in this position increased the Km by a factor of 20 or greater relative to the optimal Plk1 peptide. The residues that comprise this hydrophobic pocket are similarly conserved in the Plk1 homologs of other species, although the Saccharomyces cerevesiae and Saccharomyces pombe homologs have an Ile replacement for Leu211, and in fission yeast and Drosophila melanogaster there is an Ile in place of Val222. Furthermore, strong deselection for Pro in the Ser/Thr+1 position of the motif would direct the substrate peptide chain outside of the kinase’s substrate-binding cleft and preclude favorable interactions with any additional C-terminal residues, because unlike kinases of the Cdk family, which function with a cyclin partner, Plk1 does not function in complex with a binding partner that could restore these contacts.
In the case of Aurora B, the weak selection for Arg that is observed in the Ser/Thr-3 position probably arises from an electrostatic interaction with Glu177, whereas very strong selection for Arg in the Ser/Thr-2 is likely due to an electrostatic interaction with Glu220 and Glu281. All three of these Glu residues form an extended acidic patch in the kinase active site, and all are conserved in Aurora kinase sequences from yeast to man (fig. S4), implying that selection for basic residues and exclusion of acidic residue in the Ser/Thr-3 and Ser/Thr-2 positions of the substrate motif is an evolutionarily important feature. Consistent with this model, replacement of the Ser/Thr-2 Arg by an Asp resulted in a 5-fold increase in Km and over a two-fold drop in Vmax relative to the optimal Aurora B peptide. Met and other hydrophobic amino acids selected in the Ser/Thr+1 position appear to fit into a hydrophobic pocket formed by Trp237 and Met249 on the activation loop and Leu256 of the active site cleft. These residues are conserved in vertebrate, worm, fly, and yeast Aurora kinases, although Met249 is replaced by Leu in D. melanogaster and S. pombe and by Val in S. cerevesiae. Thus, despite similar selection for hydrophobic residues in the Ser/Thr+1 position, evolutionary conservation of the acidic and basic Ser/Thr-2 selection pockets may ensure substrate exclusivity between Plk1 and the Aurora kinases.
For Nek2, the selection for Phe and other hydrophobic amino acids in the Ser/Thr-3 position likely arises from the presence of a spatially flat hydrophobic surface formed by two alanines, Ala95 and Ala145. Small residues at this location are found in all Nek2 homologs, but are rarely found in this part of the substrate-binding cleft in the other mitotic kinases (see Plk1 and Aurora B in Fig. 7B) or in PKA, where these positions are occupied by Phe and Glu. Selection for Arg in the Ser/Thr-2 position of the motif likely arises from an electrostatic interaction with Glu208, and the selected Ile and other hydrophobic amino acids in the Ser/Thr+1 position of the peptide likely fit into a hydrophobic pocket formed by Phe176 in the activation loop and Pro180 and Met183 of the active site. The Arg and His selection that we observed in the Ser/Thr+2 position of the substrate motif may arise from electrostatic interactions with Glu48. The identity of these specificity-determining residues is also conserved among mammals, zebrafish, frog, and yeast Nek2 family members (fig. S5).
Substrate enrichment, colocalization, and motif exclusion--A unified model for mitotic kinase selectivity
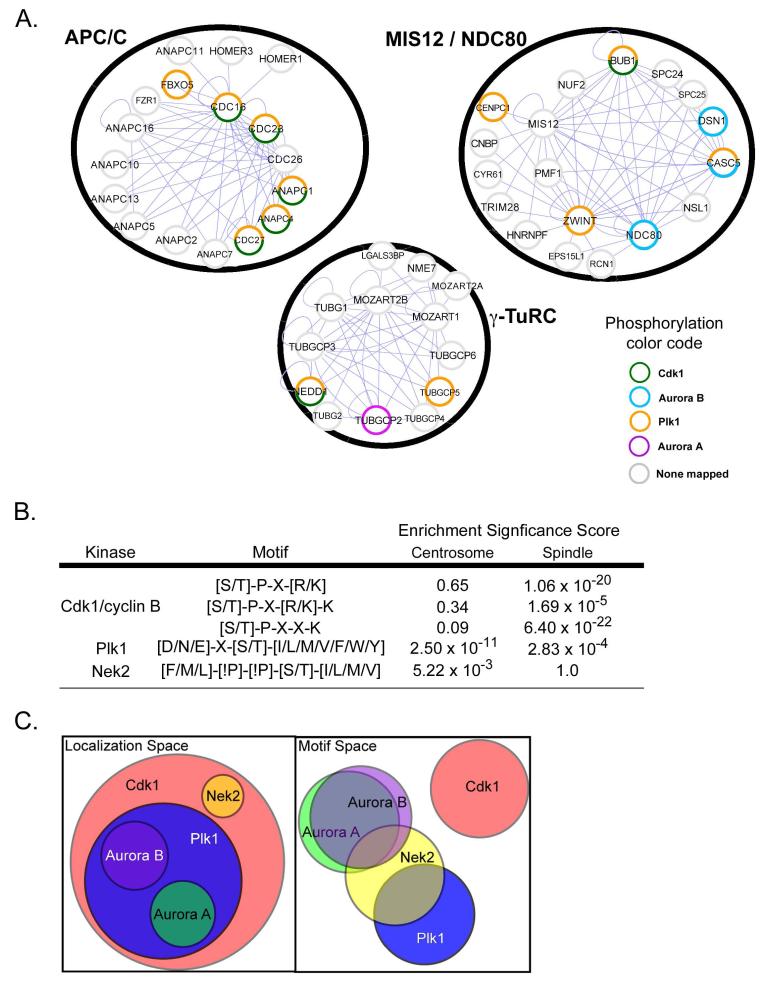
The modeling results suggested that the structural elements responsible for selection or exclusion of specific amino acids within mitotic kinase phosphorylation motifs reflect evolutionarily conserved features that may be critically important in substrate selection during cell division, when multiprotein mitotic complexes undergo phosphorylation at exclusive sites by the cooperative action of multiple distinct kinases. Figure 8A, for example, shows three representative mitotic protein complexes that were experimentally identified in a systematic mass-spectrometry screen (91), with proteins in each complex annotated for previously mapped mitotic phosphorylation sites (17, 92-100). These complexes comprise the γ-tubulin ring complex (γ-TuRC) important for spindle assembly, the anaphase-promoting complex (APC/C) important for chromosome segregation, and the outer kinetochore complex MIS12/NDC80 important for binding kinetochores to spindle microtubules (101). In each case, multiple proteins within each mitotic complex are targeted by one or more different mitotic kinases.
Fig. 8. Orthogonal mitotic kinase motif and localization ‘spaces’ provide substrate site specificity.
(A) A selection of mitotic protein complexes identified by Hutchins et al. (122) are shown within black circles and annotated based on the literature for the presence of mapped mitotic phosphorylation sites by the indicated kinases. Note that multiple mitotic kinases phosphorylate discrete sites within single complexes. Blue lines indicate direct intra-cluster interactions. (B) Subproteomes associated with the spindle and centrosome were scored for the occurrence of the Cdk1, Nek2 and Plk1 motifs and compared to the expected number of sites at those locations based on the entire proteome. The enrichment significance score indicates the statistical significance of overenrichment based on a hypergeometric probability distribution. (C) Mitotic kinase functionality is represented by Venn diagrams of ‘localization space’ and ‘motif space’. In ‘localization space’, each circle represents the total subcellular locales available to that kinase. In ‘motif space’, each circle represents the sequences that can be phosphorylated by the kinase. In this representation, a major mitotic kinase can overlap with every other kinase in at most one of these two spaces.
During cell division, the localization of Cdk1, Plk1 and the Aurora A and B kinases overlap at specific mitotic substructures, particularly along the outer portions of the centrosomes, at the kinetochores, and along the mitotic spindle. This contrasts with Nek2, which localizes to the proximal centriole within the core of the centrosome (24) (Fig. 1). To examine whether substrates for these kinases might be enriched at the subcellular structures where these kinases reside, we performed a bioinformatic analysis for enrichment of potential phosphorylation sites for Cdk1, Plk1, Nek2, and Aurora kinases on the subproteome found at the centrosome (102) and the spindle apparatus (103), relative to the entire human proteome (International Protein Index-version 3.23) (see Materials and Methods for details). All three variations of the Cdk1/cyclin B motif revealed by PS-OPLS were statistically over-represented in proteins found on the mitotic spindle and none of the Cdk1 motifs were over-represented on the centrosome subproteome (Fig. 8B). The Plk1 motif defined by PS-OPLS was significantly enriched on proteins at both the centrosome and spindle apparatus, whereas the Nek2 motif was enriched on proteins at the centrosome, but not on the spindle subproteome, as might be expected for a centrosomal kinase. The Aurora motif was not significantly enriched at the centrosome or the spindle, which may not be surprising because there are many basophilic kinases with similar motifs, for example, PKA and protein kinase G (PKG), that control processes other than mitosis. The enrichment of potential substrates at subcellular structures where Cdk1, Plk1 and Nek2 localize during mitosis supports the intuitive idea that substrates are likely to undergo extensive phosphorylation in regions of the cell where the local concentration of the kinase is highest.
Whereas motifs for individual kinases appeared to be significantly enriched in the subcellular proteomes appropriate to those kinases, we wondered about the prevalence of co-occurence of motifs for multiple kinases in the same protein. We found that in both the entire proteome and the nuclear subproteome, the co-occurrence of motifs for all pairs of mitotic kinases was significantly enriched (see Materials and Methods, table S2), indicating perhaps that different sites on the same protein are commonly phosphorylated by distinct kinases in the normal course of mitosis. Because the sites phosphorylated by Cdk1, Plk1, and the different Aurora kinases are mutually exclusive, this observation is consistent with the co-localization of multiple mitotic kinases at centrosomes, kinetochores, or along the mitotic spindle, or various combinations thereof. For Nek2, motif co-occurrence suggests that some substrates may move from the inner centrosome to other mitotic structures in order to be phosphorylated by the appropriate kinase at the appropriate time.
Discussion
Mitosis is an intricate and highly regulated process in which the temporal order of events is controlled by the action of specific protein kinases, including the master regulator Cdk1 and the other major mitotic kinases, Aurora A, Aurora B, Nek2, and Plk1. Proper progression through mitosis requires that these kinases phosphorylate specific sites on specific proteins in an organized and presumably non-overlapping manner. We used PS-OPLS, which in contrast to studies using amino acid substitutions within a single peptide corresponding to a known substrate phosphorylation site, allows every amino acid within each position in a sequence motif to be tested independently of the rest of the sequence. The method therefore identifies not only amino acids that are positively selected in each position, but also those that are disfavored or deselected in each position. Subsequent kinetic studies with individual peptides verified the PS-OPLS motifs, and provided insights into the relative role of flanking residues in peptide binding or turnover, or both, at the kinase active site.
The Cdk1/cyclin B motif revealed by PS-OPLS expands upon the generally accepted S/T-P-X-R/K consensus motif to now include a Lys in the Ser/Thr+4 position. This expanded motif explains ~25% of the reported Cdk1 substrates detected with a chemical genetics screen for Cdk1/cyclin B in HeLa cell extracts (57). A mass spectrometry-based screen for mitotic phosphoproteins that used a weighted bioinformatics analysis to determine abundant mitotic phosphorylation motifs showed many phosphorylation sites containing the sequence S/T-P-X-X-K, leading the authors to propose that this constituted a substrate motif for a new mitotic kinase that had not yet been identified (104). Our PS-OPLS results suggest that this motif matches that of human Cdk1/cyclin B.
Our structural modeling results for Cdk1/cyclin B suggested that selection for Lys in the Ser/Thr+4 position in the motif arises from a pair of acidic residues within cyclin B both of which are conserved in mammals, worms, and frogs, and one of which is conserved in flies. No such acidic residues, however, are found in the corresponding positions of Clb2, the budding yeast mitotic cyclin, suggesting that selection for basic residues in this position of the phosphorylation motif is unique to metazoans. A mass spectrometry screen of in vivo Cdk1/Clb2 substrates did not reveal enrichment for Lys in the Ser/Thr+4 position of the mapped phosphorylation sites in substrates from S. cerevesiae (105), which contrasts with what was observed for Cdk1/cyclin B substrates in human cells (49).
The Plk1 motif identified by PS-OPLS revealed strong selectivity for Asn, in addition to Asp and Glu, in the Ser/Thr-2 position. We identified phosphopeptides containing Asn in the Ser/Thr-2 position from three proteins, Scc1, Bub1, and p31comet, for which phosphorylation in nocodozole-treated cells was abrogated by Plk1 inhibition. Scc1 is a subunit of the cohesin complex that prevents chromosome segregation until it is cleaved by separase at the metaphase-anaphase transition (106). Although Scc1 is phosphorylated at multiple sites by Plk1, which enhances the cleavage of Scc1 by separase, all the phosphorylation sites had not been mapped (106). The location of the phosphorylation site at Ser138 that we found is near one of the separase cleavage sites and may increase Scc1 cleavage. Phosphorylation of Bub1 by Cdk1 recruits Plk1 to kinetochores in vivo and enhances Plk1 phosphorylation of Bub1 in vitro, although the Plk1 phosphorylation sites on Bub1 were not known (97). We identified Ser399 as a Plk1-dependent Bub1 phosphorylation site in vivo that matches the Asn-containing Plk1 motif, although we do not know if this site (or those on Scc1 and p31comet) is a direct or indirect Plk1 target. Maintenance of the spindle checkpoint until attachment of all of the chromosome kinetochores involves an interaction between Mad2 and Cdc20 (107). p31comet binds Mad2 causing Mad2 to release Cdc20 (108). Although Plk1 is required for the recruitment of Mad2 to the kinetochore for the spindle assembly checkpoint, the Plk1-dependent phosphorylation of p31comet suggests Plk1 may have other roles as well (26). A mass-spectrometry screen for Plk1 substrates on the early mitotic spindle identified many spindle-associated proteins with Plk1-dependent phosphorylation sites containing Asn in the Ser/Thr-2 position, providing independent validation of our PS-OPLS results (98).
The strong similarity in optimal phosphorylation motifs that we observed between Aurora A and B is not surprising because these kinases are similar in their primary sequences, and the functions of both kinases are performed by a single kinase in yeast (109). PS-OPLS motif determination for Aurora A and B suggested that Arg in the Ser/Thr-2 position is likely to be an important determinant for substrate selection, and that Lys is a poor substitute for Arg despite their similarity in charge. Although this finding agrees well with most mapped Aurora A and B substrates (Fig. 4C, D), some mapped sites contain a Lys in the Ser/Thr-2 position, including the regulatory Thr210 site on the activation loop of Plk1 that is phosphorylated by Aurora A during mitotic entry (78, 110). Phosphorylation of Thr210 by Aurora A during release from a DNA damage checkpoint requires the formation of a complex between Aurora A and the G2/M regulatory protein hBora (78, 110), suggesting that a targeting subunit (hBora), rather than intrinsic kinase selectivity, may be important for phosphorylation of this particular site.
The Aurora A motif that we determined is consistent with previous reports (74, 111). Both of those studies revealed the importance of an Arg residue in the Ser/Thr-2 position, and the Ser/Thr+1 hydrophobic residue within single peptide substrates, as well as the strong negative selectivity for a Pro residue in the Ser/Thr+1 position. The optimal phosphorylation motif revealed by PS-OPLS screening of Aurora B has some differences from the consensus motif [R/K]-X-[S/T]-[I/L/V] derived from ten phosphorylation sites on kinetochore proteins that are phosphorylated by the yeast Aurora ortholog Ipl1 (109) and from a K-R-S-[S/T]-S motif mapped in the evolutionarily conserved Aurora B substrate INCENP (72, 76), likely because these prior motifs were based on a small number of phosphorylated peptides rather than an global assessment of site preferences provided by PS-OPLS.
The motif that we identified for Nek2 is the first reported and revealed strong negative selection against Pro in the Ser/Thr+1 position, similar to what we observed for Plk1 and the Aurora kinases, indicating that Nek2 will not phosphorylate sites targeted by Cdk1 and vice versa. Direct comparison of the Plk1, Aurora A, Aurora B, and Nek2 motifs (Fig. 6A, B) revealed strong mutual exclusivity of Aurora A and Plk1 substrates on the basis of amino acid residues in the Ser/Thr-2 position. In contrast, the strongest positive selection within the Nek2 motif was in the Ser/Thr-3 and Ser/Thr+2 positions. The optimal peptide for Nek2 contained an Arg in the Ser/Thr-2 position, which was also present in the Aurora kinase motif, and optimal Nek2 peptides were phosphorylated by Aurora B (Fig. 7A). Nek2 may also phosphorylate some of the same sequences as Plk1, if such sequences had hydrophobic amino acids in the Ser/Thr-3 position to satisfy the Nek2 motif. However, the optimal Plk1 peptide, which contained a His in this position, was a poor Nek2 substrate in vitro.
The positions in the Nek2 motif showing the strongest negative selection, aside from Ser/Thr+1 Pro, were acidic residues and phosphoThr residues in the Ser/Thr-3, +1, and +2 positions, suggesting that Nek2 phosphorylation sites should be mutually exclusive with sites phosphorylated by casein kinase 1 (CK1), which shows a strong selection for acidic or phosphoSer/Thr residues in the Ser/Thr-3 position of its substrates (112). The association of CK1δ with centrioles at the centrosomal core is mediated by the scaffolding protein AKAP450 (113, 114) and we predict that motif exclusivity between Nek2 and CK1δ may establish distinct phoshorylation sites for these kinases on centriolar substrates.
For the mitotic kinases that we studied, the motif data and structural modeling results suggest an evolutionarily conserved mode of mitotic kinase substrate targeting in which the presence or absence of a Pro in the Ser/Thr+1 position functions as a binary switch allowing phosphorylation of a site by either Plk1, Aurora A, Aurora B, or Nek2 if a residue other than proline is present in this position, or by Cdk1 if a Pro is present. This proline specificity has been suggested as a mode of regulation between Cdk1 and Aurora A and between the CMGC proline-directed kinase group, of which Cdk1 is a member, and the basophilic CAMK and AGC kinase groups, to which Aurora A and Aurora B are closely related but are not members (74, 115, 116).
Integrating the motif and colocalization data suggests that specificity of phosphorylation by these kinases is enforced by a combination of non-overlapping localizations and both positively and negatively selected sequence motifs. Each kinase possesses a positively selected motif, and also displays an ‘anti-motif’ whereby the motifs of other kinases are specifically disfavored. Plk1 and the Aurora kinases do not phosphorylate the same sites despite overlapping localizations because their motifs are mutually exclusive. Despite partially overlapping motifs, Nek2 and Plk1 and Nek2 and the Aurora kinases would not phosphorylate the same sites because of non-overlapping localizations, because Nek2 localizes to the proximal centrioles within the core of the centrosome, whereas Aurora A and Plk1 are localized at the periphery of the pericentriolar material (36), Thus, our data suggest that kinases exist in two functionally orthogonal spaces: ‘localization space’ and ‘motif space’ (Fig. 8C). ‘Localization space’ reflects all of the subcellular locales where the kinase can reside. ‘Motif space’ contains the optimal sequence motifs of all S/T kinases. In this context, each major mitotic kinase overlaps with every other kinase in, at most, one of space, either localization or motif space. For example, Cdk1 overlaps all of the other kinases in ‘localization space’, but not in ‘motif space’ and Nek2 overlaps Plk1 and Aurora Kinases in ‘motif space’, but not in localization spaces’. Thus, the combination of positive and negative amino acid motif selection and spatial exclusivity that we observe appears to underlie the cooperative nature of mitotic kinase signaling. This potentially provides both a systems-level mechanism for regulating substrate phosphorylation, and a coordinated evolutionary pressure to maintain discriminatory substrate motifs and localizations for major mitotic kinases and their substrates.
Materials and Methods
Kinase Protein Production and Purification
Recombinant full-length human wild-type C-terminally His6-tagged Aurora A, Xenopus laevis wild-type Aurora B (residues 60-361):INCENP (residues 790-847) complex, and full-length human T175A Nek2A proteins were purified from Escherichia coli as described previously (88, 117, 118). Nucleotide sequence encoding the kinase domain of human Plk1 (amino acids 38-346) was cloned as an N-terminally His6-tagged fusion construct into a modified version of the pET28a (Novagen) bacterial expression vector. The resulting fusion protein contains an N-terminal His6-tag followed by maltose binding protein (MBP) and a Tev protease cleavage site between MBP and the kinase domain. A Thr210 mutation to Asp was introduced with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) to increase specific activity. The protein was expressed in E. coli Rosetta 2 (Novagen) cells and purified by Ni-NTA affinity chromatography The kinase domain was further purified by affinity chromatography on amylose beads, cleaved with Tev protease, re-applied to Ni-NTA to remove the cleaved His6-MBP tag, and finally purified by gel filtration on a Superose 12 column. The Cdk1/Cyclin B kinase complex, which consisted of active human recombinant full-length C-terminally His6-tagged Cdk1 and glutathione-S-transferase (GST)-tagged human full-length cyclin B, was purchased from Millipore.
Phosphorylation Motif Determination by Peptide Library Array Screening
PS-OPLS was performed following Hutti et al. (42). Briefly, solution-phase kinase reactions were performed in parallel on 198 separate biotinylated, partially degenerate oriented peptide libraries (Anaspec, Inc) arrayed in a 384 well microtiter plate in a 20 row X 9 column format (fig. S1). Each peptide library contains a C-terminal biotin tag, a equal mixture of serine and threonine at the orienting phospho-acceptor residue, a single second fixed amino acid located between the Ser/Thr-5 and Ser/Thr+4 positions, and a mixture of amino acids at all other positions. Individual libraries contain any of the 18 non-Ser/Thr natural amino acids, as well as phosphothreonine or phosphotyrosine, in the second fixed position, corresponding to the 20 rows. Scanning down each column of the array moves the position of the fixed amino acid from Ser/Thr-5 to Ser/Thr+4 relative to the fixed phospho-acceptor residue. As an example, the peptide library with Lys fixed in the Ser/Thr-4 position has the following sequence: Y-A-X-K-X-X-X-S/T-X-X-X-X-A-G-K-K-biotin, where amino acids are represented in 1-letter code, and X is an equal mixture of all 17 natural amino acids excluding Cys, Ser, and Thr to prevent oxidation effects and eliminate secondary phosphorylation events. S/T denotes a 1:1 mixture of Ser and Thr. Kinase reactions were performed at 30°C in a total volume of 16 μL containing 31.25 μM peptide library, 100 μM ATP, and 200 μCi of [32P]-γ-ATP, in 150 mM NaCl (500 mM NaCl for Nek2 kinase reactions), 10 mM MgCl2, 1 mM DTT, 0.1% Tween 20, and 50 mM Tris, pH 7.5. For Aurora A and Aurora B, reactions were performed for 6 hours with 0.25 and 0.792 μg of protein per reaction, respectively. For Nek2, reactions were performed for 8 hours with 2.4 μg of protein per reaction. For Plk1, reactions were performed for 3 hours with 0.5 μg of protein per reaction. For Cdk1/Cyclin B, reactions were performed for 4 hours with 0.08 μg of protein complex per reaction. Following incubation, 2 μL of each reaction were simultaneously transferred to a Streptavidin-coated membrane (Promega SAM2 biotin capture membrane) using a 384-slot pin replicator (VP Scientific). The membrane was washed three times with 140 mM NaCl, 0.1% SDS, 10 mM Tris, pH 7.4, three times with 2 M NaCl, twice with 2 M NaCl containing 1% H3PO4, and once with water. The extent of peptide library phosphorylation was determined by imaging the membrane with a phosphorimager (Molecular Dynamics).
In Vitro Kinase Assays
Kinase assays for kinetic parameter determination were performed at 30°C in 90 μL of kinase reaction buffer (50 mM Tris, pH 7.5, 150 mM NaCl [500 mM NaCl for Nek2 assays], 10 mM MgCl2, 100 μM ATP, 9 μCi [32P]-γ-ATP, and 1 mM DTT). Each reaction contained 0.003 μg of Aurora B:INCENP complex, 3.6 μg of T175A Nek2 protein, or 0.054 μg of T210D Plk1 kinase domain. The sequences of the optimal substrate peptides for Aurora B, Nek2, and Plk1 were ARRHSMGWAYKKKK, WFRMSIRGGYKKKK, GHDTSFYWAAYKKKK, respectively. These optimal peptides were determined by taking the most highly selected amino acid from each position, Ser/Thr-4 to Ser/Thr+3, of the PS-OPLS blot and using Ser as the phospho-acceptor residue, at position 0. A C-terminal tyrosine was added to each peptide to allow determination of concentration of peptide solutions by UV spectrophotometry, and four C-terminal lysines were appended to increase solubility and electrostatic interaction with phosphocellulose paper. Additional peptides with single amino acid changes from the optimal peptides were as indicated in Figs. 3B, 4B, and 5B. Concentrations of peptides were as indicated. 5 μL of each reaction were spotted on phosphocellulose at 0, 3, 6, 9, 12, and 15 minutes in duplicate. The phosphocellulose paper was washed 4 times with 0.5% phosphoric acid, transferred to vials containing scintillation cocktail and counted. From these kinase assays, Km, Vmax, and Vmax/Km values were determined by curve fitting assuming Michaelis-Menten kinetics. For each concentration of peptide, care was taken to ensure that less than 5% of the total substrate was phosphorylated and that the reaction rate was linear with respect to time.
Kinase assays for comparison of activity of each kinase against the optimal peptides of each kinase were performed at 30°C in 25 μL of kinase reaction buffer. Each reaction contained 0.008 μg of Aurora B:INCENP complex, 1 μg of T175A Nek2 protein, or 0.046 μg of T210D Plk1 kinase domain. Amounts of each kinase were chosen so that reactions of each kinase with its optimal peptide would yield approximately equivalent amounts of 32P-radiolabel incorporation. Concentrations of optimal peptide were set to the Km of the kinase in the reaction. Reactions were performed in triplicate and 5 μL of each reaction was spotted on phosphocellulose at 0 minutes and 1 hour. The phosphocellulose paper was washed 4 times with 0.5% phosphoric acid, and analyzed by scintillation counting.
Mass Spectrometry Identification of Plk1-Dependent Mitotic Phosphorylation Sites containing Asn in the Ser/Thr-2 Position
HeLa cells were cultured in DMEM supplemented with 10% Fetal Bovine Serum (Gibco / Invitrogen), plus 0.2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Sigma-Aldrich). In some cases, cells were transfected with LAP-tagged murine bait proteins (Mad2l1 or Bub1) and selected with 500 μg/ml G418 (Gibco). Cells were arrested in mitosis by addition of 100 μg/ml nocodazole for 18 hr prior to harvesting. For mitotic cells in which the activity of endogenous Plk1 was inhibited, nocodazole-arrested cells were further incubated with 250 nM of BI4834 (a gift from Boehringer Ingelheim, Vienna, Austria) for an additional 2 hrs. Cell pellets were harvested, snap-frozen in liquid nitrogen and stored at −80°C. Thawed pellets were resuspended in one pellet-volume of extract buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl, 5 mM EDTA, 20 mM β-glycerophosphate, 10 mM NaF, 10% glycerol, and 0.1% NP-40) containing 1 μM okadaic acid, 0.2 mM NaVO4, 1 mM DTT, 0.1 mM PMSF and 10 μg/ml each of leupeptin, pepstatin and chymostatin, dounce homogenized, and clarified by centrifugation at 14,000 rpm for 15 min. Mitotic cohesin and kinetochore protein complexes were immunoprecipitated with antibodies against the endogenous cohesin regulator PDS5A or tandem affinity purified for LAP-tagged Mad2l1 and Bub1, respectively as described (91, 119). Following extensive washing, bound proteins were eluted using 0.2 M glycine pH 2.0, neutralized, digested in solution using trypsin or subtilisin, and analyzed by MS.
Digests were analysed using a UltiMate 3000 Nano-LC system (Dionex Benelux, Amsterdam, The Netherlands) equipped with a trap column for sample desalting and concentration. Samples were loaded onto an analytical C18 column (PepMap C18, 75 μm ID × 150 mm length, 3 μm particle size, 100 Å pore size, Dionex) for separation. Mobile phase A contained 5% acetonitrile and 0.1% formic acid, while mobile phase B contained 80% acetonitrile and 0.08% formic acid. Following a 10 min wash in 0.1% TFA, peptides were eluted using a linear gradient from 20% to 50% mobile phase B in 180 min at a flow rate of 300 nl/min. Mass spectrometric analyses were conducted on a hybrid linear ion trap/Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (LTQ-FT Ultra, Thermo Fisher Scientific) with a 7-Tesla superconducting magnet, equipped with a nano-electrospray ionization (ESI) source. Full-scan measurements (m/z range 400-1800) were conducted in the ICR cell, yielding a survey scan with resolution of 100,000 and a typical mass accuracy of <2 ppm. Collision-induced dissociation (CAD) fragmentation and spectrum acquisition were performed in the linear ion trap using the multi-stage activation (MSA) method of Schroeder et al. (120). MS RAW files were analysed by database searching using Mascot (121) for phosphopeptides containing Asn in the Ser/Thr-2 position. The following parameters were used for the database search: carboxymethylation (+58.0055 u) of cysteine was set as fixed and oxidation (+15.9949 u) of methionine and phosphorylation (+79.966331 u) as variable modifications. Mass tolerances of the parent ion and the fragments were set to 10 ppm and 0.80 Da, respectively. Each mass spectrum of an Asn-containing phosphopeptide identified by MASCOT was then manually inspected and annotated.
Structural Modeling
The X-ray crystal structures of Cdk2/cyclin B (PDB identifier 2JGZ), and the kinase domains of Aurora B in complex with an INCENP fragment (PDB identifier 2BFX), Nek2 (PDB identifier 2JAV), and human Plk1 (PDB identifier 2OU7) were used as base models. For the Plk1 model, the activation loop segment was modeled based on the corresponding region from the zebrafish crystal structure (PDB identifier 3D5W). For the Plk1 and Aurora B models, peptides were manually docked into the substrate binding cleft based on the structure of the substrate GSK3-beta peptide in the active site cleft of Akt (PDB identifier 106L). For the Cdk1 model, the structure of CyclinA/Cdk2 containing a substrate peptide in the active site cleft (PDB identifier 2CCI) was used for manually docking the peptide, whereas for Nek2, the peptide structure from the PKA catalytic subunit:AMP-AlF4−:substrate peptide co-complex was used for docking. Molecular surface representations of the Aurora B, Cdk1, Nek2, and Plk1 active sites were created using PyMOL and shaded by electrostatic potential using surface projections of charge calculated with DelPhi software (122).
Centrosome and Spindle Enrichment Bioinformatic Analysis
We obtained a list of proteins associated with the centrosome from Andersen et al. (102) and a list of spindle proteins from Sauer et al. (103). Sequences for these proteins were downloaded directly from NCBI Entrez Protein using NCBI Entrez Utilities (http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html). The human proteome was downloaded as version 3.23 of the file ipi.HUMAN.fasta from the International Protein Index (http://www.ebi.ac.uk/IPI/). We generated a list of the amino acid sequences surrounding each Ser or Thr residue within the sequences of each protein in each of these three data sources. The human proteome yielded 1,850,231 unique sites from 66,619 proteins. The centrosome data and spindle data yielded 35,425 unique sites from 524 proteins and 34,909 unique sites from 277 proteins, respectively. For each motif of interest (Fig. 8B), statistical significance of enrichment of that motif in the centrosomal or spindle proteomes relative to the full proteome was calculated using the hypergeometric distribution:
where N is the number of Ser/Thr sites in the human proteome, n is the number of Ser/Thr sites in the centrosomal or spindle proteome, m is the number of motif sites in the human proteome, and k is the number of motif sites in the centrosomal or spindle proteome. This corresponds to the probability of seeing as many instances or more of the motif by chance as are seen in the centrosomal or spindle proteome if a dataset of the same size as the centrosomal or spindle proteome is drawn at random from the human proteome.
Kinase Phosphorylation Motif Logos
Phosphorylation content of individual peptide libraries in the PS-OPLS blots was quantified by phosphorimage analysis using ImageQuant 5.2 software (Molecular Dynamics). Background correction was performed by subtracting an adjacent region of the blot of the same size that did not overlap with any of the phosphorylated library spot positions. Sequence logos were generated from the log2 values of the intensities of background corrected, normalized peptide library spots using POSTSCRIPT files generated by Visual Basic code adapted from Stephen Shaw’s original PSSM logo code (123), which contains POSTSCRIPT code adapted from Tom Schneider’s MAKELOGO (version 8.69, http://www.bio.cam.ac.uk cgi-bin seqlogo logo.cgi) (124).
Enrichment of Multiple Kinase Motifs in the Same Protein
Predicted high-stringency sites on proteins for CyclinB/Cdk1, Plk1, Aurora A and B, and Nek2 were generated from PS-OPLS results using Scansite (http://scansite.mit.edu) (125). Sites for Aurora Kinase A and B were combined into a single list, as were sites for each of the two Cdk1/cyclinB motifs (one for Ser/Thr+3 R/K and one for Ser/Thr+4 K). Predicted sites were identified by scanning all human proteins from SWISS-PROT (version 42.7), or limited to those containing “nucleus” or “nuclear” in the description or keywords fields. For each pair of kinases, the statistical significance of co-occurance was calculated according to a hypergeometric distribution.
Supplementary Material
Acknowledgements
We gratefully acknowledge members of the Yaffe laboratory for technical assistance, Kristen Naegle for assistance with Bioinformatics, Stephen Shaw for providing PSSM logo code, and Duaa Mohammad and Chris Ellson for critical reading of the manuscript.
Funding: This work was supported by NIH grants GM-60594, GM-68762, ES-015339, and CA-112967 to MBY, a Ludwig Institute for Cancer Research Graduate Fellowship to JA, and the European Commission as part of the Sixth Framework Programme Integrated Project grant ‘MitoCheck’ (LSHG-CT-2004-503464) to BH, JRAH, and JMP; Jane Coffin Childs Fund, Canadian Institutes of Health Research to DL; The Tobacco Research Foundation of Virginia to PTS.
Footnotes
Competing interests: None
References and Notes
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Ubersax JA, Ferrell JE., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 3.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberts B. Molecular Biology of the Cell. Garland Science; New York: 2002. pp. 1027–1062. [Google Scholar]
- 5.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 7.Pines J, Rieder CL. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 2001;3:E3–6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- 8.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 9.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 10.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Bentley AM, Normand G, Hoyt J, King RW. Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol. Biol. Cell. 2007;18:4847–4858. doi: 10.1091/mbc.E06-06-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- 13.Lamb NJ, Fernandez A, Watrin A, Labbe JC, Cavadore JC. Microinjection of p34cdc2 kinase induces marked changes in cell shape, cytoskeletal organization, and chromatin structure in mammalian fibroblasts. Cell. 1990;60:151–165. doi: 10.1016/0092-8674(90)90725-t. [DOI] [PubMed] [Google Scholar]
- 14.Larsson N, Marklund U, Gradin HM, Brattsand G, Gullberg M. Control of microtubule dynamics by oncoprotein 18: dissection of the regulatory role of multisite phosphorylation during mitosis. Mol. Cell. Biol. 1997;17:5530–5539. doi: 10.1128/mcb.17.9.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ookata K, Hisanaga S, Bulinski JC, Murofushi H, Aizawa H, Itoh TJ, Hotani H, Okumura E, Tachibana K, Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J. Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- 17.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 18.Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 19.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 20.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 21.Mori D, Yano Y, Toyo-oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, Wynshaw-Boris A, Hirotsune S. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol. Cell. Biol. 2007;27:352–367. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandall S, Severin F, McLeod IX, Yates JR, 3rd, Oegema K, Hyman A, Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J. Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- 24.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahonen LJ, Kallio MJ, Daum JR, Bolton M, Manke IA, Yaffe MB, Stukenberg PT, Gorbsky GJ. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 28.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 29.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 32.van Vugt MA, van de Weerdt BC, Vader G, Janssen H, Calafat J, Klompmaker R, Wolthuis RM, Medema RH. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 2004;279:36841–36854. doi: 10.1074/jbc.M313681200. [DOI] [PubMed] [Google Scholar]
- 33.Wong OK, Fang G. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J. Cell Biol. 2005;170:709–719. doi: 10.1083/jcb.200502163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, Osada H. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11663–11668. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, Le Guellec R, Couturier A, Doree M, Philippe M, Prigent C. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J. Cell Sci. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- 37.van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- 38.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 41.Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 43.Higashi H, Suzuki-Takahashi I, Taya Y, Segawa K, Nishimura S, Kitagawa M. Differences in substrate specificity between Cdk2-cyclin A and Cdk2-cyclin E in vitro. Biochem. Biophys. Res. Commun. 1995;216:520–525. doi: 10.1006/bbrc.1995.2653. [DOI] [PubMed] [Google Scholar]
- 44.Holmes JK, Solomon MJ. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 45.Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 46.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 47.Zarkowska T, U S, Harlow E, Mittnacht S. Monoclonal antibodies specific for underphosphorylated retinoblastoma protein identify a cell cycle regulated phosphorylation site targeted by CDKs. Oncogene. 1997;14:249–254. doi: 10.1038/sj.onc.1200824. [DOI] [PubMed] [Google Scholar]
- 48.Beretta L, Dobransky T, Sobel A. Multiple phosphorylation of stathmin. Identification of four sites phosphorylated in intact cells and in vitro by cyclic AMP-dependent protein kinase and p34cdc2. J. Biol. Chem. 1993;268:20076–20084. [PubMed] [Google Scholar]
- 49.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bregman DB, Pestell RG, Kidd VJ. Cell cycle regulation and RNA polymerase II. Front. Biosci. 2000;5:D244–257. doi: 10.2741/bregman. [DOI] [PubMed] [Google Scholar]
- 51.Dohadwala M, da Cruz e Silva EF, Hall FL, Williams RT, Carbonaro-Hall DA, Nairn AC, Greengard P, Berndt N. Phosphorylation and inactivation of protein phosphatase 1 by cyclin-dependent kinases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6408–6412. doi: 10.1073/pnas.91.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrari G, Rossi R, Arosio D, Vindigni A, Biamonti G, Montecucco A. Cell cycle-dependent phosphorylation of human DNA ligase I at the cyclin-dependent kinase sites. J. Biol. Chem. 2003;278:37761–37767. doi: 10.1074/jbc.M304462200. [DOI] [PubMed] [Google Scholar]
- 53.Goss VL, Hocevar BA, Thompson LJ, Stratton CA, Burns DJ, Fields AP. Identification of nuclear beta II protein kinase C as a mitotic lamin kinase. J. Biol. Chem. 1994;269:19074–19080. [PubMed] [Google Scholar]
- 54.Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–577. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- 55.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 56.Darieva Z, Bulmer R, Pic-Taylor A, Doris KS, Geymonat M, Sedgwick SG, Morgan BA, Sharrocks AD. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature. 2006;444:494–498. doi: 10.1038/nature05339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin HR, Ting NS, Qin J, Lee WH. M phase-specific phosphorylation of BRCA2 by Polo-like kinase 1 correlates with the dissociation of the BRCA2-P/CAF complex. J. Biol. Chem. 2003;278:35979–35987. doi: 10.1074/jbc.M210659200. [DOI] [PubMed] [Google Scholar]
- 58.Matsumura S, Toyoshima F, Nishida E. Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J. Biol. Chem. 2007;282:15217–15227. doi: 10.1074/jbc.M611053200. [DOI] [PubMed] [Google Scholar]
- 59.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 60.Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat. Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 62.Toyoshima-Morimoto F, Taniguchi E, Nishida E. Plk1 promotes nuclear translocation of human Cdc25C during prophase. EMBO Rep. 2002;3:341–348. doi: 10.1093/embo-reports/kvf069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Leuken R, Clijsters L, van Zon W, Lim D, Yao X, Wolthuis RM, Yaffe MB, Medema RH, van Vugt MA. Polo-like kinase-1 controls Aurora A destruction by activating APC/C-Cdh1. PLoS One. 2009;4:e5282. doi: 10.1371/journal.pone.0005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yarm FR. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Shi X, Paddon H, Hampong M, Dai W, Pelech S. B23/nucleophosmin serine 4 phosphorylation mediates mitotic functions of polo-like kinase 1. J. Biol. Chem. 2004;279:35726–35734. doi: 10.1074/jbc.M403264200. [DOI] [PubMed] [Google Scholar]
- 66.Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 67.Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 2004;5:167–171. doi: 10.1038/sj.embor.7400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 70.Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 71.Ban R, Irino Y, Fukami K, Tanaka H. Human mitotic spindle-associated protein PRC1 inhibits MgcRacGAP activity toward Cdc42 during the metaphase. J. Biol. Chem. 2004;279:16394–16402. doi: 10.1074/jbc.M313257200. [DOI] [PubMed] [Google Scholar]
- 72.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouche JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, Ducommun B. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 74.Ferrari S, Marin O, Pagano MA, Meggio F, Hess D, El-Shemerly M, Krystyniak A, Pinna LA. Aurora-A site specificity: a study with synthetic peptide substrates. Biochem. J. 2005;390:293–302. doi: 10.1042/BJ20050343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawajiri A, Yasui Y, Goto H, Tatsuka M, Takahashi M, Nagata K, Inagaki M. Functional significance of the specific sites phosphorylated in desmin at cleavage furrow: Aurora-B may phosphorylate and regulate type III intermediate filaments during cytokinesis coordinatedly with Rho-kinase. Mol. Biol. Cell. 2003;14:1489–1500. doi: 10.1091/mbc.E02-09-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J. Biol. Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 78.Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 79.Minoshima Y, Kawashima T, Hirose K, Tonozuka Y, Kawajiri A, Bao YC, Deng X, Tatsuka M, Narumiya S, May WS, Jr., Nosaka T, Semba K, Inoue T, Satoh T, Inagaki M, Kitamura T. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 80.Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakai H, Urano T, Ookata K, Kim MH, Hirai Y, Saito M, Nojima Y, Ishikawa F. MBD3 and HDAC1, two components of the NuRD complex, are localized at Aurora-A-positive centrosomes in M phase. J. Biol. Chem. 2002;277:48714–48723. doi: 10.1074/jbc.M208461200. [DOI] [PubMed] [Google Scholar]
- 82.Sugiyama K, Sugiura K, Hara T, Sugimoto K, Shima H, Honda K, Furukawa K, Yamashita S, Urano T. Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene. 2002;21:3103–3111. doi: 10.1038/sj.onc.1205432. [DOI] [PubMed] [Google Scholar]
- 83.Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- 84.Wu JC, Chen TY, Yu CT, Tsai SJ, Hsu JM, Tang MJ, Chou CK, Lin WJ, Yuan CJ, Huang CY. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J. Biol. Chem. 2005;280:9013–9022. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- 85.Yasui Y, Urano T, Kawajiri A, Nagata K, Tatsuka M, Saya H, Furukawa K, Takahashi T, Izawa I, Inagaki M. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 86.Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK, Huang CY. Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol. Cell. Biol. 2005;25:5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 2001;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rellos P, Ivins FJ, Baxter JE, Pike A, Nott TJ, Parkinson DM, Das S, Howell S, Fedorov O, Shen QY, Fry AM, Knapp S, Smerdon SJ. Structure and regulation of the human Nek2 centrosomal kinase. J. Biol. Chem. 2007;282:6833–6842. doi: 10.1074/jbc.M609721200. [DOI] [PubMed] [Google Scholar]
- 89.Chen Y, Riley DJ, Zheng L, Chen PL, Lee WH. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 2002;277:49408–49416. doi: 10.1074/jbc.M207069200. [DOI] [PubMed] [Google Scholar]
- 90.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 91.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A, Lawo S, Pelletier L, Stark H, Nasmyth K, Ellenberg J, Durbin R, Buchholz F, Mechtler K, Hyman AA, Peters JM. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 93.Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C, Hieter P, Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol. Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- 95.Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell Proteomics. 2011;10:M110 004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Y, Wu F, Ward T, Yan F, Wu Q, Wang Z, McGlothen T, Peng W, You T, Sun M, Cui T, Hu R, Dou Z, Zhu J, Xie W, Rao Z, Ding X, Yao X. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, Chen Q, Feng J, Hou J, Yang F, Liu J, Jiang Q, Zhang C. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 2009;122:2240–2251. doi: 10.1242/jcs.042747. [DOI] [PubMed] [Google Scholar]
- 101.Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, Maiolica A, Stark H, Musacchio A. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 103.Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol. Cell. Proteomics. 2005;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- 104.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 110.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohashi S, Sakashita G, Ban R, Nagasawa M, Matsuzaki H, Murata Y, Taniguchi H, Shima H, Furukawa K, Urano T. Phospho-regulation of human protein kinase Aurora-A: analysis using anti-phospho-Thr288 monoclonal antibodies. Oncogene. 2006;25:7691–7702. doi: 10.1038/sj.onc.1209754. [DOI] [PubMed] [Google Scholar]
- 112.Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 113.Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J. Mol. Biol. 2002;322:785–797. doi: 10.1016/s0022-2836(02)00857-4. [DOI] [PubMed] [Google Scholar]
- 114.Keryer G, Witczak O, Delouvee A, Kemmner WA, Rouillard D, Tasken K, Bornens M. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol. Biol. Cell. 2003;14:2436–2446. doi: 10.1091/mbc.E02-09-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu G, Fujii K, Belkina N, Liu Y, James M, Herrero J, Shaw S. Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J. Biol. Chem. 2005;280:10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 116.Huang H, Li L, Wu C, Schibli D, Colwill K, Ma S, Li C, Roy P, Ho K, Songyang Z, Pawson T, Gao Y, Li SS. Defining the specificity space of the human SRC homology 2 domain. Mol. Cell. Proteomics. 2008;7:768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 117.Satinover DL, Leach CA, Stukenberg PT, Brautigan DL. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8625–8630. doi: 10.1073/pnas.0402966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, Stukenberg PT, Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 119.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, Pelletier L, Kittler R, Hua S, Naumann R, Augsburg M, Sykora MM, Hofemeister H, Zhang Y, Nasmyth K, White KP, Dietzel S, Mechtler K, Durbin R, Stewart AF, Peters JM, Buchholz F, Hyman AA. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 121.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 122.Nicholls A, Bharadwaj R, Honig B. Grasp: graphical representation and analysis of surface properties. Biophys. J. 1993;64:A166. [Google Scholar]
- 123.Fujii K, Zhu G, Liu Y, Hallam J, Chen L, Herrero J, Shaw S. Kinase peptide specificity: improved determination and relevance to protein phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.