Abstract
The molecular mechanisms determining transmissibility and prevalence of drug-resistant tuberculosis in a population were investigated through whole genome sequencing of 1,000 prospectively-obtained patient isolates from Russia. Two-thirds belonged to the Beijing lineage, which was dominated by two homogeneous clades. MDR genotypes were found in 48% of isolates overall and 87% of the major clades. The most common rifampicin-resistance rpoB mutation was associated with fitness-compensatory mutations in rpoA or rpoC, and a novel intragenic compensatory substitution was identified. The proportion of MDR cases with XDR-tuberculosis was 16% overall with 65% of MDR isolates harboring eis mutations, selected by kanamycin therapy, which may drive the expansion of strains with enhanced virulence. The combination of drug resistance and compensatory mutations displayed by the major clades confer clinical resistance without compromising fitness and transmissibility, revealing a biological contribution to the tuberculosis program weaknesses driving the persistence and spread of M/XDR-tuberculosis in Russia and beyond.
Introduction
Tuberculosis is the second leading cause of death from an infectious disease, after HIV. In 2011, there were an estimated 8.7 million new cases and 1.4 million deaths from the disease1. The increasing prevalence of drug resistance is a major public health concern that threatens progress made in controlling drug-sensitive tuberculosis2. Globally, 4% of new cases and 20% of previously-treated cases are estimated to have multidrug-resistant (MDR) tuberculosis, defined as resistance to at least rifampicin and isoniazid with the highest proportions in Eastern Europe and Central Asia1. MDR cases require prolonged costly treatment with toxic second-line drugs, and rates of treatment failure and mortality are high2–4. Extensively drug-resistant (XDR) strains, which are MDR with additional resistance to any fluoroquinolone and at least one of the second-line injectable agents (kanamycin, amikacin or capreomycin), have now been found in every region of the world1. Globally, the proportion of MDR-tuberculosis cases with XDR-tuberculosis has reached 9%. In the UK, 26 XDR cases were reported between 1995 and 2011; 45% (9/20) of the patients with known country of birth originated from Eastern Europe5. Recent years have seen an ominous accumulation of reports of ‘totally’ drug-resistant strains, which are not susceptible to any tested drugs6.
Prevalence of drug-resistant tuberculosis is dependent on the rate of acquisition of resistance mutations (acquired resistance) and transmission of drug-resistant strains (primary resistance). In Mycobacterium tuberculosis, drug resistance arises through chromosomal mutation, typically resulting in a fitness cost seen as a reduced growth rate in vitro7. Fitness cost generally inversely correlates with the mutation’s frequency in clinical isolates8. Compensatory mutations mitigating the deleterious effect of resistance mutations are also important in determining the transmissibility of specific genotypes9. Studies using molecular epidemiological clustering rates to assess the transmission cost of resistance genotypes report varying results10,11 suggesting that fitness costs may be affected by epistasis, that is the phenotypic effect of a mutation depends on the presence or absence of other mutations in the same genome12.
Whole genome sequencing (WGS) offers the power to track the evolutionary mechanisms that promote development and transmission of drug resistance within pathogen populations with unparalleled resolution. Evidence for adaptive selection in response to antibiotic therapy can be found by identifying homoplasies (mutations arising independently multiple times within a phylogeny) or loci that are the subject of frequent mutation13–15. By identifying all changes that occur in a genome it is possible to uncover co-occurring polymorphisms that contribute to resistance phenotypes or signify epistatic effects12.
The high incidence of MDR and XDR-tuberculosis in Samara3,16, a region of over three million citizens in Russia, afforded the opportunity to study the emergence and spread of antibiotic resistance within a population. In this prospective study, the largest of its kind yet reported, we used WGS to investigate the molecular mechanisms of resistance, fitness compensation and positive epistasis that together determine the transmissibility and prevalence of drug-resistant strains. Comparative analysis of XDR isolates from the UK addressed the origin of highly drug-resistant tuberculosis in this low-prevalence country.
Results
Population structure
During a two-year period (2008-2010), 2,348 M. tuberculosis isolates were prospectively-collected from individual patients living in Samara, Russia. The genomes of 1,000 isolates were sequenced. Comparison of patient epidemiological data indicated this sample was representative of the entire population and covered the distribution of tuberculosis patients across the whole region (Figure 1, Supplementary Table 1). For comparison, we selected 28 sequences from a study of over 2,000 London-based tuberculosis patients, originating from 90 different countries, as representatives of the global population17,18. We included five phenotypically-XDR strains isolated from UK-patients in 2011, as well as an isolate from Estonia representing the clone that dominates the M. tuberculosis population of this country19.
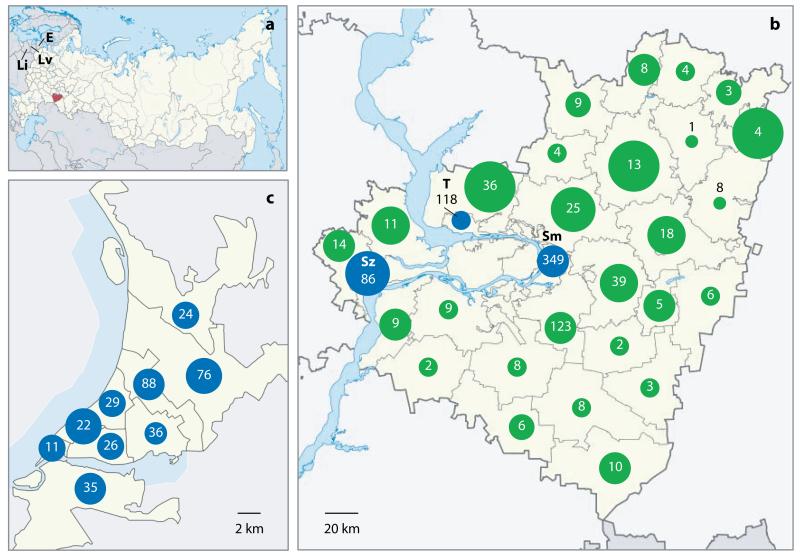
Figure 1. Coverage of the tuberculosis patient population.
The location of Samara Oblast in Russia (red) and the Baltic States (Lithuania (Li), Latvia (Lv) and Estonia (E)) are shown in (a). The number of sequenced patient-isolates from each territory (green) and city (blue; Samara City (Sm), Togliatti (T) and Syzran (Sz)) of Samara Oblast (b) or district of Samara City (c) are shown inside circles. The area of each circle reflects coverage of the region (the number of isolates sequenced relative to the number of tuberculosis cases notified).
Mapping reads for each isolate against the reference sequence, H37Rv, revealed a total of 32,445 single nucleotide polymorphisms (SNPs) in non-repetitive regions of the genome, including 238 associated with drug resistance. These variable sites were used to reconstruct a maximum-likelihood phylogeny (Figure 2, Supplementary Data 1). Tree topology was consistent with the global phylogenetic structure of M. tuberculosis sensu stricto comprising four main lineages20,21. Of the 1,000 Samaran isolates, 642 belonged to the Beijing lineage, 355 to the EuroAmerican lineage, two to the CAS lineage and a single isolate to the EAI lineage, reflecting the proportions seen in the whole patient population.
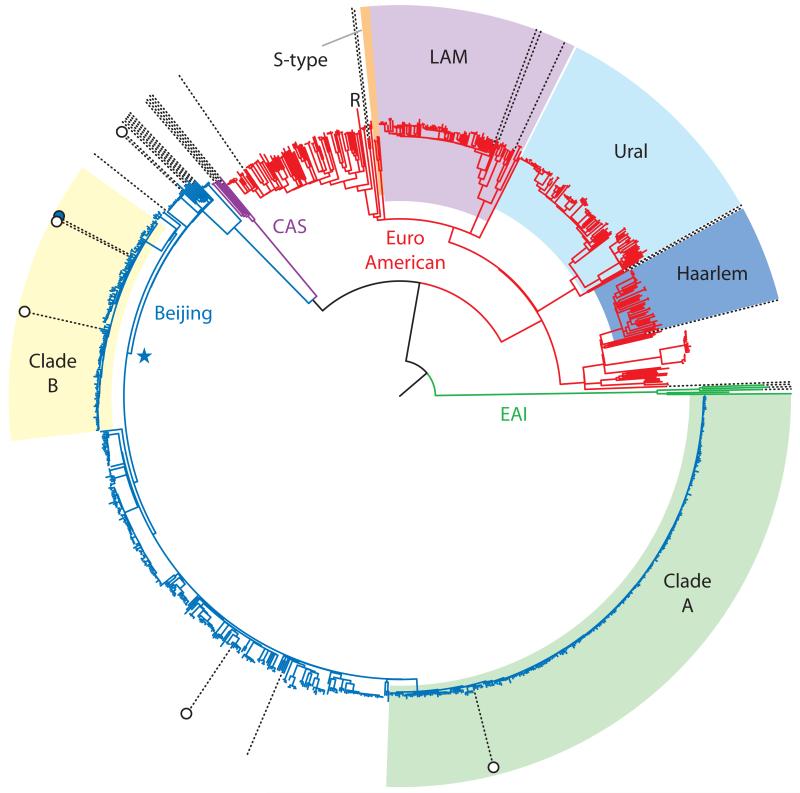
Figure 2. Maximum likelihood phylogeny of 1,035 M. tuberculosis isolates based on 32,445 variable sites.
The four M. tuberculosis lineages: Beijing, CAS, EuroAmerican and EAI, are indicated. The EuroAmerican SNP-defined sublineages23 and the major Beijing clades are shaded. The ancestral node of the Beijing East European sublineage is indicated with a star. Radial dotted lines show the positions of isolates from the UK; those with an XDR phenotype are marked with white circles. The Estonian strain is indicated by a filled blue circle. The position of the reference sequence, H37Rv, is marked ‘R’. The East European sublineage, Clade A and Clade B had 100% bootstrap support (Supplementary Data 1).
In comparison to the disparate Beijing isolates from the UK, the majority of Samaran Beijing sequences formed a monophyletic group, we term the ‘East European’ sublineage18 (Supplementary Figure 1), consistent with a single relatively recent expansion of this lineage in the region. Bayesian population genetic clustering22 defined the two largest clades nested within the Beijing lineage: clade A and clade B, comprising 264 and 119 isolates, respectively (Supplementary Figure 2). In addition to these clades, the majority of other Beijing isolates were members of smaller clusters; 60% (387/642) of all Beijing isolates differed from their last common ancestor by five or less SNPs (Supplementary Figure 3). The Western part of Samara is geographically-isolated by the River Volga. Comparison of the isolate population in the West to the rest of the region revealed a significant reduction in the proportion of clade A (18% vs 28%, p=0.04; Supplementary Table 2, Supplementary Figure 4).
The EuroAmerican lineage showed a significantly higher nucleotide diversity (π=0.0042, SD 0.00018, 95% CI 0.0038-0.0045) than the Beijing lineage (π=0.0022, SD 0.00039, 95% CI 0.0015-0.0030). Fewer EuroAmerican isolates were members of closely-related clusters; 39% (137/355) differed from their last common ancestor by five or fewer SNPs (p<0.001). Although, global diversity within the EuroAmerican lineage is not yet well characterized, Homolka et al.23 identified eight SNP-based sublineages that were broadly concordant with molecular fingerprint-based classification. We identified four of these sublineages in Samara (Haarlem, LAM, Ural and S-type), whilst 36% isolates could not be classified by this scheme (Figure 2).
Short genetic distances between isolates have been used to infer the likelihood of direct transmission24. In this dataset, four pairs of patients with sequenced isolates shared households. Within three households the patient isolates were almost identical with zero, two and three SNPs separating each pair, consistent with intra-household transmission or infection from a common source24. Patients in the fourth household were infected with unrelated isolates (183 SNPs difference). In addition to the household pair, 30 further pairs of isolates, three clusters of three isolates and one cluster of five isolates had zero SNP differences. Patients with identical isolates were resident in the same region of Samara in only 20 of these 35 clusters. Mapping addresses revealed that patients with identical strains lived up to 136km apart (Supplementary Figure 5).
XDR-tuberculosis isolates from four UK patients, who originated from the Baltic States, were members of the East European sublineage with two belonging to clade B and one to clade A, suggesting that infections may have been acquired during stays in their country of origin or within sympatric communities in the UK (Figure 2). In support of this conjecture, the fifth XDR isolate, from a Chinese patient, was not a member of the East European sublineage. The Estonian isolate was also member of clade B. Correlation of VNTR fingerprint data indicated that this is the same strain that predominates across Northwestern Russia18,19,25. The East European isolates were remarkably closely-related with isolates from different countries separated by as little as 13 SNPs (Supplementary Table 3).
Prevalence of MDR genotypes
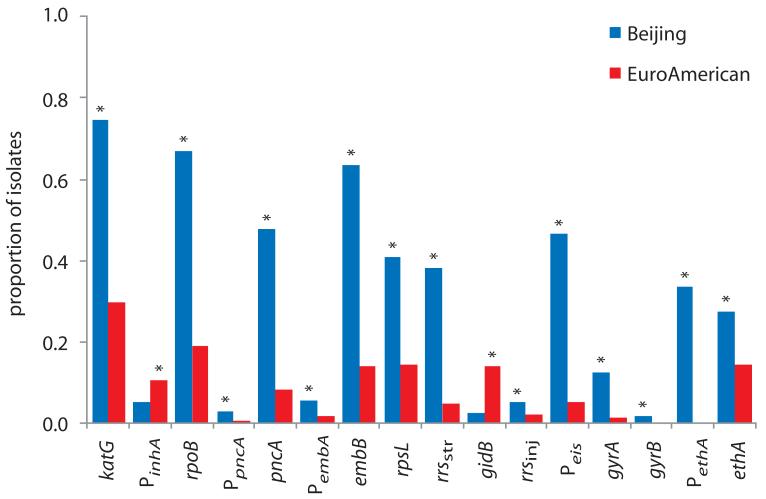
A maximum-likelihood phylogeny of the 1,000 Russian isolates was reconstructed and illustrated with drug resistance genotypes (Figure 3, Supplementary Tables 4-6). The most commonly mutated drug resistance locus, katG codon 315, which confers high-level resistance to isoniazid26, was substituted in 74% (478/642) of Beijing isolates and 30% (106/355) of EuroAmerican isolates (p<0.001; Figure 4, Supplementary Table 7). A novel nonsense SNP at codon 668 also mediated resistance, consistent with the requirement of KatG for activation of the pro-drug. All (478) of the Beijing katG mutants carried the S315T substitution, whilst 11% (12/106) of EuroAmerican isolates harbored one of three alternative substitutions in codon 315 (p<0.001).
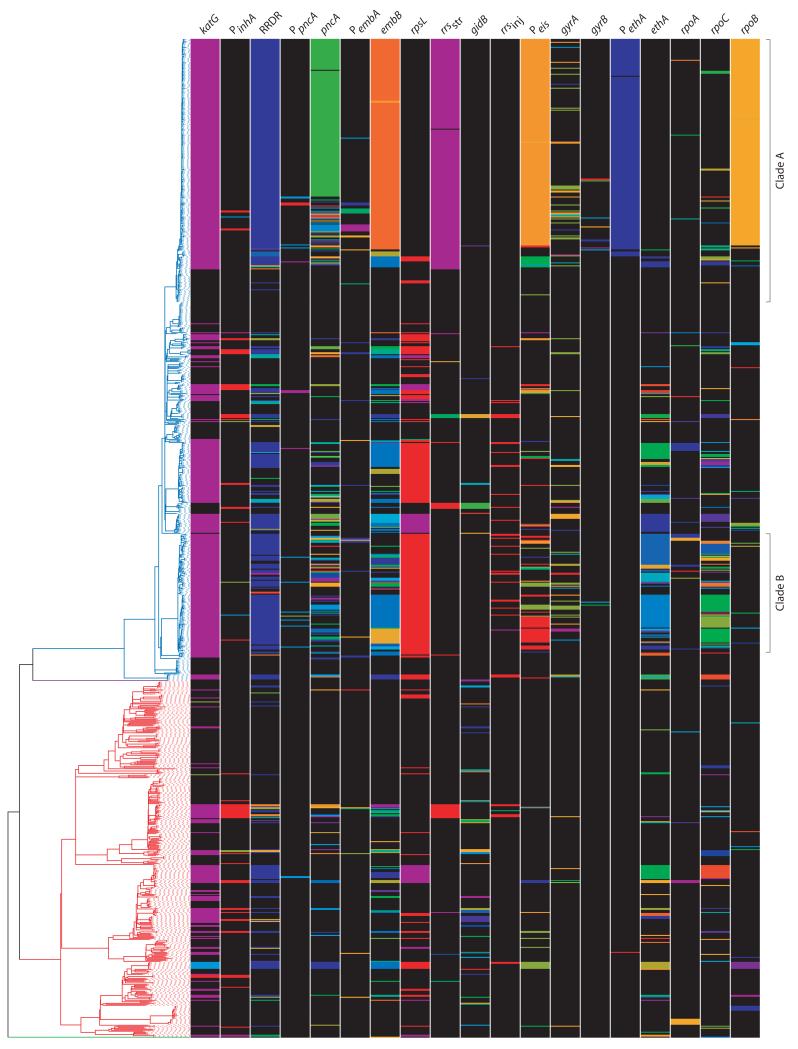
Figure 3. Phylogenetic distribution of drug resistance and compensatory genotypes.
The phylogeny of 1,000 Russian isolates is depicted on the left; lineages are colored as Figure 2. The first 16 columns depict drug resistance loci. ‘P’ denotes a promoter region. Within the 16S rRNA gene, rrsstr refers to the 530 stem-loop and 915 regions involved in streptomycin resistance49 and rrsinj to downstream regions associated with resistance to the second-line injectables36. Colored bands represent different polymorphisms and include previously identified and novel mutations described in the text. The last three columns show nsSNPs in the RNA polymerase genes, rpoABC, excluding those shown in the RRDR. The genotypes illustrated are provided in full in Supplementary Table 4.
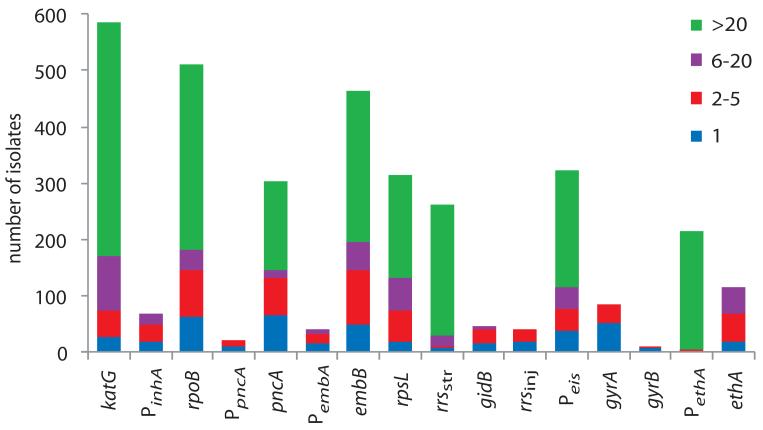
Figure 4. Prevalence of drug resistance mutations and association with lineage.
The proportion of isolates harboring polymorphisms at each drug resistance locus was categorized by lineage. Asterisks indicate significant differences between lineages (Supplementary Table 7). Data is based on the polymorphisms detailed in Supplementary Table 4.
A total of 70 isolates had mutations in the promoter of the fabG1-inhA operon, which confer low-level cross-resistance to isoniazid and its structural analogs, ethionamide and prothionamide27. Based on phylogenetic reconstruction, for 44 isolates the inhA mutation arose in a katG mutant. It is improbable that isoniazid therapy would select for mutations conferring low level resistance in the presence of a high-level resistance SNP indicating that the majority of these promoter SNPs were acquired in response to ethionamide/prothionamide therapy.
Mutations within the 81bp rifampicin resistance-determining region (RRDR) of rpoB are accurate predictors of rifampicin resistance in M. tuberculosis28. Within this region we identified 20 non-synonymous SNPs (nsSNPs) and two small deletions, which affected 70% (430/642) of Beijing and 19% (67/355) of EuroAmerican isolates (p<0.001). The most common rifampicin-resistance genotype, S450L, was found in 90% (390/435) of Beijing isolates with RRDR mutations and 67% (45/67) of EuroAmerican isolates (p<0.001).
Based on these genotypes, 66% (422/642) of Beijing and 17% (61/355) of EuroAmerican isolates have a predicted MDR phenotype (p<0.001). The proportion of isolates that were MDR in clades A and B was significantly higher than for the rest of the Beijing lineage (332/383 vs 90/259, p<0.001).
Compensatory mutations in MDR isolates
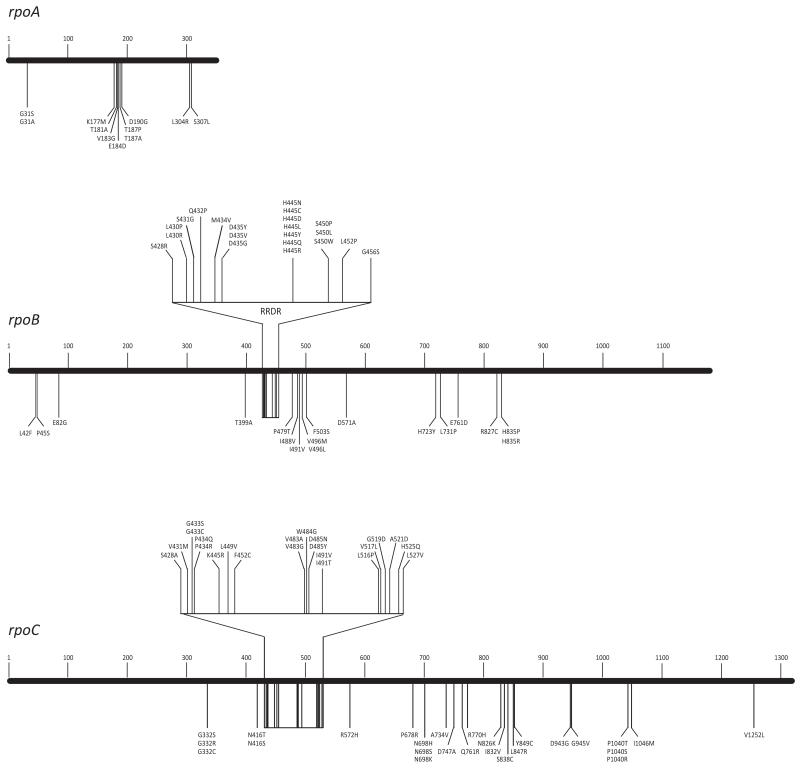
We investigated the occurrence of compensatory mutations in RpoA and RpoC18 in isolates carrying rifampicin-resistance mutations. We identified 14 different nsSNPs in rpoA, 11 of which were found in isolates containing the rpoB S450L mutation (equivalent to Escherichia coli S531L), emerging significantly more frequently in this genetic background (16/435 vs 4/565, p=0.001). Mutations in the most commonly substituted residue, T187, were acquired independently at least seven times, providing strong evidence that rpoA is the subject of positive selection pressure. Clustering of SNP sites within three small regions suggesting these sites are important for interaction with the rifampicin-binding pocket (Figure 5). Eighteen of the 58 nsSNPs identified in rpoC were homoplasic. The rpoC nsSNPs were significantly more likely to arise in isolates harboring RpoB S450L than those with WT RRDR or other resistance mutations (76/435 vs 11/565, p<0.001; Supplementary Table 8). In total, 36 of the 59 amino acid substitutions in RpoA or RpoC found associated with the mutation RpoB S450L have not been previously reported18,29.
Figure 5. Distribution of rifampicin resistance and compensatory amino acid substitutions in the RNA polymerase genes, rpoBCA.
Resistance mutations are clustered in the RRDR region of rpoB. All other substitutions depicted are putative compensatory mutations that co-occurred with rpoB S450L.
Overall, 47% (170/390) of isolates containing the S450L mutation had a putative compensatory mutation in rpoA or rpoC. However, they were remarkably infrequent in clade A (Figure 3, Supplementary Figure 6); 89% (150/169) of non-clade A Beijing isolates harboring S450L had an nsSNP in rpoA or rpoC, compared to 9% (20/221) of clade A isolates (p<0.001). This was unexpected given the obvious epidemiological success of this clade. Thus, we predicted that the large distal cluster of clade A isolates carrying RpoB S450L harbored alternative compensatory mutations that restored fitness. By inspection of the phylogeny we deduced the branch on which this putative mutation likely occurred; one of the four SNPs on this branch resulted in the RpoB substitution E761D (Supplementary Figure 6). Intragenic compensatory mutations in rpoB harboring resistance mutations have been observed in experimentally-evolved E. coli30, Pseudomonas aeruginosa31 and Salmonella enterica32. We surmise that the E761D substitution provides an analogous fitness benefit in M. tuberculosis strains carrying rpoB S450L.
In addition to the E761D substitution and excluding RRDR polymorphisms, a further 26 nsSNPs were identified in rpoB. Sixteen of these co-occurred with S450L (Figure 5) and were significantly more likely to be found in isolates without alternative compensatory mutations (24/37 vs 2/396, p<0.001). Multiple substitution events in codons 496 and 835 provide affirmation that regions of rpoB other than RRDR are under selective pressure. Including all putative compensatory SNPs, 97% (421/435) of isolates carrying rpoB S450L harbored additional rpoABC mutations, which did not appear in any isolates with wild-type RRDR and may mitigate the deleterious effect of this resistance mutation.
Sherman et al.33 proposed that loss of catalase-peroxidase function, in isoniazid-resistant KatG mutants, was compensated by upregulation of alkyl hydroperoxidase, AhpC. We identified four polymorphic sites within the ahpC regulatory region, including two homoplasies. Of nine isolates harbouring the ahpC SNPs, four carried KatG S315T, four had a rare mutation (S315G or W668*) and one had wild-type katG. The ahpC SNPs were significantly more likely to arise in isolates with unusual katG mutations (2/12 vs 3/572, p=0.004), supporting the theory that S315T has low or no fitness cost34.
Prevalence of XDR genotypes
We ascertained that 17% (71/422) of Beijing MDR and 7% (4/61) of EuroAmerican MDR isolates had genotypes predicting an XDR phenotype (p=0.046).
Mutations in the eis promoter that confer kanamycin resistance35 were found in 66% (317/483) of all MDR isolates. Eight different sites were polymorphic, including five homoplasies. The eis mutations were significantly more common in isolates belonging to clades A or B compared to the rest of the Beijing lineage (275/332 vs 21/90, p<0.001). Mutations in the drug target, rrs, which confer resistance to amikacin and capreomycin, as well as kanamycin36, were found in only 40 isolates, four of which had a pre-existing SNP in the eis promoter.
Fluoroquinolones target the DNA gyrases, GyrA and GyrB, and resistance is conferred by mutations within the quinolone resistance-determining regions (QRDRs) that interact with the drugs37. Eighty-six isolates harbored mutations within the QRDR of gyrA and 11 Beijing isolates had SNPs within the gyrB QRDR. Substitutions in gyrAB arose relatively more often in isolates with rrs mutations than those with eis promoter mutations (15/40 vs 62/317, p=0.009; Supplementary Figure 7).
In addition to those isolates with fluoroquinolone-resistant genotypes, we noted a significant number of isolates with ambiguous basecalls within the QRDRs (Supplementary Table 9). This phenomenon was almost never observed at other drug resistance loci. For 18 isolates with ambiguous gyrA genotypes, ambiguity was often apparent at more than one of the codons (90, 91 and 94) that most commonly confer resistance (Supplementary Figure 8). Inspection of raw sequencing reads mapping to this region revealed that substitutions did not co-occur on a single read, indicating multiple fluoroquinolone-resistant clones co-existed in the patient. Fluoroquinolone-treatment of non-tuberculosis infections with could drive acquisition of fluoroquinolone-resistance in chronically-infected tuberculosis patients38. In this case, we would expect to see resistance in non-MDR isolates; however, we found no resistant or heterogeneous QRDR genotypes in non-MDR isolates indicating that the fluoroquinolone exposure was tuberculosis therapy.
Adaptive selection at other drug resistance loci
Repeated independent acquisition of SNPs, revealed by phylogenetic homoplasy, provides strong evidence of selection13 (Supplementary Table 10). Farhat et al.14 recently identified 22 genomic regions that were novel targets of positive-selection in drug-resistant strains (excluding those in repetitive regions) including four that harbored homoplasies in our dataset. Using a complementary approach, Zhang et al.15 identified 98 novel regions that were enriched for SNPs in drug-resistant versus drug-sensitive strains, including 11 where we identified homoplasies.
EmbB harbored homoplasic mutations at codons 306, 406 and 497 that are commonly associated with ethambutol-resistance, although discordance with susceptibility testing is reported39. Homoplasic mutations were identified in five further embB codons (Table 1). Surprisingly, the most frequent homoplasic embB substitution was D354A, which affected a large cluster of clade A isolates, plus two unrelated isolates. This unusual mutation was associated with phenotypic ethambutol resistance in 50% (83/166) of isolates tested. Multiple SNP acquisitions in the region upstream of embAB provide evidence that operon upregulation also confers resistance40. Promoter and coding SNPs frequently co-occurred; isolates with two mutations were more often phenotypically resistant (27/28 vs 199/375, p<0.001; Supplementary Table 5) offering an explanation for the poor concordance between embB 306 mutations and phenotypic resistance39.
Table 1. Ethambutol resistance mutations.
| Locus | Substitution | Reference | SNP | Number of isolates |
Number of acquisitionsb |
Additional mutationsc |
|---|---|---|---|---|---|---|
| PembAB | −16 | C | T/A/G | 15/7/1 | 9/3/1 | M306I(4); D354(6) |
| −15 | C | G | 1 | 1 | - | |
| −12 | C | T | 9 | 8 | M306V(1); D354(2); Q497R(1) | |
| −11 | C | A | 1 | 1 | - | |
| −8 | C | A | 7 | 1 | D354(7) | |
| embB | M306Va/La | A | G/C | 114/4 | 29/2 | −12(1); D354(2) |
| M306Ia | G | A/C/T | 23/18/11 | 14/9/2 | Q497R(4); −16 (4) | |
| Y319S/C | A | C/G | 3/3 | 1/2 | - | |
| D354A | A | C | 213 | 3 | −8(7); −12(2); −16(6); M306V(2) | |
| E378A | A | C | 2 | 2 | - | |
| G406Da/A | G | A/C | 16/16 | 6/4 | - | |
| Q497K | C | A | 9 | 3 | - | |
| Q497Ra | A | G | 27 | 11 | −12(1); M306I(4) | |
| H1002R | A | G | 3 | 3 | - | |
| D1024N | G | A | 3 | 2 | - |
High confidence mutation in TB Drug Resistance Mutation Database (see URLs)
Number of times the mutation independently arose
Number of isolates with additional mutation is given in brackets
The pyrazinamide resistance gene, pncA41, was the most variable gene in the genome (Table 2, Supplementary Figure 9). In addition, it’s promoter harbored five different mutations. GidB was the second most polymorphic gene, indicating that it is a target of selective pressure and supporting its proposed role in streptomycin-resistance42. Discounting the common sublineage-defining SNPs43, gidB mutations were relatively more common in EuroAmerican versus Beijing isolates (47/355 vs 16/642, p<0.001) and were less likely to be concomitant with rpsL or rrs (1/47 vs 13/16, p<0.001). This skewed distribution suggests gidB mutations have a lineage-specific effect. EthA, which catalyzes activation of the pro-drug ethionamide44, was also amongst the most highly-variable genes in the population. A homoplasic SNP 7bp upstream of ethA was independently acquired at least twice, both times in clade A. Positive selection of this SNP suggests it functions clinically in resistance, though only 33% (56/172) of the promoter mutants tested phenotypically resistant. In total, 84% (409/483) of all MDR isolates carried mutations in ethA or it’s upstream region.
Table 2. Mutations in highly-polymorphic genes implicated in drug resistancea.
| Locus | Drug resistance |
Gene length (bp) |
nsSNPs | Nonsense SNPs |
Indels b |
Large deletions |
|---|---|---|---|---|---|---|
| pncA | pyrazinamide | 561 | 79 | 1 | 16 | 5 |
| gidB | streptomycin | 675 | 28 | 2 | 9 | 1 |
| ethA | ethionamide | 1470 | 33 | 6 | 19 | 2 |
Determined by calculating the number of nsSNPs per gene, adjusted for length
all indels resulted in frameshifts
Transmissibility of drug resistance
Previous studies relied upon fingerprint clustering to estimate the transmission dynamics of drug resistance genotypes10,11. Employing a similar principle, but with the improved resolution of WGS, we investigated transmissibility by using the phylogeny to estimate the number of isolates that independently acquired a SNP versus the number of isolates that inherited that SNP from an inferred common ancestor, indicating primary resistance.
SNPs conferring resistance to rifampicin, isoniazid, streptomycin and ethambutol were significantly more likely to be found in phylogenetic clusters, than not (p<0.05; Figure 6, Supplementary Table 11). By inferring the order of SNP acquisition events from the phylogenetic tree, we determined that for 97% (481/495) of isolates with RRDR SNPs a mutation in KatG S315 occurred prior to or on the same branch as the RRDR SNP. Hence, phylogenetic clustering of rpoB SNPs is essentially a surrogate for clustering of MDR genotypes.
Figure 6. Transmissibility of drug resistance genotypes.
The number of isolates within clusters sharing a genotypic marker was estimated by maximum likelihood reconstruction of the polymorphisms onto the phylogeny. A cluster size of one suggests acquired resistance while larger clusters are indicative of primary transmitted resistance.
Pyrazinamide-resistance genotypes were the most often acquired; the modal number of isolates harboring each pncA coding or promoter polymorphism was one (65 of 106 clusters) and only one mutation was shared by a phylogenetic cluster of more than seven isolates. The common pncA nsSNP, encoding the conservative substitution I6L, was found in 157 clade A isolates. This SNP was not associated with phenotypic pyrazinamide resistance in vitro (Supplementary Table 5), however, no secondary pncA mutations were identified in these isolates.
SNPs in gyrA were significantly less likely to be found in clusters (p=0.006; Supplementary Table 11); the largest cluster contained five isolates and 59% (52/88) of resistant isolates did not cluster. In contrast, eis promoter SNPs were typically found in large clusters, the most notable a cluster of 207 clade A isolates.
Discussion
In the largest bacterial WGS project reported to date, we provide a region-wide snapshot of the M. tuberculosis population in Samara, Russia. Circulating strains belonged mainly to two lineages: Beijing and EuroAmerican. In concordance with other Russian-based studies, the Beijing lineage was dominant and accounted for two-thirds of isolates16,25. Relative to the Beijing lineage, EuroAmerican isolates were phylogenetically-diverse and the tree topology supports the division of this lineage into multiple sublineages23. Samaran Beijing isolates were essentially monophyletic with respect to isolates representing the global population, forming a group we term the East European sublineage, which was dominated by two clades of extremely limited diversity. Short genetic distance between tuberculosis isolates has been used to infer transmission links24. In this population, we found large geographic distances within even identical clusters suggesting that they did not always reflect transmission events or that the importance of casual contact may be underestimated.
Genotypes conferring drug resistance were extremely common; 48% of isolates had an MDR genotype and 16% of these were XDR. In comparison to a small microbiologically-based study conducted in Samara in 200145, the proportion of MDR isolates resistant to amikacin has remained relatively stable (7.2% vs 8.5%) whereas the frequency of fluoroquinolone resistance has risen substantially from 4.3% to 23.8%. However, fluoroquinolone resistance was significantly more likely to be acquired than other resistance genotypes indicating that gyrAB mutants may have impaired transmission fitness, impeding the spread of XDR clones. Current rates of resistance support the continued use of fluoroquinolones plus amikacin or capreomycin, rather than kanamycin, for MDR therapy. Whilst a fluoroquinolone plus prothionamide had high efficacy for MDR treatment in Lithuania, which has a comparable MDR-tuberculosis problem and similar historical treatment strategy4, the frequency of ethA mutations in Russian isolates means thioamides may be ineffective in this population.
Drug resistance has previously been associated with the Beijing lineage16. Here we provide evidence that Beijing isolates were more likely to harbor the isoniazid-resistance genotype, katG S315T, that has a negligible fitness cost34 and the rifampicin-resistance genotype, rpoB S450L, that we find strongly associated with putative compensatory mutations within the RNA polymerase genes. Other studies have shown that rpoC mutations restored fitness in competitive growth assays29 and were significantly more common in isolates belonging to a fingerprint-cluster than in non-clustered isolates9. These observations explain, at least in part, the predominance of Beijing MDR isolates. The widespread use of kanamycin in MDR therapy may further exacerbate spread of MDR strains by selecting strains with eis mutations that both confer resistance35 and increase bacterial multiplication in host macrophages46 by disrupting the protective immune response47. Interestingly, eis mutations were significantly associated with the dominant Beijing clades.
The epidemiological success of clade A was particularly striking and the ‘comb-like’ structure of the tree, apparent in the distal portion of the clade, suggested a highly-infectious clone. The incomplete dispersion of clade A to Western Samara supports its recent and rapid spread. From this we deduced the presence of unidentified fitness-enhancing mutations, which led to the discovery of novel intra-rpoB compensatory mutations associated with rifampicin-resistance. In addition, we surmised that a novel SNP upstream of ethA likely confers low-level clinical ethionamide resistance, not detected in vitro, through promoter disruption or enhanced binding of the EthR repressor48. Typically, the most common drug resistance SNPs are associated with the least fitness cost8 implying that successful clones would carry these SNPs. Thus, the discovery of this novel ethA promoter SNP and the rare embB nsSNP D354A, in clade A was unexpected.
The majority of clade A isolates had a conservative I6L substitution in the pyrazinamide-resistance gene, pncA, which does not confer resistance in vitro and is the only pncA nsSNP found in a large cluster. The deduced frequency of acquired pyrazinamide resistance in the sequenced population, plus the small cluster size associated with primary resistance, suggests that mutants with non-functional PncA have impaired transmission efficiency. Given the prevalence of pyrazinamide resistance in the population, it is difficult to reconcile the epidemiological success of clade A with a pyrazinamide-sensitive phenotype. We speculate that the I6L substitution results in intermediate PncA activity that manifests as pyrazinamide sensitivity in vitro but clinical resistance in vivo, and retains sufficient nicotinamidase activity for efficient transmission.
We propose that the unusual combination of drug resistance and compensatory mutations acquired by clade A comprise a ‘perfect storm’ providing clinical drug resistance without compromising fitness and transmissibility.
Preventing tuberculosis transmission relies on accurate and rapid diagnosis. Molecular methods that reduce the time taken to detect drug-resistant tuberculosis strains expedite the institution of effective therapy and efficient infection-control measures, thus minimizing the infectious period. WGS offers the potential for rapid unambiguous determination of all existing, clinically-significant drug resistance mutations and the reducing costs have neutralized a major argument over the value of targeted sequencing versus WGS. We have reported the existence of double mutations at some resistance loci, as well as the lineage- and clade-specific association of certain mutations which are suggestive of undiscovered epistatic interactions. These observations indicate that drug-resistance may be more multi-factorial than previously appreciated which, in some cases, may explain discordance between phenotypes and genotypes. As more resistance loci are identified, and the phenotypic effects of multiple mutations and strain background are elucidated, the public health value of routine WGS for diagnosis of drug resistance will increase, although it may vary depending on the prevalence and likely exposure to resistant strains.
We have reported, with others, the extensive prevalence of MDR Beijing isolates in Russia and former Soviet states of Eastern Europe1,4,16, which shared a common treatment and BCG vaccination strategy. In addition to programmatic and clinical weaknesses, we have identified plausible biological explanations contributing to the devastating MDR-tuberculosis situation in the region. The current dominance of clade B across Eastern Europe19,25 and the isolation of both clade B and clade A in the UK, indicate that the situation throughout the European Union could follow that in the East.
Methods
Study population and whole genome sequencing
From October 2008 to 2010, 2,348 patients with pulmonary disease and culture-proven tuberculosis were recruited from all 18 civilian tuberculosis dispensaries located across Samara. M. tuberculosis isolates were prospectively archived at the Samara Tuberculosis Service, Samara, Russia. Anonymized epidemiological data was stored on a password-protected ACCESS database. Informed consent was obtained from all patients. The study was approved by the Samara Medical Ethics Committee, the Queen Mary Research Ethics Committee and the University of Cambridge Research Ethics Committee.
Isolates were cultured on Middlebrook media for 4–6 wk at 37°C. Sweeps of colonies were harvested, lysed by vortexing with glass beads and genomic DNA purified using a DNeasy Blood and Tissue kit (Qiagen). Paired-end multiplex libraries with a mean insert size of 200bp were prepared as previously described50. Sequencing was performed at the Wellcome Trust Sanger Institute on the Illumina Genome Analyzer GAII or the HiSeq 2000 platform generating reads of 54-bp, 75-bp or 100-bp.
Sequence analysis
Sequence reads were aligned to the corrected H37Rv reference genome18,51 with SMALT (see URLs) and GATK indel realignment applied52. Pindel53 was used to predict the positions of indels and structural variants; these were visually checked in the mapping files. Candidate SNPs were identified using SAMtools54. At each mapped position, alleles were considered to be valid if supported by greater than 70% of mapped reads, including at least 5 in each direction and a minimum mapping quality of 45. SNPs located within repetitive regions were excluded from analysis18.
Mixed basecalls in non-repetitive regions of the genome were considered valid if mapping quality was ≥45, both calls were supported by at least five reads on each strand, and P-values for strand bias, base quality bias, map quality bias and tail distance bias were ≥0.001.
To assess data consistency, nine isolates that were sequenced with 54-bp paired-end reads were re-cultured and 100-bp paired-end sequence generated. For each of these technical replicates, there were no inconsistencies in bases called at variant sites that passed quality filters in both sequences.
Phylogenetic and population genetic analyses
A maximum likelihood phylogeny was reconstructed with RAxML55 using a general time-reversible model with gamma correction for among site rate variation. Calculation of 100 bootstrap replicates provided support for nodes on the tree. The phylogenetic tree was visualized with FigTree (see URLs tree.bio.ed.ac.uk/software/figtree). Ancestral sequences were reconstructed onto each node of the phylogeny using PAML56. From these ancestral sequences, SNPs were reconstructed onto branches of the tree.
To statistically define the population structure we used the software BAPS (Bayesian Analysis of Population Structure)57,58, in particular its module hierBAPS22, which delineates the population structure using nested clustering. Three nested levels of molecular variation were fitted to the data using 10 independent runs of the stochastic optimization algorithm with the a priori upper bound of the number of clusters varying over the interval 50-300 across the runs.
To estimate the nucleotide diversity π59 for the EuroAmerican and Beijing clades, the functions available in the PGEToolbox60 were used in parallel on a cluster computing environment. The very large number of sequences in each clade would require excessive amounts of computing resources when analyzing all the sequences in a single process, and hence, to allow for more economical calculations, 50 random subsets of 100 strains were sampled from each clade and the inference was performed by using 100 bootstraps for each of them, as described by Cai et al.60, and averaging the results. Confidence intervals for the π estimates were computed using the normal distribution approximation with the standard deviation derived by the bootstrap procedure.
Molecular fingerprinting and microbiological testing
Isolates were characterized by spoligotyping according to standard methods61.
Susceptibility to the first-line drugs rifampicin, isoniazid, streptomycin, ethambutol and pyrazinamide was determined using the absolute concentration method on Lowenstein-Jensen slopes62 or using the automated Mycobacterial Growth Indicator Tube (MGIT) 960 system (Becton Dickinson)63. For MDR isolates, susceptibility to the second-line drugs - amikacin, capreomycin, ofloxacin, moxifloxacin and prothionamide - was also determined using the MGIT system61.
A quality assurance procedure was implemented to ensure that isolate metadata corresponded to the appropriate sequence. Data for isolates belonging to the SNP-defined Beijing lineage that did not exhibit the characteristic Beijing spoligotype and, conversely, isolates in other SNP-defined lineages that shared the Beijing spoligotype were excluded from further analysis (n=45). The specificity of katG S315T for prediction of isoniazid resistance is >99%64; thus, microbiological data for isolates with this genotype but a sensitive phenotype were also excluded (n=58).
Statistical methods
Simple descriptive statistics were used to compare the sample population patient data with the remaining population and to characterize the prevalence of mutations and geographic distribution of sublineages; 95% confidence intervals (SD) were established. In addition, the sampled population was evaluated by attributive risk analysis. The significance of differences between studied groups of variables was calculated using two-sample tests of proportions: Pearson chi-square or Fisher’s exact test when any expected group size was less than 5. Statistical tests were two-sided at α=0.05. The analysis was done using STATA (version 12.1, StataCorp).
Supplementary Material
Acknowledgements
We are grateful to members of the Public Health England National Mycobacterium Reference Laboratory and the Samara Regional Tuberculosis Laboratory for bacteriological work, particularly Dr Madeline Stone, Dr Ximena Gonzalo and Agnieszka Broda. We would also like to thank the Samara Tuberculosis Service particularly Dr Ivan Fedorin and also Dr Vadim Kulichenko. We thank Dr Richard Hooper for expert statistical advice, Prof Sven Hoffner for bacteriological advice and Dr Stephen Bentley for sequencing advice. This study was supported by the European Union Framework Programme 7 (Grant number 201483; TB-EUROGEN) with sequencing funded by the Wellcome Trust (Grant number 098051) and EUROGEN. SN is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science (095198/Z/10/Z) and is also supported by the European Research Council Starting Grant 260477.
Footnotes
URLs
TB Drug Resistance Mutation Database: www.tbdreamdb.com
SMALT: www.sanger.ac.uk/resources/software/smalt
FigTree: tree.bio.ed.ac.uk/software/figtree
Accession codes
Raw sequence data have been submitted to the European Nucleotide Archive under accession number ERP000192.
Conflicts of Interest
All authors declare that they have no conflicts of interest.
References
- 1.World Health Organisation Global tuberculosis report. 2012 [Google Scholar]
- 2.Gandhi NR, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 3.Balabanova Y, et al. Survival of civilian and prisoner drug-sensitive, multi- and extensive drug-resistant tuberculosis cohorts prospectively followed in Russia. PLoS ONE. 2011;6:e20531. doi: 10.1371/journal.pone.0020531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balabanova Y, et al. Survival of drug resistant tuberculosis patients in Lithuania: retrospective national cohort study. BMJ Open. 1:2011. doi: 10.1136/bmjopen-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.London Health Protection Agency Tuberculosis in the UK: annual report on tuberculosis surveillance in the UK. 2012 [Google Scholar]
- 6.Udwadia ZF. MDR, XDR, TDR tuberculosis: ominous progression. Thorax. 2012;67:286–288. doi: 10.1136/thoraxjnl-2012-201663. [DOI] [PubMed] [Google Scholar]
- 7.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 8.Böttger EC, Springer B. Tuberculosis: Drug resistance, fitness, and strategies for global control. Eur. J. Pediatr. 2007;167:141–148. doi: 10.1007/s00431-007-0606-9. [DOI] [PubMed] [Google Scholar]
- 9.De Vos M, et al. Putative compensatory mutations in the rpoC gene of rifampicin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob. Agents Chemother. 2012;57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dye C, Williams BG, Espinal MA, Raviglione MC. Erasing the world’s slow stain: Strategies to beat multidrug-resistant tuberculosis. Science. 2002;295:2042–2046. doi: 10.1126/science.1063814. [DOI] [PubMed] [Google Scholar]
- 11.Cohen T, Sommers B, Murray M. The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect. Dis. 2003;3:13–21. doi: 10.1016/s1473-3099(03)00483-3. [DOI] [PubMed] [Google Scholar]
- 12.Borrell S, Gagneux S. Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 2011;17:815–820. doi: 10.1111/j.1469-0691.2011.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkhill J, Wren BW. Bacterial epidemiology and biology - lessons from genome sequencing. Genome Biol. 2011;12:230. doi: 10.1186/gb-2011-12-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2013;45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 2013;45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 16.Drobniewski F, et al. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA. 2005;293:2726–2731. doi: 10.1001/jama.293.22.2726. [DOI] [PubMed] [Google Scholar]
- 17.Brown T, Nikolayevskyy V, Velji P, Drobniewski F. Associations between Mycobacterium tuberculosis strains and phenotypes. Emerg. Infect. Dis. 2010;16:272–280. doi: 10.3201/eid1602.091032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casali N, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruuner A, et al. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 2001;39:3339–3345. doi: 10.1128/JCM.39.9.3339-3345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagneux S, et al. Variable host–pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homolka S, et al. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS ONE. 2012;7:e39855. doi: 10.1371/journal.pone.0039855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokrousov I, et al. Mycobacterium tuberculosis population in Northwestern Russia: an update from Russian-EU/Latvian border region. PLoS ONE. 2012;7:e41318. doi: 10.1371/journal.pone.0041318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 28.Telenti A, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–651. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 29.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 2011;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall AR, Griffiths VF, MacLean RC, Colegrave N. Mutational neighbourhood and mutation supply rate constrain adaptation in Pseudomonas aeruginosa. Proc. R. Soc. B Biol. Sci. 2010;277:643–650. doi: 10.1098/rspb.2009.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 2012;85:142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 33.Sherman DR, et al. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 34.Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 2002;70:4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaunbrecher MA, Sikes RD, Metchock B, Shinnick TM, Posey JE. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2009;106:20004–20009. doi: 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maus CE, Plikaytis BB, Shinnick TM. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2005;49:3192–3197. doi: 10.1128/AAC.49.8.3192-3197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruri F, et al. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J. Antimicrob. Chemother. 2012;67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginsburg AS, Grosset JH, Bishai WR. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 2003;3:432–442. doi: 10.1016/s1473-3099(03)00671-6. [DOI] [PubMed] [Google Scholar]
- 39.Shen X, et al. Association between embB codon 306 mutations and drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007;51:2618–2620. doi: 10.1128/AAC.01516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy SV, et al. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2000;44:326–336. doi: 10.1128/aac.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 42.Wong SY, et al. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011;55:2515–2522. doi: 10.1128/AAC.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spies FS, et al. Streptomycin resistance and lineage-specific polymorphisms in Mycobacterium tuberculosis gidB gene. J. Clin. Microbiol. 2011;49:2625–2630. doi: 10.1128/JCM.00168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2003;47:3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balabanova Y, et al. Multidrug-resistant tuberculosis in Russia: clinical characteristics, analysis of second-line drug resistance and development of standardized therapy. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:136–139. doi: 10.1007/s10096-004-1268-4. [DOI] [PubMed] [Google Scholar]
- 46.Wu S, et al. Activation of the eis gene in a W-Beijing strain of Mycobacterium tuberculosis correlates with increased SigA levels and enhanced intracellular growth. Microbiology. 2009;155:1272–1281. doi: 10.1099/mic.0.024638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin D-M, et al. Mycobacterium tuberculosis Eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engohang-Ndong J, et al. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 2004;51:175–188. doi: 10.1046/j.1365-2958.2003.03809.x. [DOI] [PubMed] [Google Scholar]
- 49.Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 1993;9:1239, 1246. doi: 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 50.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 52.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 57.Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang J, Hanage WP, Fraser C, Corander J. Identifying currents in the gene pool for bacterial populations using an integrative approach. PLoS Comput. Biol. 2009;5:e1000455. doi: 10.1371/journal.pcbi.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nei M. Molecular Evolutionary Genetics. Columbia University Press; 1987. [Google Scholar]
- 60.Cai JJ. PGEToolbox: A Matlab Toolbox for Population Genetics and Evolution. J. Hered. 2008;99:438–440. doi: 10.1093/jhered/esm127. [DOI] [PubMed] [Google Scholar]
- 61.Kamerbeek J, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canetti G, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull. World Health Organ. 1963;29:565–578. [PMC free article] [PubMed] [Google Scholar]
- 63.Kruuner A, Yates MD, Drobniewski FA. Evaluation of MGIT 960-based antimicrobial testing and determination of critical concentrations of first- and second-line antimicrobial drugs with drug-resistant clinical strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:811–818. doi: 10.1128/JCM.44.3.811-818.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur. Respir. J. 2008;32:1165–1174. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.