Abstract
Centrosome duplication is licensed by the disengagement, or ‘uncoupling’, of centrioles during late mitosis. However, arrest of cells in G2 can trigger premature centriole disengagement. Here, we show that premature disengagement results from untimely activation of the APC/C leading to securin degradation and release of active separase. APC/C activation during G2 arrest is dependent on Plk1-mediated degradation of the APC/C inhibitor, Emi1, but Plk1 also has a second APC/C-independent role in promoting disengagement. Importantly, APC/C and Plk1 activity also stimulate centriole disengagement in response to hydroxyurea or DNA damage-induced cell cycle arrest and this leads to centrosome amplification. However, the re-duplication of disengaged centrioles is dependent on Cdk2 activity and Cdk2 activation coincides with a subsequent inactivation of the APC/C and re-accumulation of cyclin A. Release from these arrests leads to mitotic entry but, due to the presence of disengaged and/or amplified centrosomes, formation of abnormal mitotic spindles that lead to chromosome missegregation. Thus, oscillation of APC/C activity during cell cycle arrest promotes both centrosome amplification and genome instability.
INTRODUCTION
Maintenance of genome stability requires centrosome duplication to be tightly coupled to cell cycle progression (Mazia, 1987). This ensures accurate segregation of chromosomes through formation of a bipolar mitotic spindle with one centrosome at each pole. As cells segregate their centrosomes, along with chromosomes, during mitosis, then each daughter cell will inherit one centrosome. This must be duplicated once and once only in the following cell cycle to maintain this fidelity. Loss of coupling between centrosome duplication and the cell cycle can lead to centrosome amplification, a common hallmark of cancer cells that is thought to promote tumour progression (Basto et al., 2008; Ganem et al., 2009).
Recent insights have begun to shed light on how centrosome duplication is coupled to the cell cycle (reviewed in (Bettencourt-Dias and Glover, 2007; Loncarek and Khodjakov, 2009; Nigg and Raff, 2009; Strnad and Gonczy, 2008; Tsou and Stearns, 2006a). Firstly, both centrosome duplication and DNA replication are under the control of Cdk2. This ensures that both processes are only initiated upon entry into S-phase when this protein kinase becomes active upon binding first cyclin E and later cyclin A (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999). Physically, centrosome duplication involves the replication of the two centrioles, which form the core of the centrosome and upon which the pericentriolar material (PCM) is assembled. Significant progress has now been made in identifying the core components required for new centriole biogenesis; these include the SAS-4/CPAP, SAS-5/Ana2 and SAS-6 proteins (Delattre et al., 2006; Dobbelaere et al., 2008; Kleylein-Sohn et al., 2007; Pelletier et al., 2006; Strnad and Gonczy, 2008). Furthermore, structural studies have revealed how the oligomerization of SAS-6 can define the 9-fold symmetry of centrioles (Kitagawa et al., 2011; van Breugel et al., 2011). However, much still remains to be learnt about how centriole duplication is initiated both by Cdk2, and another crucial regulatory kinase, Plk4 (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Pelletier et al., 2006).
A second control mechanism that ensures centrosome duplication is coupled to the cell cycle occurs during mitosis. As cells enter mitosis, they possess two centrosomes each composed of two centrioles. Initially, these are all connected with the two new centrioles, referred to as procentrioles, tightly attached to the sidewall of their parental centrioles in an orthogonal arrangement, and the older two parental centrioles (also known as the mother and daughter), bridged more distantly through their proximal ends via an extended fibrous linker. Separation of the parental centrioles occurs at the G2/M transition through, firstly, displacement of linker proteins in a process called centrosome disjunction and, secondly, the action of microtubule-based motor proteins, most notably Eg5, that can crosslink and slide microtubules in an anti-parallel manner. This leads to assembly of a bipolar spindle with each spindle pole containing a centriole pair (Nigg and Raff, 2009; Walczak and Heald, 2008). Separation of the procentriole from its parental centriole, an event known as centriole disengagement, occurs later in mitosis after anaphase onset. That this is coincident with sister chromatid separation falls in line with recent data suggesting that both events are under the control of the enzyme, separase (Thein et al., 2007; Tsou and Stearns, 2006b). This cysteine protease cleaves the Scc1/Rad21/kleisin subunit of cohesin to initiate sister chromatid separation (Nasmyth, 2002). Both separase and cohesin subunits have been localized to centrosomes (Chestukhin et al., 2003; Gimenez-Abian et al., 2010; Gregson et al., 2001; Guan et al., 2008; Kong et al., 2009; Nakamura et al., 2009; Wong and Blobel, 2008), while cleavage of engineered cohesin rings promotes unscheduled centriole disengagement (Schockel et al., 2011). Together, these data raise the exciting possibility that the same biochemical mechanism promotes both sister chromatid separation and centriole disengagement.
In addition to separase, centriole disengagement is dependent on the mitotic kinase, Plk1. Genetic knockout in human cells demonstrated the requirement for separase in centriole disengagement; however, in these cells disengagement was only delayed and eventually occurred in a manner that was dependent on Plk1 (Tsou et al., 2009). Intriguingly, this dual dependency of centriole disengagement on Plk1 and separase mirrors the regulation of sister chromatid separation in which Plk1 promotes dissociation of cohesin from chromosome arms in prophase, while separase cleaves the remaining cohesin at centromeres in anaphase (Gimenez-Abian et al., 2004).
Hence, the centrosome duplication and DNA replication cycles are coupled by a requirement for the same regulators not only in S-phase, but also in M-phase. Furthermore, centriole disengagement is a prerequisite for a new round of centriole duplication, with procentrioles having to be displaced from the sidewall of the parental centrioles to free up space for growth of another procentriole (Loncarek et al., 2008; Tsou and Stearns, 2006b). Thus, centrioles cannot reduplicate until they have undergone disengagement, thereby separating in time the licensing event (disengagement) from the biogenesis event (duplication). Recently, a third coupling mechanism has been proposed whereby new procentrioles require a Plk1-dependent modification to make them competent for duplication. This requires passage through mitosis and exposure to Plk1 activity and prevents the growth of so-called “granddaughter” centrioles from daughters (or procentrioles) within the same cell cycle (Loncarek et al., 2010; Wang et al., 2011).
Here, we set out to explore the mechanisms regulating centrosome organization in G2 by arresting cells with the highly selective Cdk1 inhibitor, RO-3306 (Vassilev et al., 2006). In line with recent findings, we found that Cdk1 inhibition in G2 promotes premature centriole disengagement (Loncarek et al., 2010; Steere et al., 2011). Mechanistically, we found that this was dependent on both Plk1 and separase, the latter being activated as a result of loss of Emi1-mediated inhibition of the APC/C (Ma et al., 2009). Whilst Plk1 promotes degradation of Emi1, our data support a model whereby Plk1 has a second, more direct, role in promoting centriole disengagement, independent of the APC/C (Loncarek et al., 2010). Importantly, we found that other conditions that arrest cells in S or G2, including hydroxyurea (HU) or ionizing radiation (IR)-mediated DNA damage, also promote APC/C- and Plk1-dependent centriole disengagement. Moreover, the subsequent inactivation of the APC/C upon prolonged arrest contributes to a Cdk2-dependent reduplication of centrioles, as well as endoreduplication of DNA (Ma et al., 2009). This study therefore reveals that oscillation of APC/C activity provides an additional mechanism for coupling the centrosome duplication and DNA replication cycles.
RESULTS
Centriole disengagement coincides with APC/C activation during G2 arrest
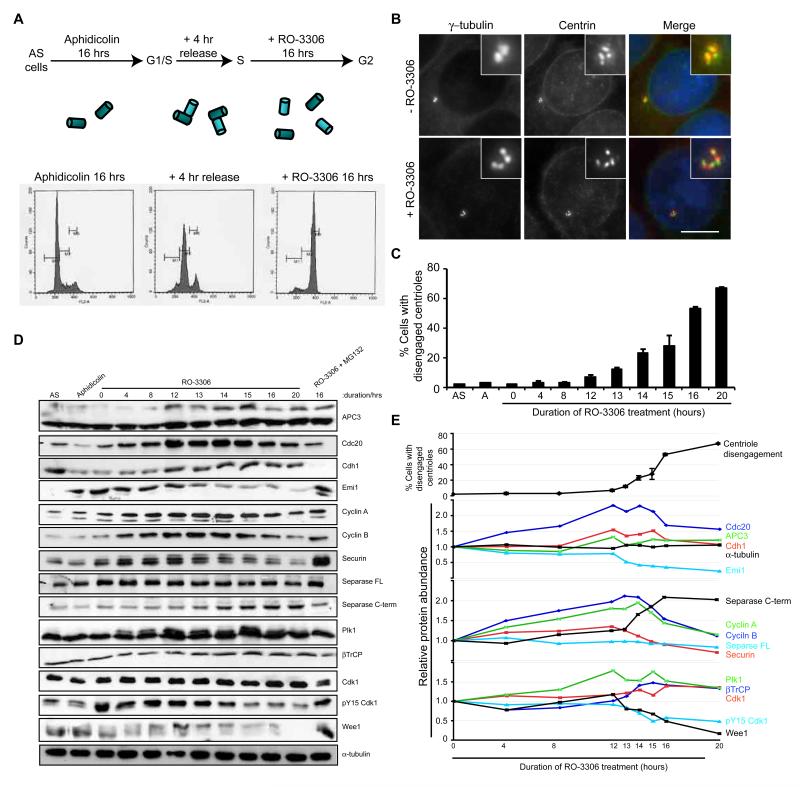
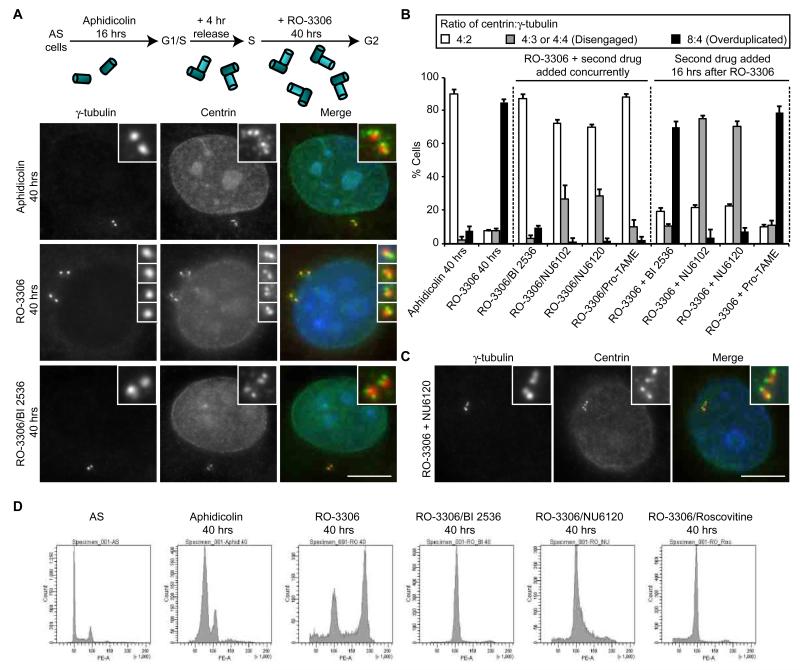
To examine centrosome organization during G2 arrest, HeLa cells were presynchronised in S-phase with aphidicolin and released for 4 hours before addition of the Cdk1-specific inhibitor, RO-3306 (Fig. 1A). Centrosomes were analysed by immunofluorescence microscopy with antibodies against the PCM marker, γ-tubulin, immediately after the 4 hour release from aphidicolin or after an additional 16 hours RO-3306 treatment (Fig. 1B). Whereas 98% cells that had not been treated with RO-3306 showed two dots typical of an S or G2 cell, the majority (52%) of RO-3306 treated cells showed three or four γ-tubulin stained spots. Co-staining with antibodies against the centriolar component, centrin2, revealed the presence of four centrioles as expected for S or G2 cells confirming that this was the result of premature centriole disengagement (Fig. 1B), consistent with results observed by Loncarek et al. (2010). Similar results were obtained after RO-3306 treatment of U2OS or hTERT-RPE1 cells (Fig. S1A, B), or following treatment of HeLa cells with a structurally distinct Cdk1 inhibitor, roscovitine, or depletion of Cdk1 by RNAi (Fig. S1C-E). Hence, centriole disengagement is a general response to loss of Cdk1 activity and arrest of cells in G2. Strikingly, when we examined the timing of centriole disengagement, we found that this became apparent from 12 hours post RO-3306 treatment. This closely correlated with reduced expression of the APC/C inhibitor, Emi1, reduced expression of the APC/C substrates, cyclin A, cyclin B, Cdc20 and securin, and the appearance of hyperphosphorylated APC3 (Cdc27) (Fig. 1C-E). These observations are all consistent with activation of the APC/C. RNAi-mediated depletion of Cdk1 also led to loss of securin (Fig. S1F). Together, these data fall in line with the demonstration that Cdk1 inhibition leads to activation of the APC/C (Ma et al., 2009) and led us to propose that premature centriole disengagement during G2 arrest is a consequence of untimely activation of the APC/C.
Figure 1. Centriole disengagement in G2 arrest occurs coincident with APC/C activation.
A. Schematic of the synchronisation protocol used in all experiments unless stated otherwise (AS, asynchronous). Upon release from aphidicolin arrest, cells contain two pairs of engaged centrioles (cylinders), while subsequent G2 arrest with the Cdk1 inhibitor, RO-3306, leads to premature disengagement. Flow cytometry profiles for HeLa cells, used in all experiments unless indicated, are shown. B. Immunofluorescence microscopy (IF) of cells treated as in A, +/- RO-3306, and stained for γ-tubulin (red) and centrin (green). Merge panels include DNA (blue). Scale bar, 10 μm. C. Quantification of centriole disengagement with time of RO-3306 treatment (AS, asynchronous; A, aphidicolin arrest). Data shown as mean ± sd; n=3, >100 cells counted. D. Extracts from cells treated as in C were immunoblotted for the proteins indicated. A sample was also prepared from cells treated with RO-3306 and the proteasome inhibitor, MG132, for 16 hours following synchronisation. E. Plots showing the relative protein abundance from the blots in D from 0 to 20 hours RO-3306 treatment as compared to rate of centriole disengagement. The abundance of each protein was quantified relative to the 0 hour sample.
Premature centriole disengagement depends upon Plk1-mediated activation of the APC/C and separase
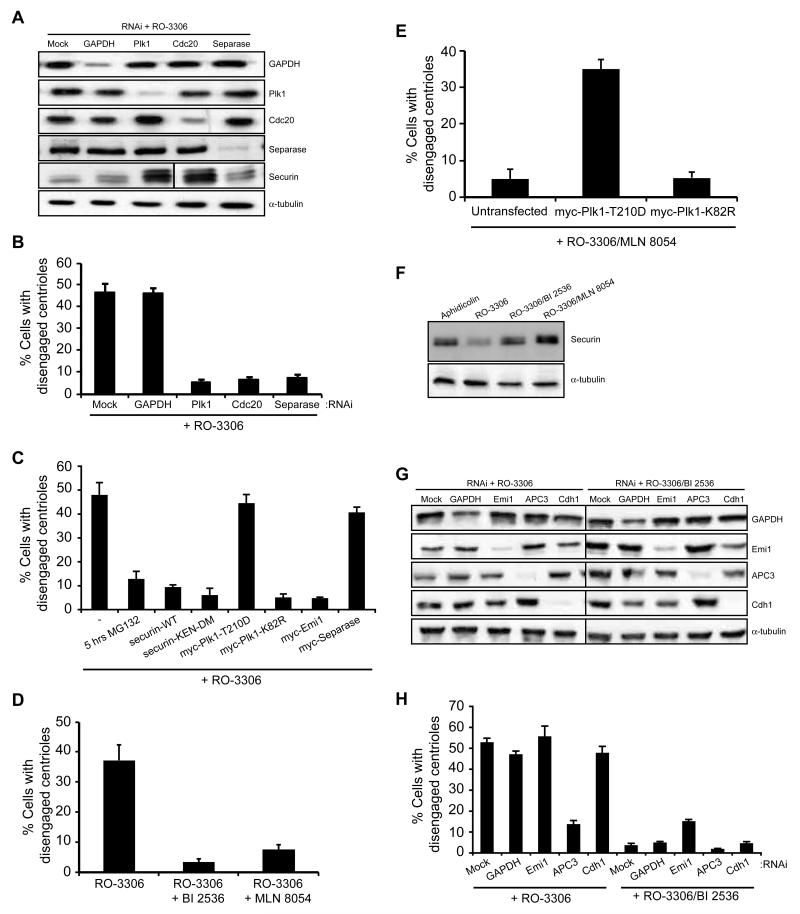
Loss of securin expression following RO-3306-induced G2 arrest was also accompanied by elevated expression of an autocatalytic C-terminal cleavage product of separase, indicative of separase activation (Fig. 1D and E). We therefore determined whether premature centriole disengagement was a result of untimely APC/C-mediated activation of separase. Depletion of separase itself or Cdc20, a co-activator of the APC/C, blocked disengagement in RO-3306 arrested cells, while Cdc20 depletion also caused accumulation of securin consistent with APC/C inhibition (Fig. 2A, B). Furthermore, addition of the proteasome inhibitor, MG132, or expression of wild-type or non-degradable (KEN-DM) securin prevented premature centriole disengagement (Fig. 2C). These data provide strong support to the hypothesis that separase is a major activator of centriole disengagement (Schockel et al., 2011; Thein et al., 2007; Tsou and Stearns, 2006b), and demonstrate that APC/C-dependent separase activation is responsible for premature disengagement in G2-arrested cells.
Figure 2. Premature centriole disengagement is dependent upon activation of the APC/C and Plk1.
A. Immunoblot of proteins indicated following transfection with siRNAs against GAPDH, Plk1, Cdc20 or separase 12 hours prior to synchronization and treatment as in Figure 1A. B. Centriole disengagement in cells treated as in A. C. Centriole disengagement in cells synchronised and treated with RO-3306 for 16 hours, plus MG132 for the final 5 hours, or following overexpression of the constructs indicated. D. Centriole disengagement in cells treated with RO-3306 with or without the Plk1 inhibitor, BI 2536, or Aurora A inhibitor, MLN 8054, as indicated for 16 hours. E. Centriole disengagement in cells transfected as indicated and treated with RO-3306 and MLN 8054 for 16 hours. F. Immunoblot of securin and -tubulin in synchronized cells treated as indicated for 16 hours. G. Immunoblot of proteins indicated following transfection with siRNAs against GAPDH, Emi1, APC3 or Cdh1 12 hours prior to synchronization and treatment with RO-3306 or RO-3306 plus BI 2536. H. Centriole disengagement in cells treated as in G. Data in B, C, D, E and H shows mean ± sd; n=3, >100 cells counted.
Besides separase, centriole disengagement is regulated by Plk1 (Loncarek et al., 2010; Tsou et al., 2009). Here, we found that siRNA-mediated depletion of Plk1, or expression of a catalytically-inactive mutant (K82R) prevented premature disengagement in cells treated with RO-3306 (Fig. 2A-C). Similarly, chemical inhibition of either Plk1 or Aurora A blocked RO-3306-induced disengagement, while expression of an activated Plk1 mutant (T210D) overcame the block imposed by the Aurora A inhibitor (Fig. 2D, E; and S2A, B). This is consistent with Plk1 activation being dependent on Aurora A-mediated phosphorylation of Plk1 at T210 (Macurek et al., 2008; Seki et al., 2008). One explanation for the role of Plk1 is that it acts as an upstream activator of the APC/C through promoting the βTrCP-mediated degradation of the APC/C inhibitor, Emi1 (Hansen et al., 2004; Moshe et al., 2004). Cells treated with RO-3306 and either the Plk1 or Aurora A inhibitor did indeed maintain high levels of securin, unlike cells treated with RO-3306 alone (Fig. 2F), whilst ectopic expression of Emi1 could block premature disengagement in RO-3306 treated cells (Fig. 2C). Additionally, depletion of Emi1 had no impact on the level of disengagement seen following RO-3306 treatment, but in the presence of the Plk1 inhibitor a modest rescue of disengagement was seen (Fig. 2G and H). Thus, we conclude that Plk1-dependent activation of the APC/C and release of separase drives centriole disengagement in G2-arrested cells. Depletion of APC3 confirmed a role for the APC/C by preventing premature centriole disengagement, whilst, in contrast to Cdc20 depletion, knockdown of Cdh1 did not prevent disengagement (Fig. 2G and H). Thus, the APC/C is activated primarily though Cdc20 rather than Cdh1 during RO-3306-mediated G2 arrest.
Plk1 also mediates an APC/C-independent pathway of centriole disengagement
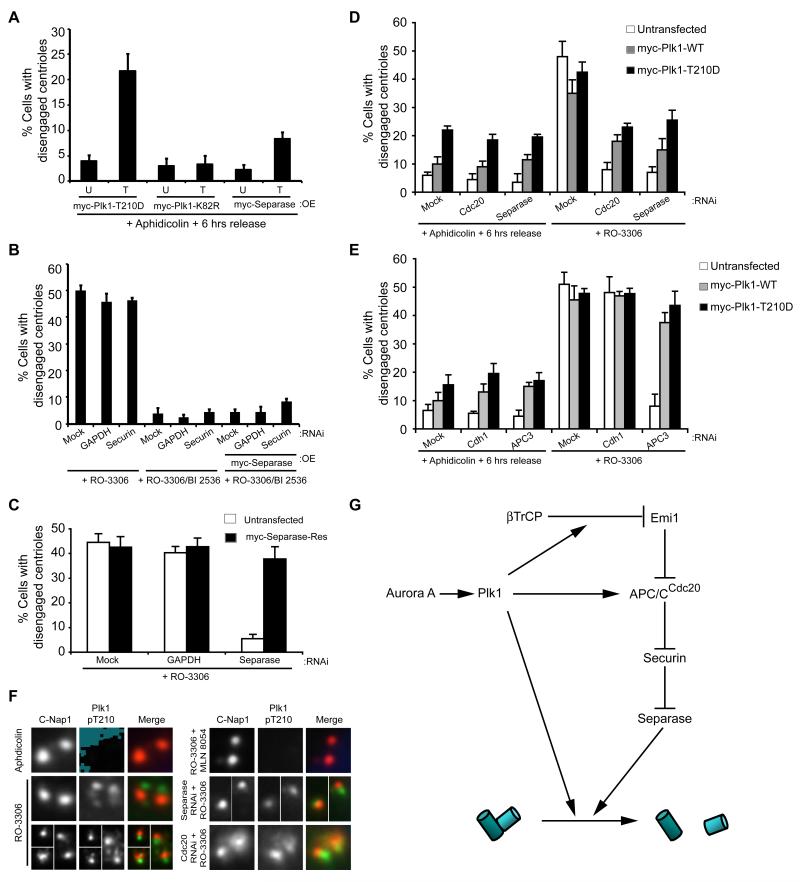
Both Plk1 and separase play essential roles in promoting loss of sister chromatid cohesion during mitosis. However, in this case, Plk1 acts independently through phosphorylation of cohesin subunits in prophase (Peters et al., 2008). As cohesin has also been proposed to mediate centriole cohesion (Schockel et al., 2011), we wished to know whether Plk1 may have an additional APC/C-independent role in centriole disengagement. We first asked whether expression of activated Plk1 or separase could promote centriole disengagement in cells synchronized in S/G2, but in the absence of a Cdk1 inhibitor when APC/C activity is low. Under these conditions, overexpression of Plk1, and to a lesser extent separase, could drive centriole disengagement (Fig. 3A). However, overexpression of separase did not induce disengagement in the presence of the Plk1 inhibitor, even in cells also depleted of securin (Fig. 3B and S3A). This was despite the fact that an RNAi-resistant version of separase was fully able to rescue disengagement in cells depleted of endogenous separase (Fig. 3C and S3B). In contrast, overexpression of wild-type or activated Plk1 could promote centriole disengagement in RO-3306-arrested cells depleted of Cdc20, separase or APC3 (Fig 3D, E and S3C, D).
Figure 3. Plk1 also has an APC/C-independent role in centriole disengagement.
A. Centriole disengagement following 6 hours release from aphidicolin arrest without RO-3306 treatment in cells that were untransfected (U) or transfected (T) with constructs indicated. B. Centriole disengagement in cells that were mock, GAPDH or securin depleted whilst also, where indicated, transfected with myc-separase. After 12 hours, cells were synchronised, released and treated with RO-3306 alone or plus BI 2536. C. Quantification of centriole disengagement in mock, GAPDH or separase depleted HeLa cells, +/- transfection with myc-separase-Res (an RNAi resistant separase construct), and treated with RO-3306 for 16 hours. D. Centriole disengagement in mock-depleted cells or cells depleted of Cdc20 or separase and transfected as indicated. After 12 hours, cells were synchronized and released without RO-3306 for 6 hours, or with RO-3306 for 16 hours. E. Centriole disengagement in mock-depleted cells or cells depleted of Cdh1 or APC3 and transfected as indicated. After 12 hours, cells were synchronized and released without RO-3306 for 6 hours, or with RO-3306 for 16 hours. F. IF of cells treated with either aphidicolin for 16 hours, or synchronised and treated with RO-3306 alone or plus MLN 8054, or RO-3306 following separase or Cdc20 depletion. Cells were stained for C-Nap1 (red) and Plk1-pT210 (green). G. Schematic indicating how Plk1, which is activated by Aurora A, may contribute to both APC/C-dependent and -independent centriole disengagement. Data in A-E show means ± sd; n=3, >100 cells counted. OE, overexpression.
We then looked at localization of activated endogenous Plk1 by immunofluorescence microscopy with Plk1-pT210 antibodies. Consistent with Loncarek et al. (2010), we found that activated Plk1 was present at disengaged centrioles in RO-3306 treated cells (Fig. 3F). In contrast, it was absent in the presence of the Aurora A inhibitor as expected. Importantly, though, activated Plk1 was present on centrosomes that were not disengaged in RO-3306-arrested cells depleted of Cdc20 or separase. This provides further evidence that the block to disengagement imposed by these treatments is not an indirect effect on Plk1 activation and that the presence of endogenous levels of activated Plk1 at centrosomes in G2-arrested cells is not sufficient for disengagement. Taken together, these data lead us to propose that Plk1 promotes centriole disengagement through both APC/C-dependent and independent pathways (Fig. 3G).
HU and IR induce separase- and Plk1-dependent centriole disengagement
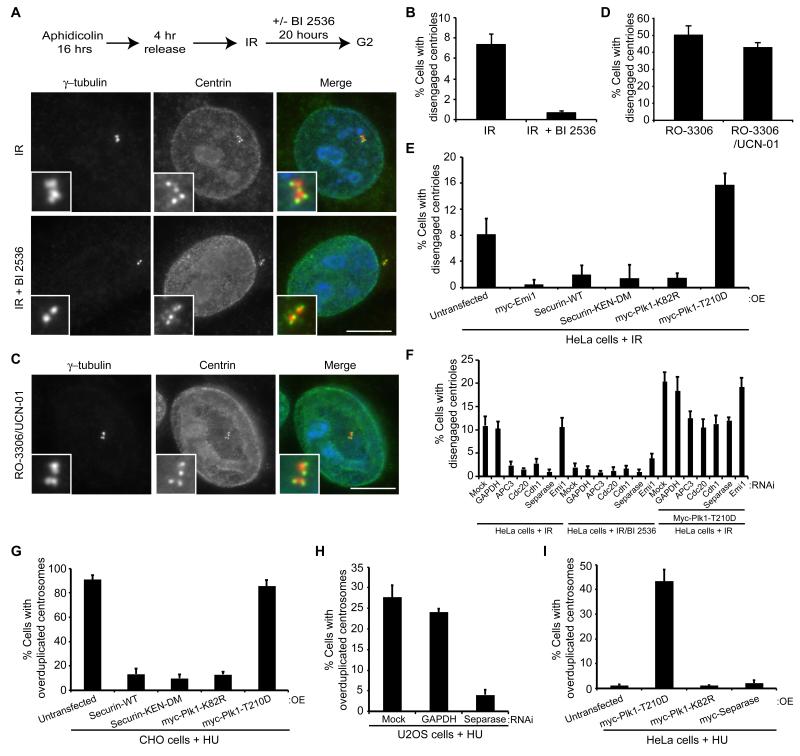
It is well established that exposure of G2 cells to IR activates the DNA damage checkpoint and leads to cell cycle arrest (Kastan and Bartek, 2004). DNA damage also triggers centrosome amplification (Bourke et al., 2007; Dodson et al., 2004; Sato et al., 2000). The mechanism by which amplification occurs is not clear, although recent work suggests that this may follow centriole disengagement (Saladino et al., 2009), whilst DNA damage during mitosis also leads to inappropriate centriole splitting (Hut et al., 2003). As this scenario is reminiscent of the response we observed to Cdk1 inhibition, we sought to confirm that IR induced centriole disengagement and examine to what extent this was dependent on Plk1 and separase. HeLa cells were synchronized and exposed to IR (5 Gy). Following a further 20 hour incubation, cells were fixed and stained with γH2AX antibodies to confirm the presence of DNA damage (Fig. S4A), as well as γ-tubulin and centrin antibodies to assess centriole engagement (Fig. 4A, B). IR exposure did promote centriole disengagement, although at levels substantially lower than following RO-3306 treatment. This is consistent with RO-3306 not simply acting as a DNA damaging agent. Indeed, although RO-3306 introduced DNA damage in a limited number of HeLa cells as measured by γH2AX staining, this did not correlate with the large number of cells in which centriole disengagement was observed (Fig. S4B-D). Furthermore, inhibiting the Chk1-dependent DNA damage response pathway with UCN-01 did not prevent centriole disengagement in response to RO-3306 (Fig. 4C, D).
Figure 4. IR- or HU-induced cell cycle arrest leads to Plk1- and APC/C-dependent centriole disengagement.
A. Schematic of treatment protocol (IR, 5 Gy) and IF of cells post-IR only or IR plus BI 2536 stained for γ-tubulin (red) and centrin (green). Merge panels include DNA (blue). Scale bar, 10 μm. B. Centriole disengagement in cells post-IR only or IR plus BI 2536. C. IF of synchronized cells treated with RO-3306 and the Chk1 inhibitor, UCN-01, for 16 hours and stained as in A. Scale bar, 10 μm. D. Centriole disengagement in cells treated as in C. E. Centriole disengagement 20 hours post-IR treatment in cells transfected with constructs indicated. F. Centriole disengagement 20 hours post-IR treatment in mock-depleted cells or cells depleted of GAPDH, APC3, Cdc20, Cdh1, Separase or Emi1 and transfected as indicated. G. Centrosome overduplication during HU arrest in CHO cells transfected with constructs indicated. H. Centrosome overduplication in HU-arrested U2OS cells depleted as indicated. I. Centrosome overduplication in HU-arrested HeLa cells transfected with constructs indicated. Data in B and D-I show means ± sd; n=2, >200 cells counted. OE, overexpression.
A likely explanation for why centriole disengagement occurs at much lower levels in response to DNA damage than RO-3306 treatment is that Plk1 is inhibited by the DNA damage checkpoint (Smits et al., 2000). Moreover, it has been demonstrated that APC/C-Cdh1 becomes activated following DNA damage in order to maintain a G2 arrest, leading to reduced Plk1 levels and degradation of cyclins A2 and B1 (Bassermann et al., 2008; Sudo et al., 2001). However, treatment with the Plk1 inhibitor, BI 2536, suppressed even the low level of centriole disengagement that occurred following IR exposure, suggesting that residual Plk1 activity was still present in these cells (Fig. 4A, B). Indeed, Plk1 has been shown to retain activity in the short-term following DNA damage, possibly phosphorylating Rad51 (Yata et al., 2012). In this case, to maintain activation of the APC/C, Emi1 is down-regulated via a p21WAF1 pathway (Lee et al., 2009; Wiebusch and Hagemeier, 2010). In our hands, expression of Emi1, securin or inactive Plk1 (Fig. 4E), or depletion of APC3, Cdc20, Cdh1 or separase (Fig. 4F) blocked IR-induced centriole disengagement consistent with a crucial role for the APC/C in this response. Furthermore, expression of constitutively active Plk1 promoted disengagement following IR even when components of the APC/C pathway were depleted (Fig. 4F and S5), again supporting an additional APC/C-independent role for Plk1.
Importantly, expression of securin or inactive Plk1 or depletion of separase blocked HU-induced centrosome amplification (i.e. cells with >4 centrioles) in CHO or U2OS cells, respectively, whilst expression of activated Plk1 drove centrosome amplification in HU-arrested HeLa cells, which do not normally undergo amplification (Fig. 4G-I). Hence, we propose a general mechanism whereby cell cycle arrest in S or G2 promotes Plk1-dependent centriole disengagement and subsequent centrosome amplification in part through activation of the APC/C and separase.
Oscillation of APC/C activity promotes centrosome amplification
As indicated above, centrosome amplification in response to DNA damage might arise through duplication of disengaged centrioles (Saladino et al., 2009). We therefore tested whether the disengaged centrioles that arise from RO-3306 treatment undergo duplication if the arrest is maintained for an extended period. After 40 hours in RO-3306, most cells still exhibited 4 γ-tubulin-stained spots, but now each of these was associated with two centrin dots such that the cells typically had an 8:4 ratio of centrin to γ-tubulin (Fig. 5A, B). This indicated that disengaged centrioles do undergo a further round of duplication upon long-term G2 arrest. Flow cytometry revealed that cells also underwent endoreduplication (Fig. 5D), as expected (Ma et al., 2009). Importantly, concurrent inhibition of Plk1 and Cdk1 prevented overduplication of centrioles consistent with Plk1 being required for disengagement, whereas addition of the Plk1 inhibitor 16 hours after the Cdk1 inhibitor blocked neither disengagement nor duplication (Fig. 5A, B). Similarly, chemical inhibition of the APC/C with the small molecule, pro-TAME (Zeng et al., 2010), blocked disengagement and overduplication if added at the same time as RO-3306, but inhibited neither if added 16 hours after RO-3306 (Fig. 5B). Hence, we conclude that Plk1 and APC/C activity are not required for re-duplication once disengagement has occurred.
Figure 5. Disengaged centrioles undergo reduplication during prolonged G2 arrest.
A. Schematic of treatment protocol and IF of cells treated with aphidicolin, RO-3306 or RO-3306 plus BI 2536 for 40 hours, and stained for γ-tubulin (red) and centrin (green). Merge panels include DNA (blue). Scale bar, 10 μm. B. Quantification of the ratio of centrin:γ-tubulin dots following 40 hours treatment with aphidicolin, RO-3306 alone, or RO3306 plus BI 2536, the Cdk2 inhibitors, NU6102 or NU6120, or the APC/C inhibitor, pro-TAME, added concurrently, or 16 hours after RO-3306. Data show means ± sd; n=3, >100 cells counted. C. IF of cells treated with RO-3306 for 40 hours, but with NU6120 added 16 hours after the start of RO-3306 treatment. Cells were stained as in A. Scale bar, 10 μm. D. Flow cytometry of asynchronous (AS) HeLa cells and those treated with aphidicolin for 40 hours, or pre-synchronised and treated with RO-3306 or RO-3306 plus BI 2536, NU6120 or roscovitine for 40 hours.
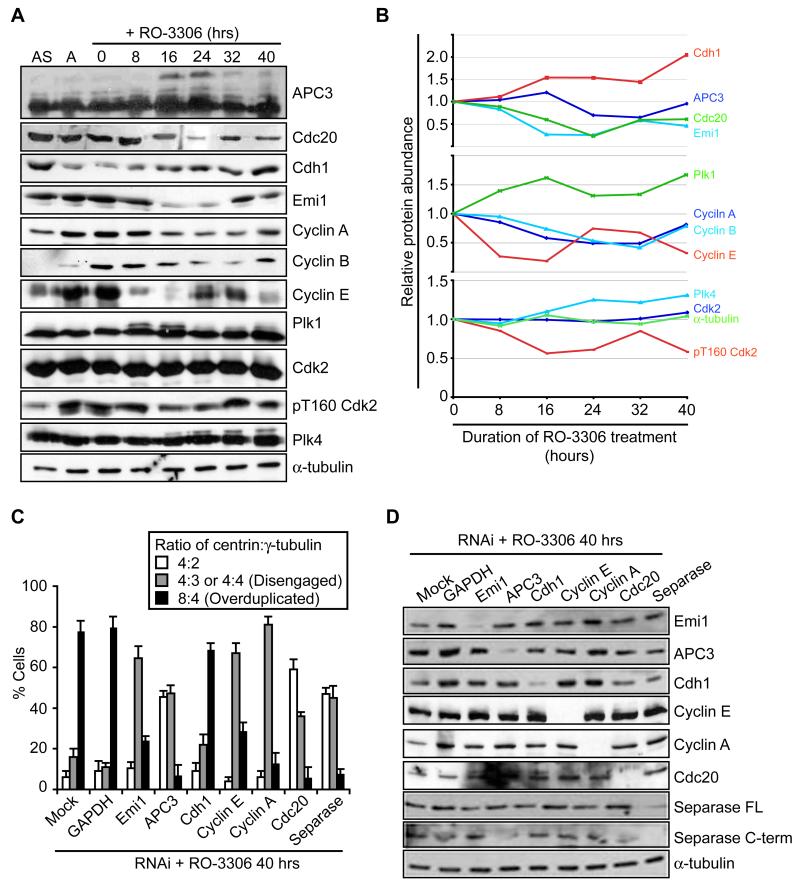
Conversely, if Cdk2 activity was inhibited, either concurrent with Cdk1 inhibition or 16 hours after addition of the Cdk1 inhibitor, then disengagement occurred, but not overduplication (Fig. 5B, C). This was shown with two different Cdk2 inhibitors (NU6102 and NU6120; Hardcastle et al., 2004). It is noteworthy, though, that disengagement occurred more slowly in the presence of the Cdk2 inhibitor than in its absence, which may well be due to APC/C activation being partially dependent on phosphorylation by Cdk2 in the absence of Cdk1 (Ma et al., 2009). The requirement for Cdk2 for overduplication implies the need to either re-accumulate cyclin A following its initial degradation, or form a complex with cyclin E. Immunoblotting revealed the re-accumulation of cyclins A and B, along with Emi1 and Cdc20, and the loss of APC3 hyperphosphorylation at longer times of arrest, all indicative of a reduction in APC/C activity (Fig. 6A, B). Furthermore, depletion of cyclin A blocked overduplication but not disengagement consistent with a role in re-activating Cdk2 (Fig. 6C, D). Additionally, depletion of Emi1 reduced the level of overduplication, suggesting that its reappearance is important for switching the APC/C off and allowing cyclin A to accumulate once more. Based on these results, we argue that even in G2-arrested cells there is a temporal separation of disengagement from duplication that is promoted by oscillation of APC/C activity. Importantly, though, cyclin E levels were also seen to oscillate, albeit with different timing to cyclin A, and depletion of cyclin E led to a partial block to centriole overduplication (Fig. 6A-D). Hence, it seems likely that Cdk2 can form complexes with both cyclins A and E to drive centriole overduplication during long-term cell cycle arrest.
Figure 6. Oscillation of APC/C activity promotes centrosome reduplication.
A. Immunoblot for proteins indicated in asynchronous (AS) cells, aphidicolin-arrested cells (A) or cells released from the aphidicolin arrest and treated with RO-3306 for the hours stated. B. Plots showing the relative protein abundance from the blots in A from 0 to 40 hours RO-3306 treatment. The abundance of each protein was quantified relative to the 0 hour sample. C. Quantification of the ratio of centrin:γ-tubulin dots following 40 hours treatment with RO-3306 in cells that were mock-transfected or transfected with siRNAs against GAPDH, Emi1, APC3, Cdh1, cyclin E, cyclin A, Cdc20 and separase. Data show means ± sd; n=3, >100 cells counted. D. Immunoblot for proteins indicated in cells treated as in C.
Premature centriole disengagement leads to unstable spindle structures
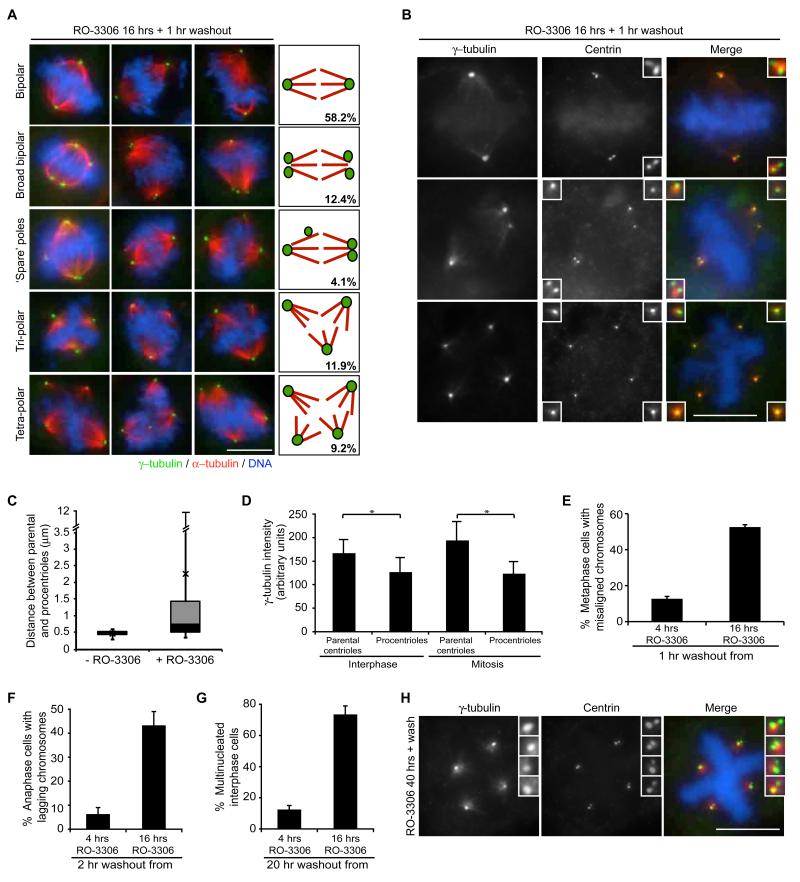
Release from RO-3306 arrest allows rapid entry into mitosis and, hence, RO-3306 has been proposed as an excellent tool for reversible synchronisation in G2 (Vassilev, 2006). However, as it also induces centriole disengagement and amplification, we revisited the consequences of entering mitosis upon drug release. Initially, HeLa cells were synchronized and arrested with RO-3306 as above, before release for 1 hour and observation by immunofluorescence microscopy. Clearly, most cells had entered mitosis confirming that this drug is readily reversible. However, a significant fraction (>40%) of abnormal spindles were observed with elevated numbers of ‘pseudo’-bipolar spindles with broad or multiple poles or even ‘spare’ poles not contributing to spindle formation, as well as more conventional tripolar and tetrapolar spindles (Fig. 7A). Staining with centrin antibodies revealed the frequent presence of isolated centrioles within the spindle and a substantial variation in the distance between centrioles within a pair (Fig. 7B, C). Electron microscopy also confirmed the presence of isolated centrioles in mitotic cells (data not shown).
Figure 7. Premature centriole disengagement leads to spindle defects in mitosis.
A. IF images and schematics illustrating mitotic spindle structures resulting from 1 hour washout from 16 hour RO-3306 treatment; mean percentages from n=3, >200 cells counted. Cells were stained for γ-tubulin (green) and α-tubulin (red). Panels include DNA (blue). B. IF of mitotic cells following 1 hour washout from 16 hour RO-3306 treatment and stained for centrin (green) and γ-tubulin (red). Panels include DNA stained with Hoechst (blue). C. Box and whiskers plot of the spread of distances between parental and procentrioles in mitotic cells released from aphidicolin arrest (-RO-3306) or RO-3306 arrest (+RO-3306). Error bars represent the minimum and maximum values, X is the mean, and the black and grey boxes represent data from the lower (Q1) to the median (Q2) quartile, and median (Q2) to the upper (Q3) quartile, respectively. Data are taken from 100 pairs of centrioles. D. Quantification of γ-tubulin intensity on parental centrioles (brightest centrin staining) and procentrioles (weaker centrin staining) in interphase and mitotic cells. Data show means of 200 centrioles ± sd. *p<0.001. E. Quantification of the percentage of bipolar metaphase cells released for 1 hour from either 4 or 16 hours RO-3306 treatment that displayed misaligned chromosomes. F. Quantification of the percentage of anaphase cells released for 2 hours from either 4 or 16 hours RO-3306 treatment that displayed lagging chromosomes. G. Quantification of the percentage of interphase cells released for 20 hours from either 4 or 16 hours RO-3306 treatment that are multinucleated. Data in E, F and G show means ± sd; n=3, >100 cells counted. H. IF of cells treated with RO-3306 for 40 hours followed by 1 hour washout and stained as in B.
Interestingly, when four γ-tubulin spots were detected, both in G2-arrested cells and cells released into mitosis, there were usually two brighter and two fainter spots. Co-staining with centrin2 confirmed that the brighter γ-tubulin spots coincided with the brighter centrin2 spots indicating that, upon centriole disengagement, the bulk of PCM is retained by the parental centriole (Fig. 7D). Indeed, the ‘spare’ poles that were not contributing to spindle formation invariably contained the procentrioles providing an explanation for the unequal microtubule organization capacity of the different poles. Moreover, even when bipolar spindles were formed, they were less efficient at chromosome congression as >50% cells released from 16 hours RO-3306 arrest exhibited misaligned chromosomes in metaphase, as compared to <15% cells released from 4 hours RO-3306 arrest (Fig. 7E, and S6A). Furthermore, >40% of anaphase cells displayed lagging chromosomes following 2 hours release from RO-3306 (Fig. 7F and S6B), whilst >70% of interphase cells in the subsequent cell cycle were multinucleated (Fig. 7G and S6C). In sharp contrast, when cells entered mitosis with reduplicated centrosomes after the extended RO-3306 arrest, almost all mitotic cells formed a robust tetrapolar spindle with two centrin spots at each pole suggesting that spindle poles composed of two centrioles have a fixed and equal microtubule organizing capacity (Fig. 7H).
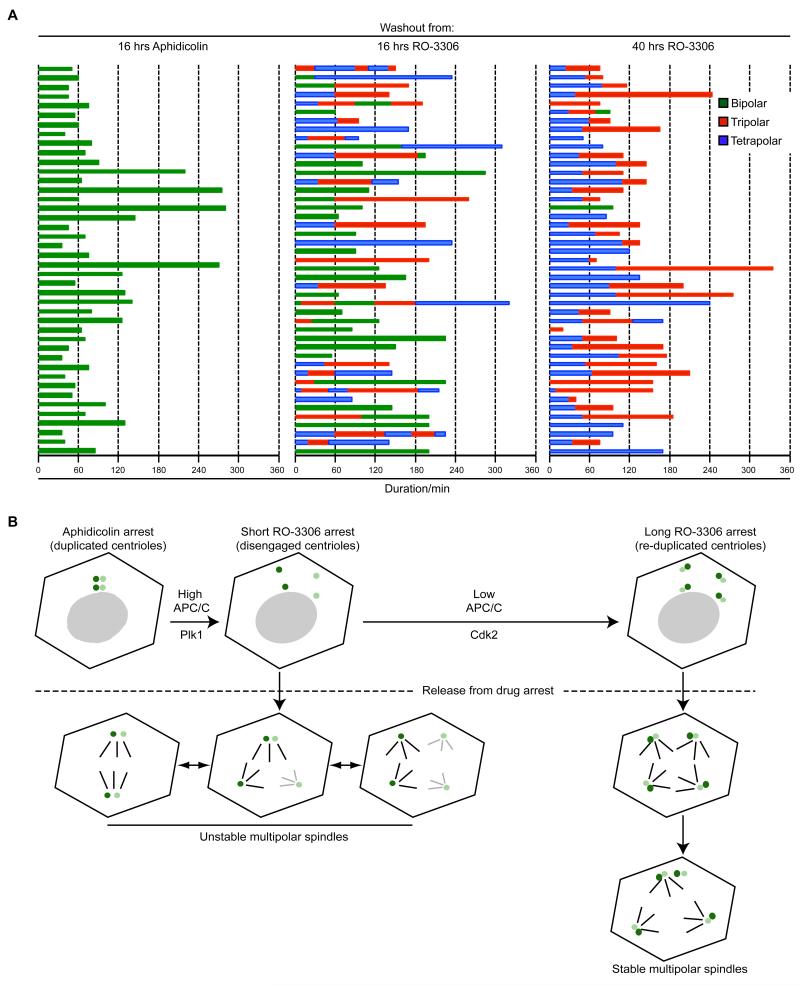
To analyse the dynamics of spindle formation in response to the different treatments, time-lapse imaging was performed with pre-synchronised HeLa cells expressing GFP-α-tubulin (Fig. 8A; and Supplementary Movies 1-6). Cells released from the aphidicolin block alone formed bipolar spindles upon mitotic entry and divided with a median time of 65 mins. On the other hand, cells entering mitosis following washout from the short-term (16 hours) RO-3306 arrest established bipolar, tripolar or tetrapolar spindles in roughly similar ratios, exchanged between these states on a frequent basis, before finally, dividing into two, three or four cells. In this case the median time in mitosis was 150 mins. Hence, cells with disengaged centrioles had difficulty in maintaining a robust spindle structure leading to prolonged mitosis. However, cells arrested with RO-3306 for sufficient time (40 hours) to allow both centriole disengagement and overduplication, almost without exception assembled a tetrapolar spindle, in line with our fixed cell observations. Interestingly, in most cases, this collapsed to a tripolar state before the cells divided with a median time in mitosis of 110 mins. We speculate that cell division with a planar tripolar spindle is more energetically favourable than division with a tetrahedral-shaped tetrapolar spindle in a 2D culture system where adhesive traction forces only occur on the ventral surface. In summary, then, spindles in which the poles all have engaged centriole pairs are more stable and promote more rapid progression through mitosis than spindles in which the poles are generated from disengaged single centrioles (Fig. 8B).
Figure 8. Engaged centriole pairs assemble stable spindle poles.
A. Live imaging was used to follow pre-synchronised HeLa:α-tubulin-GFP cells released from 16 hours aphidicolin, 16 hours RO-3306 or 40 hours RO-3306 treatment. Cells were followed for up to 7 hours following drug washout. The time that each mitotic cell spent with a bipolar (green), tripolar (red) or tetrapolar (blue) spindle was recorded for 45 cells from each condition. Each bar represents one cell. B. Schematic model showing responses of cells entering mitosis with disengaged centrioles or disengaged and reduplicated centrioles. Following a short (16 hours) RO-3306 arrest, centrioles (parental centrioles, dark green; procentrioles, light green) undergo premature disengagement as a result of high Plk1 and elevated APC/C activity. Upon drug washout and entry into mitosis these cells form a variety of spindle structures as each centriole can act as a spindle pole. However, these spindles are unstable with frequent switching between bi-, tri- and tetra-polar states, due to spindle poles with different centriole contents having different microtubule organizing capacities. In contrast, during a longer (40 hours) RO-3306 arrest, APC/C activity subsequently drops again enabling activation of Cdk2. As a result, disengaged centrioles undergo reduplication such that there are four pairs of centrioles in each cell. Upon washout, these almost invariably form tetrapolar spindles suggesting that centriole pairs have equivalent microtubule nucleating capacity. Interestingly, though, these generally resolve to tripolar spindles before division in a 2D culture system.
DISCUSSION
Centriole disengagement is a key step in the centrosome duplication cycle as it licences centrioles for the next round of duplication. Recent studies have begun to shed light on the mechanisms that regulate centriole disengagement suggesting roles for the mitotic kinase, Plk1, and the cysteine protease, separase (Schockel et al., 2011; Tsou and Stearns, 2006b; Tsou et al., 2009). Here, we demonstrate that these two enzymes also promote premature centriole disengagement and subsequent centrosome amplification in a variety of experimental and physiological situations that arrest the cell cycle in S or G2. Moreover, the activation of separase is a response to oscillating APC/C activity that occurs during cell cycle arrest (Ma et al., 2009). However, whilst Plk1 activity is required for APC/C activation, we propose that it has additional APC/C-independent role(s) in centriole disengagement. Finally, we have exploited these observations to address the consequences of premature centriole disengagement on mitotic progression, showing that disengaged centrioles form unequal spindle poles that delay mitosis and promote chromosome missegregation.
It has long been known that blocking Cdk1 activity leads to DNA endoreduplication (Hayles et al., 1994; Itzhaki et al., 1997). However, the mechanism behind this response was only recently uncovered when it was shown that loss of Cdk1 activity leads to oscillations in APC/C activity even in the absence of cell cycle progression (Ma et al., 2009). One of the key mitotic substrates of the APC/C is securin which sequesters separase to keep it inactive. We therefore postulated that separase was prematurely released and activated upon inhibition of Cdk1. Consistent with this, we found that centriole disengagement in G2-arrested cells was blocked by (i) overexpression of securin or Emi1, (ii) depletion of Cdc20, APC3 or separase, or (iii) addition of the proteasome inhibitor, MG132. Moreover, the time when centriole disengagement was first detected, around 12 hours after RO-3306 addition, coincided with the time of Emi1 and securin destruction, separase activation and APC3 hyperphosphorylation. Besides activation of the APC/C, inhibition of Cdk1 could have a second role in activating separase, as the catalytic activity of separase is inhibited by Cdk1-dependent binding of cyclin B1 (Holland and Taylor, 2006; Stemmann et al., 2001). Together, these data provide compelling evidence that centriole disengagement, whether it occurs at anaphase in an unperturbed cycle or during G2 arrest, is at least in part a result of activation of separase.
So what first activates the APC/C in response to cell cycle arrest? Clearly, one possibility is down-regulation of the interphase APC/C inhibitor, Emi1 (Di Fiore and Pines, 2007). Emi1 degradation is mediated by the SCF-βTrCP ubiquitin ligase in response to Emi1 phosphorylation by Plk1 (Hansen et al., 2004; Moshe et al., 2004). Plk1 also promotes the SCF-βTrCP-mediated degradation of Wee1 (Watanabe et al., 2004) explaining the decreases in Wee1 expression and tyrosine-15 phosphorylation of Cdk1 that were observed at this time. However, there is no evidence that cell cycle arrest directly activates Plk1; in fact, DNA damage-induced cell cycle arrest blocks Plk1 activity (Smits et al., 2000). It is possible then that the ubiquitin ligase activity of the SCF-βTrCP is elevated in RO-3306 arrested cells such that, with a certain threshold of Plk1 activity, elevated βTrCP could tip the balance in favour of Emi1 degradation and APC/C activation. Interestingly, we did observe an increase in βTrCP levels coincident with loss of Emi1 and Wee1. Emi stability, though, is also regulated by other factors, including Evi5 and Pin1 (Bernis et al., 2007; Eldridge et al., 2006), suggesting alternative routes through which loss of Cdk1 activity could lead to APC/C activation. Similarly, whilst Plk1 regulates Emi1 expression, it also phosphorylates APC/C subunits (Kraft et al., 2003), and hence can promote APC/C activation via multiple routes.
Two apparent controversies raised by this study are, firstly, that the APC/C acts in conjunction with Cdc20, rather than Cdh1, during G2 arrest, and secondly, that Cdk2 might replace Cdk1 in activating the APC/C. In relation to the first point, our data show that depletion of Cdh1 does not prevent premature centriole disengagement, whereas depletion of Cdc20 does. This strongly supports the proposal that the APC/C acts in conjunction with Cdc20 rather than Cdh1 during this form of G2 arrest. Moreover, this agrees with Ma et al. (2009) who show that Cdc20 depletion retards the degradation of APC/C substrates upon RO-3306 treatment. In contrast, we found that in response to IR both Cdc20 and Cdh1 depletion reduced centriole disengagement consistent with a proposed role for APC/C-Cdh1 during DNA damage-induced G2 arrest (Bassermann et al., 2008; Sudo et al., 2001). Hence, unsurprisingly, the nature of the G2 arrest mechanism seems to influence the cellular response. Association of Cdc20 with the APC/C is regulated by Cdk1 phosphorylation (Kraft et al., 2003). This brings us to the second controversy, namely that APC/Cdc20-mediated centriole disengagement occurs in the presence of a Cdk1 inhibitor. Importantly, concurrent addition of Cdk2 inhibitors slowed the rate of centriole disengagement implying that Cdk2 activates the APC/C. Consistent with this, Ma et al. (2009) demonstrated that Cdk2 is required for degradation of APC/C substrates, turnover of an APC/C biosensor and genome endoreduplication in the absence of Cdk1 activity. However, to our knowledge, there is no evidence that Cdk2 directly phosphorylates APC/C subunits. An alternative route through which Cdk2 could promote APC/C activation is through Emi1. Cdk2 is capable of phosphorylating Emi1, at least in vitro, reducing its binding to the APC/C (Moshe et al., 2011), whilst cyclin A was reported to be required for Emi1 turnover (Ma et al., 2009).
As indicated above, Plk1 can act upstream of separase in centriole disengagement (Tsou et al., 2009). However, our data suggest that Plk1 also has a more direct, APC/C-independent, role in centriole disengagement. Firstly, Plk1 inhibition blocked disengagement under conditions where separase should be highly active, namely when separase overexpression was combined with securin depletion. Secondly, overexpressed Plk1 was able to drive premature centriole disengagement not only in cells that were not exposed to RO-3306 but also in cells in which Cdc20, APC3 or separase were depleted. These data are consistent with observations that Plk1 can induce centriole disengagement in separase-null cells (Tsou et al., 2009), and that localization of activated Plk1 to the centrosome is required for centrosome amplification during cell cycle arrest (Inanc et al., 2010; Loncarek et al., 2010). However, endogenous levels of active Plk1 at the centrosome were not sufficient to induce disengagement in cells depleted of separase or Cdc20. Thus, as for regulation of sister chromatid cohesion, Plk1 acts both independently and in concert with separase to promote centriole disengagement.
A major goal is to identify the targets of separase and Plk1 that lead to centriole disengagement. In regulating sister chromatid cohesion, both enzymes target cohesin subunits. Plk1 phosphorylates SA2 causing dissociation of cohesin from chromosome arms in prophase, whilst it also phosphorylates Scc1 promoting its cleavage by separase at the metaphase-anaphase transition (Hauf et al., 2005; Peters et al., 2008). Frustratingly, little is understood about the molecular nature of centriole engagement. Recent EM tomography studies suggest the existence of a stalk connecting the duplicated pair of centrioles that may be composed of the SAS-5/Ana2 and SAS-6 proteins (Guichard et al., 2010; Stevens et al., 2010). These would be attractive candidates for regulation. However, there is growing evidence that cohesin subunits, and their regulators, are present at centrosomes and contribute to centriole engagement (Beauchene et al., 2010; Diaz-Martinez et al., 2010; Gimenez-Abian et al., 2010; Gregson et al., 2001; Guan et al., 2008; Kong et al., 2009; Losada et al., 2005; Nakamura et al., 2009; Thein et al., 2007; Wang et al., 2008; Wong and Blobel, 2008). Most persuasively, non-cleavable cohesin blocks centriole disengagement, while cohesin subunits engineered to contain artificial cleavage sites promote unscheduled centriole disengagement upon cleavage (Schockel et al., 2011). Thus, the weight of evidence is growing that Plk1 and separase regulate both sister chromatid separation and centriole disengagement through similar mechanisms. However, the centrosomal protein, kendrin/pericentrin, has emerged as an alternative separase target whose cleavage may also promote centriole disengagement (Matsuo et al., 2012).
Strikingly, we found that cell cycle arrest in response to either HU or IR-mediated DNA damage also led to separase-dependent premature centriole disengagement. Both these treatments are known to induce centrosome amplification but the reason has remained unclear (Balczon et al., 1995; Bourke et al., 2007). We propose that arrest in S or G2 through DNA damage checkpoint-mediated Cdk1 inactivation also leads to APC/C activation, securin degradation and activation of separase, thereby promoting centriole disengagement and centrosome amplification. This finding has wide implications as it suggests that oscillation of APC/C activity is a common response to cell cycle arrest and could have other untoward consequences, including endoreduplication and centrosome amplification, that impact genome integrity. One can speculate that the intrinsic oscillation of APC/C activity is due to the existence of proteins that are both activators and substrates of the APC/C. For example, Cdc20, is both a co-activator and substrate of the APC/C, while cyclin A, a well known APC/C substrate, is a partner of Cdk2 which, as described above, may activate the APC/C in the absence of Cdk1 (Bourke et al., 2010; Hochegger et al., 2007; Ma et al., 2009). Hence, at the simplest level, once APC/C activity increases to a certain threshold, then Cdc20 and cyclin A are degraded leading to inactivation of the APC/C; these proteins then re-accumulate leading to re-activation of the APC/C.
In reality, though, the systems level control of the cell cycle, particularly in higher eukaryotes, means that this regulation is likely to be much more complex. Analysis of cyclin E levels during the extended G2 arrest revealed that the expression of this protein also oscillated. However, it reached peak levels at a time when the APC/C was on and was degraded when the APC/C was off. Indeed, cyclin E expression is not regulated by the APC/C, but rather by the SCF in conjunction with Fbw7/Cdc4 (Koepp et al., 2001; Strohmaier et al., 2001). However, our data supports the notion that the activities of these two ubiquitin ligases are intimately linked and that a rise in the activity of one may promote loss of activity of the other (Skaar and Pagano, 2009). Of importance to our study, depletion of cyclin E did reduce centrosome overduplication during the extended arrest, albeit not to the same extent as cyclin A depletion. Assessment of Cdk2 activity through measurement of T160 phosphorylation also revealed oscillating activity. We propose that, once disengagement has occurred, both Cdk2-cyclin A and Cdk2-cyclin E have the capacity to stimulate centriole reduplication, as they do with genome endoreduplication (Ma et al., 2009). Hence, although APC/C inactivation during extended arrest may not be absolutely necessary for centriole reduplication, we argue that it occurs more efficiently in the presence of cyclin A than its absence. Plk4, which remains present in HeLa cells throughout the extended arrest, is also presumably required for centriole reduplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), although one might predict that in other cell types it would be degraded by SCF-βTrCP preventing centrosome amplification (Rogers et al., 2009).
The final conclusion from this study relates to the role of centrioles in spindle pole formation. Cells entering mitosis with disengaged centrioles assembled unstable spindles that had multiple poles with unequal microtubule organizing capacity, whereas cells entering mitosis with reduplicated centrioles formed stable spindles that had four poles of equal microtubule organizing capacity. Interestingly, this later situation allowed cells to more rapidly satisfy the spindle assembly checkpoint and complete mitosis. We interpret this to mean that engaged centriole pairs have a fixed microtubule organizing capacity that results from assembly of a defined amount of microtubule nucleating and anchoring activity. Individual centrioles can and do recruit microtubule nucleating material but the amount strictly depends on the age and maturation status of the centriole (Wang et al., 2011). What defines how much PCM is recruited to a centriole pair is an intriguing but poorly understood question, although recent studies in C. elegans and Drosophila support roles for a number of proteins in defining centrosome size (Conduit et al., 2010; Decker et al., 2011; Dix and Raff, 2007; Song et al., 2008). Intriguingly, experimentally increasing the size of the centrosome can also influence centriole configuration (Loncarek et al., 2008), highlighting the fact that these are mutually-dependent processes. It is well known that spindle poles can assemble in the absence of centrioles and establish bipolar spindles that accurately segregate chromosomes (Nigg and Raff, 2009). Whether the amount of nucleating material in “centriole-less’ poles is similar to that formed by a pair of centrioles is not clear. However, what is clear is that centriole number and arrangement are major determinants in defining spindle pole size, spindle stability and, ultimately, the fidelity of cell division.
MATERIALS AND METHODS
Cell culture and drug treatments
All media was from Invitrogen and supplemented with 10% heat-inactivated FBS, penicillin (100 IU/ml) and streptomycin (100 mg/ml). HeLa cells were cultured in MEM with 1 mM sodium pyruvate; U2OS, HeLa:α-tubulin-GFP and HeLa:centrin1-GFP cells were cultured in DMEM; CHO cells were cultured in F12-Hams; and RPE1-hTERT cells were cultured in DMEM:Ham’s F-12 with 0.25% sodium bicarbonate. All cells were cultured at 37°C in a 5% CO2 atmosphere. Cells were synchronized in G1/S with 1.6 μg/ml aphidicolin (Sigma) for 16 hours then released into fresh medium for 4 hours. To inhibit Cdk1, cells were treated with 10 μM RO-3306 (Calbiochem) or 50 μM roscovitine (Calbiochem). To inhibit Plk1 and Chk1, cells were treated with 100 nM BI 2536 (Axon Med Chem) or 2 μM UCN-01 (Sigma), respectively. 1 μM MLN 8054 was used to inhibit Aurora A. To inhibit Cdk2 10 μM NU6102 or 50 μM NU6120 was used. To inhibit the APC/C, pro-TAME was used at 12 μM. For the centrosome overduplication assay, hydroxurea (HU; Sigma) was used at 2 mM on CHO cells or 8 mM on U2OS cells for 48 hours. Overduplication was scored as cells with >4 centrioles as determined by centrin staining. MG132 (Sigma) was used at 10 μM.
RNAi and transient transfections
siRNAs were transfected using Oligofectamine or Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. siRNA oligos were from Dharmacon unless indicated. Securin was targeted with 5′-GACCCUGGAUGUUGAAUUG-3′, 5′-GCACCCGUGUGGUUGCUAA-3′, 5′-UGGGAGAUCUCAAGUUUCA-3′ and 5′-GCUGUGACAUAGAUAUUUA-3′. Cdk1 was targeted with 5′-GUACAGAUCUCCAGAAGUA-3′, 5′-GAUCAACUCUUCAGGAUUU-3′, 5′-GGUUAUAUCUCAUCUUUGA-3′ and 5′-GAACUUCGUCAUCCAAAUA-3′. Cdc20 was targeted with 5′-AAUGGCCAGUGGUGGUAAUGA-3′. Cyclin A2 was targeted with 5′-AACUACAUUGAUAGGUUCCUG-3′ and 5′-AAGGCAGCGCCCGUCCAACAA-3′. Cyclin E was targeted with 5′-AAUAAUGCAGUCUGUCCAGAC-3′. Emi1 was targeted with 5′-GAUUGUGAUCUCUUAUUAA-3′ and 5′-ACUUGCUGCCAGUUCUUCA-3′. Cdh1 was targeted with 5′-UGAGAAGUCUCCCAGUCAGUU-3′. APC3 was targeted with 5′-GGAAAUAGCCGAGAGGUAAUU-3′ and 5′-CAAAAGAGCCUUAGUUUAAUU-3′. Separase was targeted with 5′-AAGCUUGUGAUGCCAUCCUGA-3′ (Qiagen). Plk1 was targeted with 5′-CCAUUAACGAGCUGCUUAA-3′ (Ambion). Oligos against GAPDH were from Ambion (AM4631). Plasmid transfections or combined siRNA and plasmid transfection were performed using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. At 12 hours post-transfection, cells were treated with aphidicolin for 16 hours. After release from block for 4 hours, cells were treated with RO-3306 for a further 16 hours. An RNAi resistant mutant of separase (myc-Separase-Res) was generated using site-directed mutagenesis (QuikChange II, Stratagene) and the primers: 5′-CTGAGAGGAGACAAGCTTGCGACGCAATACTGAGGGCTTG-3′ and 5′-CAAGCTTGTCTCCTCTCAGCATCAGATCG-3′.
Cell extracts and immunoblotting
Cell lysates were prepared and analysed by immunoblot as previously described (Faragher and Fry, 2003). Primary antibodies used were against separase (Abcam), α-tubulin (Sigma), GAPDH (Cell Signaling), cyclin A (Santa Cruz), cyclin B (Santa Cruz), Cdk1 (Cell Signaling), Cdk1-pTyr15 (Cell Signaling), Cdk2 (Santa Cruz), securin (Abcam), myc (Cell Signaling), Cdc20 (Santa Cruz), Plk1 (Sigma), Wee1 (Santa Cruz), Emi1 (Invitrogen), β-TrCP (Invitrogen), APC3/Cdc27 (BD Bioscience), Cdk2 (Santa Cruz), Cdk2-pT160 (Cell Signaling), Cyclin E (Abcam), Plk4 (Kleylein-Sohn et al., 2007) and Cdh1 (gift from J. Gannon & T. Hunt). Blots were quantified using ImageJ (v. 1.41) by measuring the intensity of each band and subtracting the background from a corresponding area of the same lane. The 0 hour time-point of RO-3306 treatment was set at 1 and subsequent time-points measured relative to this.
Immunofluorescence microscopy and flow cytometry
Cells were extracted with 0.2% NP40 in 80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA for 30 seconds before fixation with methanol. Immunofluorescence microscopy was performed as described (Faragher and Fry, 2003) using primary antibodies against γ-tubulin (Sigma), Centrin2 N17 (Santa Cruz), C-Nap1 (Fry et al., 1998), Plk1-pT210 (BD Bioscience), γH2AX (Abcam) and α-tubulin (Sigma). Secondary antibodies used were AlexaFluor-488 and -594 goat anti-rabbit and goat anti-mouse IgGs (Invitrogen); DNA was stained with Hoechst 33258 or DAPI. Images were captured as a single focal plane on a Nikon TE300 microscope equipped with a Hamamatsu ORCA-R2 digital camera, 100x objective, NA 1.4, or Olympus BX51 microscope, 100x objective, NA 1.35, using Openlab or Volocity software (Improvision). Quantification was performed using ImageJ (v. 1.41). For flow cytometry, cells were fixed in 70% ethanol and stained with propidium iodide (40 μg/ml) in PBS containing 100 μg/ml RNase A. Cell cycle analysis was performed on a FACScan Flow Cytometer or FACScanto II (BD Biosciences) with Cell Quest or FACSDiva software, respectively.
Irradiation
Cells were synchronised by aphidicolin treatment and release before receiving 5 Gy IR. Where indicated cells were additionally treated with BI 2536. Cells were fixed and analysed 20 hours post-IR.
Live cell imaging
Time-lapse imaging was performed as described (Prosser et al., 2009) using a Leica TCS SP5 confocal equipped with a Leica DMI 6000B inverted microscope and 63x oil objective (NA 1.4). Cells were cultured in glass-bottomed dishes (no. 1.5; MatTek) and maintained on the stage at 37°C in a 5% CO2 atmosphere. Drug washout was achieved by rapid media replacement with pre-warmed OptiMEM (Invitrogen) containing 10% FBS immediately prior to the start of imaging. Z-stacks comprising of thirty 1 μm steps were acquired every 5 minutes for at least 9 hours. Stacks were processed into maximum intensity projections using LAS-AF software (Leica) and movies prepared using Image J (v. 1.41).
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful for gifts of myc-Emi1 from P. Jackson (San Francisco), securin-WT and securin-KEN-DM from M. Brandeis (Jerusalem), Plk4 antibodies, myc-Plk1-T210D and myc-Plk1-K82R from E. Nigg (Basel), myc-Plk1-WT from F. Barr (Liverpool), myc-separase from I. Nabti (London), NU6102 and NU6120 from R. Griffin (Newcastle), pro-TAME from R. King (Harvard), MLN 8054 from R. Bayliss (Leicester) and Cdh1 antibody from J. Gannon and T. Hunt (South Mimms). This work was supported by grants to A.M.F. from Cancer Research U.K., The Wellcome Trust and the Association for International Cancer Research (AICR), and Science Foundation Ireland Principal Investigator Award 10/IN.1/B2972 to C.G.M.
REFERENCES
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–15. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–67. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchene NA, Diaz-Martinez LA, Furniss K, Hsu WS, Tsai HJ, Chamberlain C, Esponda P, Gimenez-Abian JF, Clarke DJ. Rad21 is required for centrosome integrity in human cells independently of its role in chromosome cohesion. Cell Cycle. 2010;9:1774–80. doi: 10.4161/cc.9.9.11524. [DOI] [PubMed] [Google Scholar]
- Bernis C, Vigneron S, Burgess A, Labbe JC, Fesquet D, Castro A, Lorca T. Pin1 stabilizes Emi1 during G2 phase by preventing its association with SCF(betatrcp) EMBO Rep. 2007;8:91–8. doi: 10.1038/sj.embor.7400853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–63. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bourke E, Brown JA, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29:616–24. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–9. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. Processing, localization, and requirement of human separase for normal anaphase progression. Proc Natl Acad Sci U S A. 2003;100:4574–9. doi: 10.1073/pnas.0730733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, Raff JW. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20:2178–86. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, Hyman AA. Limiting Amounts of Centrosome Material Set Centrosome Size in C. elegans Embryos. Curr Biol. 2011;21:1259–67. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–9. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J Cell Biol. 2007;177:425–37. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Beauchene NA, Furniss K, Esponda P, Gimenez-Abian JF, Clarke DJ. Cohesin is needed for bipolar mitosis in human cells. Cell Cycle. 2010;9:1764–73. doi: 10.4161/cc.9.9.11525. [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol. 2007;17:1759–64. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. Embo J. 2004;23:3864–73. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell. 2006;124:367–80. doi: 10.1016/j.cell.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–89. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Diaz-Martinez LA, Beauchene NA, Hsu WS, Tsai HJ, Clarke DJ. Determinants of Rad21 localization at the centrosome in human cells. Cell Cycle. 2010;9:1759–63. doi: 10.4161/cc.9.9.11523. [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14:1187–93. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276:47575–82. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- Guan J, Ekwurtzel E, Kvist U, Yuan L. Cohesin protein SMC1 is a centrosomal protein. Biochem Biophys Res Commun. 2008;372:761–4. doi: 10.1016/j.bbrc.2008.05.120. [DOI] [PubMed] [Google Scholar]
- Guichard P, Chretien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. Embo J. 2010;29:1565–72. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–6. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15:5623–34. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle IR, Arris CE, Bentley J, Boyle FT, Chen Y, Curtin NJ, Endicott JA, Gibson AE, Golding BT, Griffin RJ, et al. N2-substituted O6-cyclohexylmethylguanine derivatives: potent inhibitors of cyclin-dependent kinases 1 and 2. J Med Chem. 2004;47:3710–22. doi: 10.1021/jm0311442. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–22. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–4. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, Hunt T, Takeda S. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178:257–68. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Taylor SS. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci. 2006;119:3325–36. doi: 10.1242/jcs.03083. [DOI] [PubMed] [Google Scholar]
- Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell. 2003;14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanc B, Dodson H, Morrison CG. A Centrosome-autonomous Signal That Involves Centriole Disengagement Permits Centrosome Duplication in G2 Phase after DNA Damage. Mol Biol Cell. 2010;21:3866–77. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki JE, Gilbert CS, Porter AC. Construction by gene targeting in human cells of a "conditional’ CDC2 mutant that rereplicates its DNA. Nat Genet. 1997;15:258–65. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–75. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Kong X, Ball AR, Jr., Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol Biol Cell. 2009;20:1289–301. doi: 10.1091/mbc.E08-04-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci U S A. 1999;96:2817–22. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim JA, Barbier V, Fotedar A, Fotedar R. DNA damage triggers p21WAF1-dependent Emi1 down-regulation that maintains G2 arrest. Mol Biol Cell. 2009;20:1891–902. doi: 10.1091/mbc.E08-08-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol. 2010;20:1277–82. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–8. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–42. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–41. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Ma HT, Tsang YH, Marxer M, Poon RY. Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis. Mol Cell Biol. 2009;29:6500–14. doi: 10.1128/MCB.00669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–23. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. Kendrin Is a Novel Substrate for Separase Involved in the Licensing of Centriole Duplication. Curr Biol. 2012 doi: 10.1016/j.cub.2012.03.048. [DOI] [PubMed] [Google Scholar]
- Mazia D. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int Rev Cytol. 1987;100:49–92. doi: 10.1016/s0074-7696(08)61698-8. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Moshe Y, Bar-On O, Ganoth D, Hershko A. Regulation of the action of early mitotic inhibitor 1 on the anaphase-promoting complex/cyclosome by cyclin-dependent kinases. J Biol Chem. 2011;286:16647–57. doi: 10.1074/jbc.M111.223339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci U S A. 2004;101:7937–42. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187:607–14. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–65. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–23. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29:1760–73. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–39. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladino C, Bourke E, Conroy PC, Morrison CG. Centriole separation in DNA damage-induced centrosome amplification. Environ Mol Mutagen. 2009;50:725–32. doi: 10.1002/em.20477. [DOI] [PubMed] [Google Scholar]
- Sato N, Mizumoto K, Nakamura M, Ueno H, Minamishima YA, Farber JL, Tanaka M. A possible role for centrosome overduplication in radiation-induced cell death. Oncogene. 2000;19:5281–90. doi: 10.1038/sj.onc.1203902. [DOI] [PubMed] [Google Scholar]
- Schockel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–72. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–8. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–24. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–6. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- Song MH, Aravind L, Muller-Reichert T, O’Connell KF. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev Cell. 2008;15:901–12. doi: 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere N, Wagner M, Beishir S, Smith E, Breslin L, Morrison CG, Hochegger H, Kuriyama R. Centrosome amplification in CHO and DT40 cells by inactivation of cyclin-dependent kinases. Cytoskeleton. 2011;68:446–458. doi: 10.1002/cm.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–26. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell. 2010;19:913–9. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–96. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–22. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Sudo T, Ota Y, Kotani S, Nakao M, Takami Y, Takeda S, Saya H. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. Embo J. 2001;20:6499–508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178:345–54. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006a;18:74–8. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006b;442:947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–54. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–9. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 2006;5:2555–6. doi: 10.4161/cc.5.22.3463. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–5. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–58. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193:727–39. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008;14:331–41. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch L, Hagemeier C. p53- and p21-dependent premature APC/C-Cdh1 activation in G2 is part of the long-term response to genotoxic stress. Oncogene. 2010;29:3477–89. doi: 10.1038/onc.2010.99. [DOI] [PubMed] [Google Scholar]
- Wong RW, Blobel G. Cohesin subunit SMC1 associates with mitotic microtubules at the spindle pole. Proc Natl Acad Sci U S A. 2008;105:15441–5. doi: 10.1073/pnas.0807660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata K, Lloyd J, Maslen S, Bleuyard JY, Skehel M, Smerdon SJ, Esashi F. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol Cell. 2012;45:371–83. doi: 10.1016/j.molcel.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.