Abstract
Accurate segregation of the replicated genome during cell division depends on dynamic attachments formed between chromosomes and the microtubule polymers of the spindle. Here we review recent advances in mechanistic analysis of microtubule attachment formation and regulation.
Introduction
Accurate genome distribution during cell division requires dynamic attachments between kinetochores, proteinaceous structures assembled on the centromeric regions of chromosomes, and spindle microtubules. Kinetochores harness the forces generated by microtubule dynamics to drive chromosome segregation and ensure chromosome biorientation—the state where sister chromatids are attached to microtubules from opposite spindle poles. Here we review recent advances in mechanistic analysis of kinetochore-microtubule attachment formation and regulation. Due to space constraints, we do not discuss the chromatin-proximal features important for building microtubule attachment sites [1–3].
Current Views of the Kinetochore-Microtubule Interface
EM tomography, super-resolution imaging, EM of purified kinetochore complexes, and atomic structures of kinetochore proteins are providing increasingly detailed views of the kinetochore-microtubule interface. EM tomography of cultured cells has revealed an amorphous interface between plus ends of spindle microtubules and the kinetochore (Fig.1A) [4,5]. The primary feature revealed by this approach is flared protofilaments at attached plus ends, which appear to connect to chromatin by slender fibrils, whose molecular composition is unclear [4]. The curvature of the protofilament flaring is distinct for kinetochore microtubules, compared to non-kinetochore spindle microtubules, consistent with a special coupling interface. EM tomography across multiple organisms has revealed similar flared protofilaments at kinetochore-attached plus ends [6].
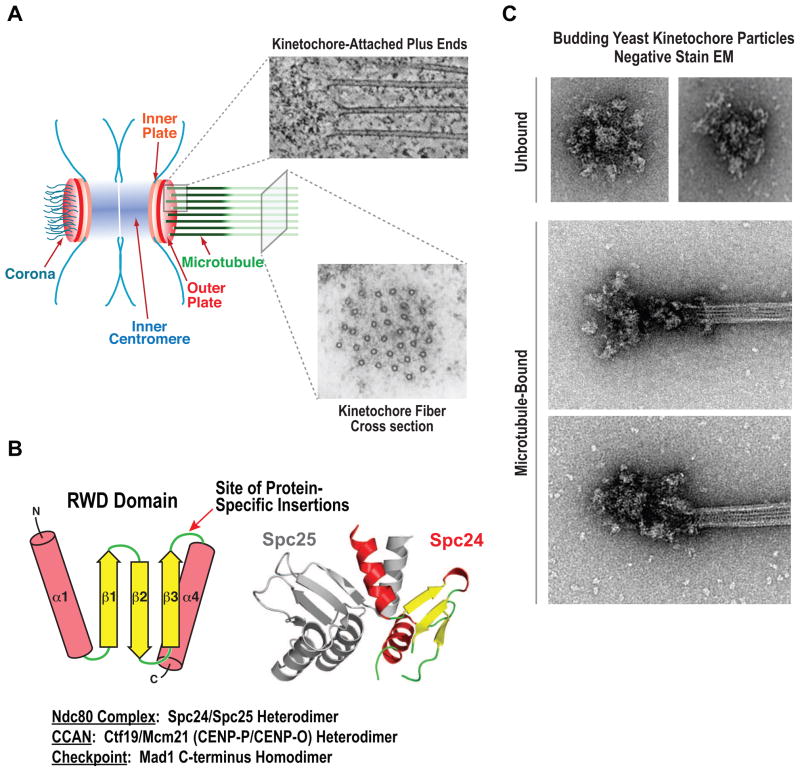
Figure 1. Different Resolution Views of Kinetochore Architecture.
(A) A slice from an EM tomogram of plus ends embedded in the outer kinetochore (top; courtesy of R. McIntosh) and a thin cross-sectional view of a cold-stable kinetochore fiber (courtesy of C. Rieder; ref. 20). The schematic on the left highlights a plate-like architecture at the kinetochore evident in older EM studies whose existence has come under debate following the development of new EM preservation methods [see ref. 5]. (B) Schematic of the RWD domain that recurs in multiple kinetochore proteins. The domain is always present in 2 copies – either as a heterodimer or as a homodimer. The structure of the Spc24/25 heterodimer from the Ndc80 complex, the first kinetochore components found to harbor this domain, is shown on the right (PDB 2FTX); Spc24 has a minimal RWD domain—the other kinetochore protein listed below harbor different insertions in the loop indicated by the arrow. (C) Negative stain EM images of purified native yeast kinetochore-like particles. Images of particles on their own and bound to microtubule ends are shown (courtesy of S. Biggins; ref 19].
Complementing the EM analysis in vivo is super-resolution imaging with probes to specific kinetochore proteins—this approach has generated positional maps of individual proteins and revealed broad conservation of the architecture and composition of the kinetochore-microtubule interface[7–10]. There has been some controversy about the number of specific molecules per microtubule attachment site measured by fluorescence microscopy—earlier work suggested approximately 8 core microtubule-binding Ndc80 complexes (see below) per kinetochore microtubule but recent work has suggested that this number may actually ~20 [11]—additional work is needed to definitively address this important question. Multivalency of attachment complexes suggested by these value ranges is now widely accepted as being critical for generating a dynamic interface with spindle microtubules although the detailed physical mechanism remains an active topic of investigation.
In addition to the EM and superresolution imaging, atomic resolution views are now available for a significant number of kinetochore parts, including the key microtubule binding complexes and the components linking these complexes to the chromatin [1,2,12]. A specific domain, referred to as the RWD domain, has emerged as a common element of functionally distinct kinetochore proteins, potentially reflecting their common origin (Fig.1B) [13–17]. While we are still some way from placing atomic structures into the picture of an intact kinetochore, the possibility of doing so has been greatly advanced by the purification of native kinetochore-like particles from budding yeast [18,19]. EM of these particles has revealed a central hub surrounded by globular domains (Fig.1C). In the presence of microtubules these particles adopt different conformations consistent with multivalent attachments being formed by the globular domains (Fig.1C). Fitting molecular views of well-studied components into the various elements observed in these striking images is an important future goal.
Though much of the work in the past decade has focused on the kinetochore itself, earlier studies showed that the kinetochore fiber, comprised of multiple stable microtubules that extend from kinetochore to the spindle pole, is highly organized (Fig.1A)[20]. Recent work suggests that the vesicle coat protein clathrin contributes to this organization, providing a mitotic function distinct from its well-studied interphase role [21–23]. Other proteins whose primary studied role is outside of mitosis, such as the Y-complex of nucleoporins and the actin-nucleating formins, also target to and function at kinetochores, highlighting an emerging importance of “moonlighting” activities in chromosome segregation [24,25].
Force Generation at the Kinetochore
Genetic analysis in multiple organisms has cemented the much-debated view that microtubule dynamics, rather than motor activity, is the primary driver of chromosome movement [26]. In support of this, in vitro experiments estimate that a single depolymerizing microtubule can generate 30–65pN force [27]. In classic experiments performed in grasshopper spermatocytes, the force needed to stall a chromosome in motion was estimated to be ~ 700 piconewtons (pN) [28]. A recent study that revisited this issue in the same experimental system using an optical trap has suggested that the stall force may be 100 times less than what was originally measured [29]. New force measurement experiments are urgently needed in vivo to resolve this large discrepancy. Regardless, the coupling interface between the kinetochore and dynamic microtubules must transduce sufficient force to the chromatin to drive chromosome movement.
The kinetochore-like particles purified from budding yeast are providing important new insights into the biophysical properties of kinetochore-microtubule interactions. Optical trapping of beads coated with these particles has revealed a catch bond‘-like force-dependent stabilization of attachments [18]. This finding suggests a first-principles model for selective stabilization of bi-oriented attachments that are being pulled towards opposite spindle poles. Tension-based modulation of microtubule dynamics has also been documented during metaphase oscillations in vertebrate kinetochores [30]. Though the identity of molecular enforcers of tension-dependent stabilization remains unclear, super-resolution imaging has shown that kinetochore conformation/organization is altered in response to microtubule dynamics [9,31,32]. In addition, recent work analyzing vertebrate kinetochore structure during metaphase oscillations has shown that the kinetochore is pliant and undergoes compression while moving poleward, potentially due to differentially positioned active and passive force-generating microtubule attachment sites [33]. As oscillations are not a universal feature of attached chromosomes, potentially the passive site positioned further out from the chromatin represents a conserved coupling point. Determining the molecular basis for force-dependent attachment stabilization and the dynamic conformational changes observed within the kinetochore are challenging but important avenues to explore in the future.
New Insights into the Primary Conserved Mediator of Kinetochore-Microtubule Interactions: The Ndc80 Complex
The 4-subunit Ndc80 complex is the primary mediator of dynamic attachments at the kinetochore [34,35]. The microtubule-binding activity of the complex resides in heterodimers of Ndc80 and Nuf2 subunits whose N termini fold into calponin homology (CH) domains. Given its central importance in chromosome segregation and ease of reconstitution, a number of structural and biophysical studies have been conducted on the Ndc80 complex. Early work revealed that microtubule-binding activity resides in the CH domains of Ndc80 and Nuf2 and in the basic N-terminal tail of Ndc80, predicted to be unstructured and targeted for phosphorylation by Aurora B kinase [36]. High resolution cryo-EM of Ndc80 complex-decorated microtubules revealed that the Ndc80 CH domain is in direct contact with the microtubule lattice (reviewed in [12]) [37,38]. Consistent with this, disruption of the interface residues on the Ndc80 CH domain abrogate microtubule binding in vivo [39,40]. A recent higher resolution cryo-EM analysis of the Ndc80-microtubule interface map points to a more complex multimodal interaction with additional points of contact involving the tail and the Nuf2 CH domain [41]. However, the precise roles of the N-terminal tail and of the Nuf2 CH domain in vivo are unclear. Tail deletion of Ndc80 in budding yeast does not affect viability whereas in human cells a similar deletion prevents kinetochore fiber formation [42–44]. Mutations in the Nuf2 CH domain appear to cause only mild defects in cultured human cells even though microtubule binding is impaired by these mutations to the same extent as Ndc80 CH domain mutations in vitro [39]. One clue into the origin of differing outcomes of similar Ndc80 complex perturbations in different species has come from biophysical experiments—e.g., unlike the budding yeast Ndc80 complex, human Ndc80 complex by itself stabilizes microtubule ends by promoting rescue [45–47]. Developing a unified conceptual framework for the mechanistic contributions of the N-terminal Ndc80 tail and the Nuf2 CH domain function is essential given the central role of the Ndc80 complex at the kinetochore.
Cooperators of the Ndc80 Complex: Different Flavors, Different Mechanisms?
An emerging theme in recent years is that the Ndc80 complex needs cooperators to generate efficient coupling of the kinetochore to dynamic microtubule ends. Surprisingly, the cooperators appear to be distinct in different species, a feature that is somewhat disconcerting and the explanation for which remains unclear. All of the cooperators described to date are characterized by their dependency on the Ndc80 complex for kinetochore localization. To date, these cooperators include bona fide dynamic end couplers like the Dam1 and Ska complexes[48–50], microtubule dynamics modulators like XMAP215 [51,52], and, somewhat surprisingly, the DNA replication factor Cdt1 [53]. The yeast Dam1 complex, which oligomerizes into rings/spirals encircling the microtubule lattice, is the best characterized Ndc80 cooperator [48,49,54–56]. However, a direct physical connection between the Dam1 and Ndc80 complexes has not been observed. Though the precise mechanism behind this cooperativity is still being investigated, a recent in vitro study suggests that the concerted action of a fibrillar element and a ring-based coupler provides the ideal coupling geometry for transducing force generated by depolymerization of a microtubule end [57].
Although metazoans lack the Dam1 complex, the 3-subunit Ska complex, which does not exhibit primary sequence similarity to Dam1 complex subunits, is emerging as a functional counterpart [58]. The Ska complex is a microtubule end coupler similar to Dam1 and can bind curved protofilament rings that mimic depolymerizing ends[50]. Structural work indicates that the Ska complex forms a W-shaped dimer with a winged helix motif, commonly found in DNA-binding proteins, imparting microtubule-binding activity to the outer arms [59]]. Similar to Dam1, Ska does not exhibit a direct interaction with the Ndc80 complex but enhances Ndc80 microtubule binding and its ability to track depolymerizing ends [50]. In vivo, kinetochore-microtubule interactions are compromised in Ska-inhibited human cancer cells but the complex is also implicated in controlling anaphase onset and is dispensable for viability in chicken cells [58,60,61]. Further work is needed to integrate the in vitro and in vivo analysis of Ska complex – Ndc80 complex cooperation.
Cdt1 in mammalian cells and fission yeast XMAP215 family members Dis1/TOG1 and Alp14 have also emerged as Ndc80 complex cooperators [51–53]. Although both Cdt1 and Dis1 are proposed to associate with the NDC80 loop, a short region that breaks the NDC80 coiled coil, different mechanisms have been proposed for how they stabilize Ndc80-mediated attachments. Based on super resolution microscopy, Cdt1 has been suggested to stabilize an extended confirmation of Ndc80 and enhance its microtubule binding. Dis1/Alp14 are members of the well-studied XMAP215 family of microtubule dynamics regulators and presumably a locally enriched pool at the kinetochore stabilizes bound microtubules [62]. In vitro studies, similar to those performed for Dam1 and Ska complexes mixed with Ndc80 complexes, will be important to understand the precise means by which Cdt1 and the XMAP215 family proteins cooperate with the Ndc80 complex. More importantly, the reason for the diversity of Ndc80 cooperators, despite the conservation of the Ndc80 complex, needs to be addressed.
Regulation of Kinetochore-Microtubule Attachments: From Mechanisms to Origins of Chromosomal Instability
During prometaphase, kinetochores initially interact laterally with the microtubule lattice [63]. These initial lateral interactions accelerate microtubule capture and help the kinetochore achieve the proper orientation to form stable end-coupled attachments that generate tension [64,65]. The formation and stability of end-coupled attachments are tightly regulated since incorrect attachments lead to lagging chromosomes and segregation errors. Multiple studies suggest that precise regulation of kinetochore-microtubule attachments involves interplay between kinases and phosphatases that control kinetochore composition and microtubule-binding properties of their kinetochore substrates.
The best-studied regulator of kinetochore-microtubule attachment stability is the Aurora B kinase, which is proposed to be the primary tension sensor at the kinetochore. Aurora B kinase promotes turnover of microtubule attachments by directly altering the microtubule binding properties of its substrates and regulating the recruitment of multiple proteins to the kinetochore [66–69]. Biophysical assays are providing valuable insight into the mechanism by which Aurora B kinase-mediated phosphorylation promotes kinetochore-microtubule turnover. Experiments employing phosphomimetic substitutions suggest that Aurora B-mediated phosphorylation does not simply detach the Ndc80 complex or yeast kinetochore-like particles from a microtubule; instead, phosphorylation reduces tip stabilization leading to disassembly of the bound microtubules and subsequent detachment [46,47].
Aurora B is enriched at the inner centromere region inbetween the sister kinetochores (Fig. 1A). The localization of the kinase between sister kinetochores, the compliance of centromeric chromatin, and the detrimental effects of forced localization of the kinase to the kinetochore led to a model in which attachment stability was a function of substrate proximity to the kinase at the centromere [70]. In this model, tension stabilized attachments by spatially displacing substrates from the influence of the kinase. This model has been challenged by analysis of a mutant that is defective in localizing Aurora B to centromeres in budding yeast, which surprisingly nonetheless exhibited normal tension-sensitive regulation of attachments [71]; earlier work perturbing one of the two known mechanisms involved in Aurora B centromere targeting in chicken cells suggest that this also may be true to some extent in vertebrates [72]. Thus, the mechanism by which Aurora B discriminates between correctly bioriented and incorrectly attached kinetochores remains an open question. Protein phosphatase 1 (PP1) has been known for a long time to genetically oppose Aurora B in budding yeast [73]. Both the outer kinetochore protein Knl1 (Spc105 in budding yeast) and the motor protein CENP-E harbor PP1 docking sites that are themselves Aurora B targets [74,75]. Progressive recruitment of PP1 following microtubule attachment to the kinetochore, by a mechanism that remains unclear, was proposed to stabilize bi-oriented microtubule attachments [74,76]. However, at least in budding yeast, the importance of PP1 to attachment stabilization appears to be limited [77]. Thus, the precise interplay between attachment, tension, Aurora B and PP1 remain unclear. We speculate that discrimination of correctly bioriented (under tension) versus incorrect (tensionless) attachments is intrinsic to the kinetochore and requires activated Aurora B. This speculation is based on the observation that bioriented kinetochores bound to dynamic microtubules are in a very different structural state compared to taxol-treated non-dynamic kinetochores [9]; this structural transition may control susceptibility of the kinetochore-microtubule interface to the action of active Aurora B. Inner centromere-targeted Aurora B likely has roles that are also important in segregation, e.g. in protecting against merotely and in centromeric cohesion [71,78,79] that may explain the widely conserved localization pattern.
In recent years, the mitotic kinase Plk1 has emerged as an additional major regulator of kinetochore-microtubule attachments [80,81]. Plk1, unlike Aurora B, has many distinct roles preceding anaphase (as well as post-anaphase functions), which makes it difficult to study and explains the greater attention that has been paid to Aurora B. Plk1 is enriched on kinetochores before biorientation, is reduced following microtubule attachment, and has multiple kinetochore substrates, identified in proteomic studies [82–84]. An appealing mechanism proposed for how Plk1 stabilizes attachments is via phosphorylation of BubR1, a dual function pseudokinase involved in checkpoint signaling and chromosome segregation [85]. Phosphorylation of a specific region in BubR1 by Plk1 promotes interaction with the PP2A-B56a phosphatase that stabilizes kinetochore-microtubule attachments, potentially via counteraction of Aurora B [86,87]. Consistent with this, a recent study reported that tethering constitutively Plk1 to the kinetochore stabilized kinetochore-bound microtubules [88]. However, as Plk1 has multiple targets enriched at kinetochores, which include stabilizers and destabilizers of microtubules, a more complex view of how Plk1 controls kinetochore-microtubule interactions is already emerging [89–92]. One challenge limiting analysis of Plk1 has been an inability to precisely perturb its localization to the kinetochore. Kinetochore recruitment of Plk1 is thought to involve Cdk1-, Aurora B- and Plk1-dependent priming of multiple kinetochore proteins [93–95]. Nonetheless, it is our opinion that the primary Plk1 docking site at the kinetochore remains to be identified.
In addition to the local regulation of attachment stability at the kinetochore-microtubule interface, global control of kinetochore-microtubule turnover has emerged as an important contributor to error-free segregation. In mammalian cultured cells, kinetochore microtubules switch from a dynamic state in prometaphase to a more stable state in metaphase; this change is dependent on degradation of Cyclin A during prometaphase [96] and is reminiscent of the well-known metaphase-anaphase transition, when there is additional stabilization of attachments following degradation of Cyclin B [97]. Thus Cyclin A appears to create a less stable kinetochore-microtubule interface during prometaphase, presumably to facilitate error correction. The mechanism by which Cyclin A-Cdk1 globally controls kinetochore-bound microtubule dynamics will be important to elucidate to integrate this new finding into the prior studies focused on control at individual kinetochores. Potentially contributing to the prometaphase-metaphase and metaphase-anaphase transitions in attachment stability is the Astrin-SKAP microtubule-binding complex that is recruited to kinetochores following chromosome bi-orientation [98–100].
Studies in human cells have also revealed important roles for motor proteins in kinetochore microtubule stability. The kinesin-8 family member Kif18, a processive plus end motor that suppresses end dynamics, confines kinetochores to the middle of the spindle by limiting the extent of dynamic transitions [101–104]. The kinesin-13 family of depolymerases, enriched between sister kinetochores, reduce the likelihood of merotelic attachments, where a single kinetochore connects to opposite spindle poles [105]. The chromokinesins Kif4 and Kid, concentrated on chromosome arms, regulate chromosome positioning by altering inter-kinetochore tension in a position-dependent manner within the spindle [104]; chromokinesins also likely move chromosomes to form an equatorial ring in prometaphase prior to establishment of kinetochore-microtubule attachments [64,106]. Finally the kinesin-7 family motor, CENP-E, concentrated on the outer kinetochore, increases the stability of attachments [107]. The combination of an N-terminal plus end motor activity, a long flexible interrupted coiled-coil tether, and a C-terminal non-motor microtubule-binding domain enable CENP-E to bidirectionally track growing and depolymerizing microtubule plus ends, a striking property that likely underlies its role at the kinetochore [107]. Overall motors play important roles in restricting kinetochore position and maintaining attachment stability.
Proper regulation of kinetochore microtubule attachments is important because misattachments lead to unequal distribution of the genome. A majority of solid tumor cells are aneuploid and exhibit elevated rates of chromosome missegregation [108]. Many cancer cells appear to have hyperstable kinetochore-microtubule attachments that underlie increased rates of missegregation [109,110]; in addition, cancer cells may be less efficient in error correction [68]. Remarkably, reducing kinetochore-microtubule attachment stability in cancer cell lines by overexpression of a kinesin-13 family microtubule depolymerase reduces chromosome missegregation [111]. While these studies highlight the importance of precise control of kinetochore-microtubule dynamics in preventing chromosomal instability, the genetic/epigenetic basis underlying loss of this control in cancer cells remains unclear and is important to elucidate.
Interplay Between Attachment Formation and Checkpoint Signaling
The mechanical events at the kinetochore are coordinated with checkpoint signaling, which ensures that the separation of all chromosomes only occurs after the last chromosome has attached to the spindle [112]. Silencing of the spindle assembly checkpoint is coordinated with microtubule attachment, which leads to dynein motor-based stripping of checkpoint proteins from the kinetochore in metazoans [113,114] Recent work suggests that the key event in this silencing mechanism is removal of the dynein adaptor protein Spindly [115]. In the absence of Spindly, but not in the presence of a Spindly mutant that fails to recruit dynein, the checkpoint is still silenced following attachment; as dynein is not ubiquitously present at kinetochores across species, the presence of a dynein-independent but microtubule attachment-dependent silencing pathway is perhaps not surprising. Two other components implicated in silencing are protein phosphatase I, whose recruitment by Knl1/Spc105 is important for checkpoint silencing in fungi and worms [77,116,117]. As constitutive tethering of PP1 to budding yeast kinetochores does not prevent checkpoint activation [77], whether PP1 responds to microtubule attachment is unclear. Microtubule binding by the PP1 docking protein KNL-1 has been proposed to contribute to checkpoint silencing in C. elegans, although this contribution appears to be genetically parallel to PP1 docking [117]. Thus, while microtubule attachment must be relayed to control checkpoint signaling at the kinetochore, the underlying molecular mechanisms remain to be clarified in future work. A major challenge here is our lack of understanding of how the checkpoint is activated at the kinetochore. As progress is made on this topic, understanding how attachment silences the checkpoint will become more feasible, potentially through the use of in vitro systems such as the kinetochore-like particles from budding yeast.
Perspective
The cataloguing of the majority of kinetochore proteins, genetic/RNAi analysis in multiple model organisms and in cultured mammalian cells, in vitro reconstitutions of complexes, biophysical assays with purified components/complexes and native assemblies, and structural approaches are cumulatively building a detailed picture of how kinetochore-microtubule interactions are formed and regulated. Still, many challenges lie ahead, notably resolving contradictions between in vitro and cellular studies, and elucidation of the complex kinase-phosphatase activity fluxes that coordinate different events at the kinetochore but are themselves under mechanical control. In addition, surprises are likely to emerge from work in different biological contexts, such as stem cell divisions that exhibit asymmetric chromatid segregation [118,119] and meiosis I, where homologues rather than sisters segregate [120,121]. Thus, the many outstanding questions and emerging new areas will keep the field occupied for some time to come.
Acknowledgments
Work in the Desai lab is supported a grant from the NIH (GM074215) and by the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hori T, Fukagawa T. Establishment of the vertebrate kinetochores. Chromosome Res. 2012;20:547–561. doi: 10.1007/s10577-012-9289-9. [DOI] [PubMed] [Google Scholar]
- 2.Maddox PS, Corbett KD, Desai A. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 2012;22:139–147. doi: 10.1016/j.gde.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.McIntosh JR, O‘Toole E, Zhudenkov K, Morphew M, Schwartz C, Ataullakhanov FI, Grishchuk EL. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J Cell Biol. 2013;200:459–474. doi: 10.1083/jcb.201209154. This paper extends electron tomographic analysis of kinetochore-microtubule attachments to five divergent species. The images reveal a consistent flared protofilament structure at the ends of kinetochore-attached microtubules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, Deluca JG, Carroll CW, Liu S-T, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deluca JG, Musacchio A. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol. 2012;24:48–56. doi: 10.1016/j.ceb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett KD, Yip CK, Ee L-S, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci US A. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitzberger F, Harrison SC. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012;13:216–222. doi: 10.1038/embor.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 2013;32:409–423. doi: 10.1038/emboj.2012.356. These two papers report atomic structures of a direct connection between CENP-T, a DNA-associated inner kinetochore protein, and the Ndc80 complex; CENP-T binds the same site on the Ndc80 complex as the Mis12 complex within the KMN network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Gonen S, Akiyoshi B, Iadanza MG, Shi D, Duggan N, Biggins S, Gonen T. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat Struct Mol Biol. 2012;19:925–929. doi: 10.1038/nsmb.2358. This paper employs EM to describe the structure of unbound and microtubule-bound native kinetochore-like particles purified from budding yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 21.Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 2011;30:906–919. doi: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood FE, Williams SJ, Burgess SG, Richards MW, Roth D, Straube A, Pfuhl M, Bayliss R, Royle SJ. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J Cell Biol. 2013;202:463–478. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BMA, Dasso M. The Nup107–160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 28.Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- 29*.Ferraro-Gideon J, Sheykhani R, Zhu Q, Duquette ML, Berns MW, Forer A. Measurements of forces produced by the mitotic spindle using optical tweezers. Mol Biol Cell. 2013;24:1375–1386. doi: 10.1091/mbc.E12-12-0901. This paper revisits the issue of how much force is needed to stall a moving chromosome. Employing optical traps, the authors suggest that the stall force may be 100 times lower than the widely quoted value measured previously using microneedles [discussed in ref. 28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, Cimini D, Cameron LA, Salmon ED. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol Biol Cell. 2012;23:1035–1046. doi: 10.1091/mbc.E11-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida KSK, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. This paper employs super-resolution imaging to analyze kinetochore substructure during directional instability. The measurements suggest the presence of two sites for force generation at different locations in the kinetochore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Deluca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 36.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Reis Dos G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for Kinetochore-Microtubule Attachment from the Structure of an Engineered Ndc80 Complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundin LJR, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell. 2011;22:1217–1226. doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. This paper extends prior cryo-EM studies to provide the highest current resolution view of the Ndc80 complex bound to microtubules. Coupled with in vitro and cellular experiments, a multi-mode binding model is proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kemmler S, Stach M, Knapp M, Ortíz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, Davis TN. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci US A. 2012;109:16113–16118. doi: 10.1073/pnas.1209615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, Asbury CL. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc Natl Acad Sci US A. 2013;110:7282–7287. doi: 10.1073/pnas.1220700110. These two studies employ purified human Ndc80 complex [46] and native yeast kinetochore particles [47] in which phosphomimetic residues have been introduced to elucidate the mechanism by which phosphorylation triggers kinetochore detachment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. These two papers employ purified complexes in vitro to characterize the cooperation between the Dam1 and Ndc80 complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. This paper employs single molecule assays, structural biology, and in vivo studies to elucidate the cooperation between the Ska complex and the Ndc80 complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Hsu K-S, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Tang NH, Takada H, Hsu K-S, Toda T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell. 2013;24:1122–1133. doi: 10.1091/mbc.E12-11-0817. These two papers show that the XMAP215 family of microtubule regulators cooperate with the Ndc80 complex in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Varma D, Chandrasekaran S, Sundin LJR, Reidy KT, Wan X, Chasse DAD, Nevis KR, Deluca JG, Salmon ED, Cook JG. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. This paper reveals an unexpected role for the DNA replication initiation factor Cdt1 in chromosome segregation. Cdt1 is proposed to associate with the Ndc80 loop region and cooperate with the Ndc80 complex to stabilize kinetochore-microtubule attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westermann S, Wang H-W, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 55.Maure J-F, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2012 doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Volkov VA, Zaytsev AV, Gudimchuk N, Grissom PM, Gintsburg AL, Ataullakhanov FI, McIntosh JR, Grishchuk EL. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc Natl Acad Sci US A. 2013;110:7708–7713. doi: 10.1073/pnas.1305821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guimaraes GJ, Deluca JG. Connecting with Ska, a key complex at the kinetochore-microtubule interface. EMBO J. 2009;28:1375–1377. doi: 10.1038/emboj.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.03.005. This paper reports the atomic structure of a truncated form of the Ska complex. Two 3-subunit complexes associate to form an extended W-shaped dimer. Missing in the structure is the microtubule-binding domain of the complex, which is described in ref. [50] [DOI] [PubMed] [Google Scholar]
- 60.Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, Gorbsky GJ. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19:1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohta S, Bukowski-Wills J-C, Sanchez-Pulido L, Alves F, de L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Magidson V, O’Connell CB, Lon arek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. This paper utilizes high temporal and spatial resolution tracking of centromeres to reveal that chromosomes form an equatorial chromosome ring during prometaphase that is proposed to facilitate the establishment of end-on kinetochore-microtubule attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shrestha RL, Draviam VM. Lateral to End-on Conversion of Chromosome-Microtubule Attachment Requires Kinesins CENP-E and MCAK. Curr Biol. 2013 doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Welburn JPI, Vleugel M, Liu D, Yates JR, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol. 2011;21:1158–1165. doi: 10.1016/j.cub.2011.06.015. This paper reports significant differences between transformed (HeLa) and non-transformed (RPE1) cells in the accumulation of Aurora B at the inner centromere when chromosomes are misaligned. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69*.DeLuca KF, Lens SMA, Deluca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. This paper employs RNAi-based replacement in human cells and phospho-specific antibodies to Aurora B target sites in the Ndc80 tail to characterize the contribution of tail phosphorylation in attachment stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu D, Vader G, Vromans MJM, Lampson MA, Lens SMA. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71**.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. doi: 10.1038/nature12057. This paper challenges the previously dominant model for tension-dependent stabilization of attachments that focused on separation of the outer kinetochore from inner centromere-localized Aurora B kinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yue Z, Carvalho A, Xu Z, Yuan X, Cardinale S, Ribeiro S, Lai F, Ogawa H, Gudmundsdottir E, Gassmann R, et al. Deconstructing Survivin: comprehensive genetic analysis of Survivin function by conditional knockout in a vertebrate cell line. J Cell Biol. 2008;183:279–296. doi: 10.1083/jcb.200806118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Posch M, Khoudoli GA, Swift S, King EM, Deluca JG, Swedlow JR. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Biol. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 79.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 80.Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters J-M. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 81.Moutinho-Santos T, Conde C, Sunkel CE. POLO ensures chromosome bi-orientation by preventing and correcting erroneous chromosome-spindle attachments. J Cell Sci. 2012;125:576–583. doi: 10.1242/jcs.092445. [DOI] [PubMed] [Google Scholar]
- 82*.Hegemann B, Hutchins JRA, Hudecz O, Novatchkova M, Rameseder J, Sykora MM, Liu S, Mazanek M, Lénárt P, Hériché J-K, et al. Systematic phosphorylation analysis of human mitotic protein complexes. Sci Signal. 2011;4:rs12. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83*.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84*.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Silljé HHW, Körner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.004457. doi: 10:M110.004457. These three mass spectrometry papers generate an important resource for analyzing phosphoregulation of kinetochore components by Plk1 and Aurora B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86**.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87**.Suijkerbuijk SJE, Vleugel M, Teixeira A, Kops GJPL. Integration of Kinase and Phosphatase Acti2vities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. These two papers reveal that the PP2A-B56 phosphatase is critical for stable kinetochore-microtubule attachment formation. The second paper shows that PP2A recruitment is via Plk1 phosphorylation of the pseudokinase BubR1. Thus, PP2A is more important for attachment regulation than suggested in prior models, which focused primarily on PP1. [DOI] [PubMed] [Google Scholar]
- 88*.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89*.Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol Biol Cell. 2012;23:2264–2274. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90*.Maia ARR, Garcia Z, Kabeche L, Barisic M, Maffini S, Macedo-Ribeiro S, Cheeseman IM, Compton DA, Kaverina I, Maiato H. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. These 3 papers highlight emerging roles for Polo-like kinase 1 (Plk1) in controlling kinetochore-microtubule attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H, Liu XS, Yang X, Wang Y, Wang Y, Turner JR, Liu X. Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore-microtubule attachments. EMBO J. 2010;29:2953–2965. doi: 10.1038/emboj.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bader JR, Kasuboski JM, Winding M, Vaughan PS, Hinchcliffe EH, Vaughan KT. Polo-like kinase1 is required for recruitment of dynein to kinetochores during mitosis. J Biol Chem. 2011;286:20769–20777. doi: 10.1074/jbc.M111.226605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang YH, Park J-E, Yu L-R, Soung N-K, Yun S-M, Bang JK, Seong Y-S, Yu H, Garfield S, Veenstra TD, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 94*.Carmena M, Pinson X, Platani M, Salloum Z, Xu Z, Clark A, Macisaac F, Ogawa H, Eggert U, Glover DM, et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 2012;10:e1001250. doi: 10.1371/journal.pbio.1001250. This paper shows that the Aurora B-containing chromosomal passenger complex activates Polo-like kinase at centromeres, highlighting the complex interplay between these two mitotic kinases that ensures error-free chromosome segregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeh T-Y, Kowalska AK, Scipioni BR, Cheong FKY, Zheng M, Derewenda U, Derewenda ZS, Schroer TA. Dynactin helps target Polo-like kinase 1 to kinetochores via its left-handed beta-helical p27 subunit. EMBO J. 2013;32:1023–1035. doi: 10.1038/emboj.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96**.Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–513. doi: 10.1038/nature12507. This paper reveals a new principle controlling kinetochore-microtubule interactions: global control of kinetochore-microtubule stability by Cyclin A-Cdk1. High Cyclin A during prometaphase reduces stability of kinetochore-microtubule interactions, which is proposed to facilitate error correction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98*.Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99*.Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100*.Chen J-S, Lu LX, Ohi MD, Creamer KM, English C, Partridge JF, Ohi R, Gould KL. Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J Cell Biol. 2011;195:583–593. doi: 10.1083/jcb.201105074. The first two papers report analysis of a kinetochore-localized microtubule-binding complex (Astrin-SKAP) that contributes to chromosome segregation. The third paper describes Nsk1, a subunit of a potentially analogous complex in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stumpff J, Dassow von G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su X, Qiu W, Gupta ML, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104*.Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. This paper describes how the plus end dynamics suppressor Kif18 and chromosome arm-enriched kinesins restrict centromeres to the middle of the spindle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107*.Gudimchuk N, Vitre B, Kim Y, Kiyatkin A, Cleveland DW, Ataullakhanov FI, Grishchuk EL. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat Cell Biol. 2013;15:1079–1088. doi: 10.1038/ncb2831. This paper combines in vitro single molecule assays and theoretical modeling to show that the kinetochore-localized kinesin CENP-E can bi-directionally track microtubule ends. In vivo inhibitions with a small molecule inhibitor suggest that this activity may help stabilize kinetochore-microtubule attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci US A. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–80. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 113.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wojcik E, Basto R, Serr M, Scaërou F, Karess R, Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 115.Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, Millar JBA. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117*.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. This paper reports a role for microtubule binding by the KNL-1 subunit of the KMN network in spindle checkpoint silencing that acts in parallel to docking of PP1 on KNL-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118*.Yadlapalli S, Yamashita YM. Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature. 2013;498:251–254. doi: 10.1038/nature12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119*.Elabd C, Cousin W, Chen RY, Chooljian MS, Pham JT, Conboy IM, Conboy MJ. DNA methyltransferase-3-dependent nonrandom template segregation in differentiating embryonic stem cells. J Cell Biol. 2013;203:73–85. doi: 10.1083/jcb.201307110. This pair of papers provide evidence for non-random segregation of sister chromatids during stem cell divisions in Drosophila and in mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339:1071–1074. doi: 10.1126/science.1232518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kakui Y, Sato M, Okada N, Toda T, Yamamoto M. Microtubules and Alp7-Alp14 (TACC-TOG) reposition chromosomes before meiotic segregation. Nat Cell Biol. 2013 doi: 10.1038/ncb2782. [DOI] [PubMed] [Google Scholar]