Conformational differences between myoglobin structures are studied. Most structural differences in whale myoglobin beyond the uncertainty threshold can be correlated with a few specific structural factors. There are always exceptions and a search for additional factors is needed. The results might have serious implications for biological insights from conformational differences.
Keywords: bioinformatics, computation, proteins, validation, conformation, function
Abstract
Validation of general ideas about the origins of conformational differences in proteins is critical in order to arrive at meaningful functional insights. Here, principal component analysis (PCA) and distance difference matrices are used to validate some such ideas about the conformational differences between 291 myoglobin structures from sperm whale, horse and pig. Almost all of the horse and pig structures form compact PCA clusters with only minor coordinate differences and outliers that are easily explained. The 222 whale structures form a few dense clusters with multiple outliers. A few whale outliers with a prominent distortion of the GH loop are very similar to the cluster of horse structures, which all have a similar GH-loop distortion apparently owing to intermolecular crystal lattice hydrogen bonds to the GH loop from residues near the distal histidine His64. The variations of the GH-loop coordinates in the whale structures are likely to be owing to the observed alternative intermolecular crystal lattice bond, with the change to the GH loop distorting bonds correlated with the binding of specific ‘unusual’ ligands. Such an alternative intermolecular bond is not observed in horse myoglobins, obliterating any correlation with the ligands. Intermolecular bonds do not usually cause significant coordinate differences and cannot be validated as their universal cause. Most of the native-like whale myoglobin structure outliers can be correlated with a few specific factors. However, these factors do not always lead to coordinate differences beyond the previously determined uncertainty thresholds. The binding of unusual ligands by myoglobin, leading to crystal-induced distortions, suggests that some of the conformational differences between the apo and holo structures might not be ‘functionally important’ but rather artifacts caused by the binding of ‘unusual’ substrate analogs. The causes of P6 symmetry in myoglobin crystals and the relationship between crystal and solution structures are also discussed.
1. Introduction
Differences between the structures of the same protein with and without a bound substrate analog are often used to infer protein mechanism, as well as for protein re-engineering (see, for example, Janin & Wodak, 1983 ▶). Plausible interpretations derived from such structural differences in a protein from one species are also often assumed to be valid for proteins from different species, despite significant interspecies sequence differences. It is clearly important to investigate the validity of such a use of protein structure differences for insight into protein function.
A rapid increase in the number of protein X-ray structures determined in the large-scale high-throughput research centers (PSI–Nature Structural Genomics Knowledgebase; http://kb.psi-structuralgenomics.org) has led to increased attention being paid to various aspects of the validation of structures. One area of such validation (see, for example, Jaskolski et al., 2007 ▶; Kleywegt, 2009 ▶) focuses on the proper use of protein crystallographic techniques, interpretation methods and their consistency. It was found that even subjective differences between researchers can lead to differences in structures of the same protein reportedly studied following the same protocols under the same conditions. Errors owing to improper use of techniques, low resolution or a crystallographer’s inexperience might dangerously accumulate from a ‘flawed structure which will be of limited interest with any serious errors merely polluting the structural archive (Berman et al., 2000 ▶)’, to ‘the worst case, when serious errors in a high-profile structure may actually obstruct the progress of science for years to come’ (Kleywegt, 2009 ▶). A number of methods to limit such calamities have been developed.
In another area of validation, Janin & Rodier (1995 ▶) were the first to pay serious attention to packing contacts as crystal artifacts which might erroneously be interpreted as functionally important interactions governing specific recognition in protein–protein complexes and oligomeric proteins. A number of differences between biologically important and purely crystallographic contacts were found in the magnitudes of the surface areas buried in the contacts and in their amino-acid compositions. Similar investigations soon followed (Carugo & Argos, 1997 ▶; Dasgupta et al., 1997 ▶) as well as their algorithmic implementation, allowing the deduction of protein quaternary structure from X-ray data (PQS; Henrick & Thornton, 1998 ▶). More recently, Bahadur et al. (2004 ▶) developed a residue propensity score and a hydrophobic interaction score to assess preferences in the compositions of the different types of interfaces, and derived indices of atomic packing, which was found to be less compact at nonspecific than at specific interfaces.
Krissinel (2010 ▶) notes that the assumption, that the contacts observed in crystals reflect natural macromolecular interactions, forms the basis for many studies in structural biology. However, the crystal state may correspond to a global minimum of free energy where biologically relevant interactions are sacrificed in favor of nonspecific contacts. From docking simulations, Krissinel estimated that 20% of all dimers in the PDB have a higher than 50% chance of being misrepresented by crystals.
It has also been shown (DePristo et al., 2004 ▶) that a number of structures can fit experimental X-ray data for a protein as well as or better than its structure in the PDB, suggesting that analyses depending on small differences in atomic positions may be flawed.
We have found that in 1014 pairs of 42 ribonuclease A and 18 sperm whale myoglobin structures, for which the authors of the X-ray studies did not report any significant structural movements, the root-mean-square distance difference (RMSDD) reached 0.44 Å (Rashin et al., 2009 ▶), indicating the range of nonbiological coordinate uncertainty (see §2).
Crystal contacts contain a high proportion of polar residues (see, for example, Janin & Rodier, 1995 ▶; Bahadur et al., 2004 ▶), and it has been pointed out (Kondrashov et al., 2008 ▶) that intermolecular hydrogen (and possibly ionic) bonds, and not the presence of crystal contacts, correlate with significant coordinate differences between sperm whale myoglobin structures. Thus, generic crystal contacts have already been invalidated as a definitive cause of conformational differences.
Another example of conformational differences induced by intermolecular crystal hydrogen bonds has been well documented for an asymmetric dimer of ribonuclease Sa (Sevcik et al., 1991 ▶; PDB entry 1sar). Also, in the RNaseSa–3′-GMP complex a hydrogen bond formed by side chains from a neighboring molecule in the crystal prevents the binding of 3′-GMP to the catalytic site of molecule B.
Thus, there are examples of the important roles played by intermolecular crystal hydrogen/ionic bonds in causing coordinate differences, changes in the crystal symmetry and inhibition of binding in the crystals of identical molecules (Kondrashov et al., 2008 ▶; Sevcik et al., 1991 ▶). Therefore, we attempt to validate only the role of specific hydrogen-bond-forming intermolecular contacts as a definitive cause of conformational differences. As a complementary part of this validation, we analyze whether specific intermolecular bonds form at the expense of intramolecular bonds.
Myoglobin, the first protein to have its tertiary structure solved experimentally, has been studied by many researchers across several different species using a plethora of techniques. Hundreds of independently determined structures, all of which are rather simple, make myoglobin an appealing model system for computational studies of protein structure. Furthermore, with large numbers of myoglobin structures now available in the PDB (Berman et al., 2000 ▶), we are ideally positioned to consider the detailed differences between the conformation of a structure under various conditions by directly comparing experimental structures. In this paper, we explore the differences between myoglobin structures to determine the effects of temperature, pH, crystal packing and hydrogen/ionic bonding, the binding of diverse ligands and the effects of sequence differences in myoglobins from different species on the resulting conformations. We find species-specific conformational differences, as well as distinct structural changes within the high-resolution structures of sperm whale myoglobin that are induced by multiple factors.
2. Materials and methods
2.1. RMSD versus RMSDD
The most commonly used characteristic of similarity between two X-ray structures of the same protein is the root-mean-square difference (RMSD), calculated as the square root of the mean-squared distances between the same Cα atoms in the two structures. Calculation of the RMSD requires that the pair of structures are somehow fitted together. Such fitting, however, depends on the method used (see, for example, Rashin et al., 1997 ▶) and thus might introduce poorly controlled uncertainty, giving the results more of a qualitative character.
An alternative characteristic of the similarity between two structures of the same molecule is the root-mean-square distance difference (RMSDD), which does not require the preliminary fitting of two structures (see, for example, Rashin et al., 2009 ▶) and thus imparts a more objective character to the results of the comparison. To calculate the RMSDD, one first calculates a matrix of distances between all Cα atoms for each of the two molecules, builds the matrix (DDM) of differences of distances (DDs) between all corresponding distances from two molecules and then takes the square root of the mean of squares of all elements of the DDM.
2.2. Application of principal component analysis (PCA) to proteins
PCA (Jolliffe, 2002 ▶) is used here for dimensionality reduction of complex data consisting of multiple sets of coordinates of Cα atoms. The goal is to rank order the contributions of groups of Cα atoms to the variance of the entire data set. All of the structures in the set are aligned with the 151 Cα atoms of PDB entry 1bz6 using combinatorial extension (Shindyalov & Bourne, 1998 ▶) prior to application of PCA and clustering using MATLAB (The MathWorks, Natick, Massachusetts, USA). While the RMSDD is invariant to structure superpositioning, PCA, which is convenient for large sets, is not, thus making it more qualitative.
Grouping the data set into five clusters was determined to be the most efficient clustering using the metrics of Horimoto & Toh (2001 ▶). The results of PCA and clustering are represented in two or three dimensions, with the PC1 axis representing the direction with the maximum contribution to the total variance, the PC2 axis corresponding to the direction of the maximum variance remaining after the variance along PC1 is removed from the total set and the PC3 axis corresponding to the direction of the maximum variance after removal of the variance along PC1 and PC2.
2.3. Validation of coordinate differences
Recently, we suggested (Rashin et al., 2009 ▶, 2010 ▶) the use of the RMSDD (see §2.1) derived from distance difference matrices (DDMs) for pairs of structures of the same molecule and the percentage, Δ, of distance differences (DDs) larger than 1 Å to define coordinate uncertainty thresholds. We will refer to this method below as DDM/RMSDD.
It has been found (Rashin et al., 2009 ▶) that 1014 pairs of structures of RNaseA and myoglobin (861 pairs of RNaseA structures and 153 pairs of sperm whale myoglobin structures) have RMSDDs of up to 0.44 Å owing to coordinate differences that were unexplained (or unjustified) by the authors. The set did not contain proteins complexed with protein inhibitors, structures with low water content, cryogenic structures or mutants. Any of these factors might reportedly lead to significant local or global conformational changes (see Rashin et al., 2009 ▶). At the same time, there are examples in the literature of functionally induced coordinate differences with an RMSDD of 0.45 Å and a Δ of 5% or greater.
The useful criteria that a DDM does not indicate a significant motion, but only coordinate uncertainty, when the RMSDD is below 0.46 Å and its Δ is less than 5% were introduced (Rashin et al., 2009 ▶). The thus introduced ‘coordinate uncertainties’ are distinct from the coordinate accuracy, coordinate errors or standard uncertainty usually referred to in the literature (see Rashin et al., 2009 ▶).
Further analysis might somewhat change these criteria (see also Supporting Information §S91).
In this work, we study using DDM/RMSDD all sperm whale myoglobin structures with native sequences, all cryogenic structures, structures determined from crystals exposed to extreme pH conditions and a few mutants invoked in the literature, as well as structures of horse and pig myoglobin. Residues 152–153, which are not visible in many structures, are excluded from all comparisons. In a few marginal cases we also exclude a couple of N-terminal residues from comparison (see below).
2.4. Analysis of intermolecular and long-range intramolecular hydrogen/ionic bonds
It has previously been found (Kondrashov et al., 2008 ▶) that myoglobin crystals with P6122 symmetry, which is ‘unusual’ for this protein, are associated with intermolecular hydrogen bonds and that such bonds can inhibit substrate binding (Sevcik et al., 1991 ▶). Therefore, we analyzed the formation of such bonds in structures of myoglobin characterized by large RMSDDs when compared with various different myoglobin structures. In the following, the term ‘bond’ will intermittently be used for simplicity for hydrogen and ionic (e.g. to Fe ion) bonds, whose specifics are clear from the context.
Since significant conformational differences between myoglobin structures could be caused by intermolecular hydrogen bonds in crystals, it is of interest to learn whether such intermolecular bonds might form at the expense of intramolecular bonds in individual molecules.
To clarify this, we tabulate long-range intramolecular and intermolecular hydrogen bonds involving side chains (with a single exception of a main-chain intermolecular bond in PDB entry 1u7s) in various structures from the myo46 set (see §2.5.2). We consider bonds to be long-range if they are formed by residues that are more than six amino acids apart along the sequence. Bonds are considered of ‘full strength’ if the contact distance between bond-forming heavy atoms is below 3.25 Å and ‘weak’ if the contact distance is between 3.25 and 3.45 Å. Both long-range intramolecular and intermolecular bonds in crystal structures were found with CONTACT from the CCP4 suite (Winn et al., 2011 ▶).
2.5. Myoglobin structures used in this study
2.5.1. Sets myo291 and myo216
A data set myo291 of 291 myoglobin structures (see Supplementary Table S1) is comprised of 222 sperm whale (Physeter catadon) structures, 52 horse (Equus caballus) structures (briefly characterized in Supplementary Table S2) and 17 pig (Sus scrofa) structures, including many mutants. This set was gathered to investigate the relationships between species, mutants and ligand diversity using PCA and clustering. It contains only structures with 151 well resolved residues. Through manual inspection of multiple sequence alignments generated by ClustalW2 (Larkin et al., 2007 ▶; Goujon et al., 2010 ▶), it was determined that a sequence motif could be used to select a maximal common substructure consisting of the 151 residues that either begin with GLSDGEW… and end with …KELGF or begin with VLSEGEW… and end with …KELGY. Only Cα atoms were considered and the first (or ‘A’) conformation was chosen if multiple conformations for the same atoms are listed in the PDB file.
Myoglobin helices are denoted as in the PDB with letters from A to H. GH denotes fragments up to eight residues long that include the interhelical region, C–D denotes residues from the beginning of the C helix to the end of the D helix and HC denotes a C-terminal fragment that sometimes includes a few residues of helix H.
A structure is denoted as ‘mutant’ by identifying variations in its sequence compared with aligned reference sequences: PDB entry 1bz6 for whale, PDB entry 1azi for horse and PDB entry 1mwc for pig.
216 whale myoglobin structures determined entirely by X-ray crystallography comprise set myo216. Among these there are seven structures with modified hemes: PDB entries 2eku, 2cmm, 1bvc, 1bvd, 2d6c, 2ekt and 1iop.
2.5.2. Set myo46
We considered the myo46 set as 46 native-like sperm whale myoglobin structures from the PDB that have been published in refereed journals: 1a6g, 1a6k, 1a6m, 1a6n, 1ajg, 1ajh, 1bz6, 1bzp, 1bzr, 1cq2, 1ebc, 1hjt, 1jp6, 1jp8, 1jp9, 1jpb, 1l2k, 1mbc, 1mbd, 1mbi, 1mbn, 1mbo, 1spe, 1swm, 1u7r, 1u7s, 1vxa, 1vxb, 1vxc, 1vxd, 1vxe, 1vxf, 1vxg, 1vxh, 1yog, 1yoh, 1yoi, 2mb5, 2mbw, 4mbn, 5mbn, 2z6s, 2z6t, 1abs, 1jw8 and 2jho (mutants with P6 symmetry are in bold).
Only three structures in the myo46 set contain a single D122N mutation (PDB entries 1abs, 1jw8 and 2mbw). We included these three mutated structures in myo46 because they have been compared with the wild-type structures in the literature. All 46 structures are briefly characterized (according to PDB and journal publications) in Supplementary Table S3.
We also studied some characteristics of additional myoglobin mutants in the P6 crystal form to find the combination of factors possibly responsible for the formation of this crystal form.
2.6. ‘Usual’ and ‘unusual’ ligands in the sixth coordination position
We paid special attention to whale myoglobin structures with unusual ligands in the sixth coordination site of the heme. Water (HOH), oxygen (O2) and carbon monoxide (CO), which can simultaneously bind to distal histidine (His64) NE2 and the heme iron (Fe), were considered ‘usual’ ligands. ‘Unusual’ ligands include non-ionic nitric oxide (NO) and negatively charged ligands such as hydroxyl (OH−) in PDB entry 1mbn and cyanide (CN−) in PDB entries 1ebc and 2jho, which can also simultaneously bind to the distal histidine (His64) NE2 and heme iron (Fe), as well as negatively charged azide (N3) in PDB entry 1swm (the reference in the PDB to the paper describing this structure is erroneous). Azide lies approximately parallel to the heme plane (as does photolysed CO) and binds to the distal histidine and the heme iron (Fe) with the same azide atom. Another ‘unusual’ ligand is the positively charged imidazole ion (IMD) as seen in PDB entry 1mbi (Lionetti et al., 1991 ▶). The distal pocket in 1mbi is significantly disrupted, with the distal histidine (His64) pushed out of the normal distal site. Furthermore, the imidazole ion binds with its NE1 atom towards the heme iron (Fe) and also binds with its NE2 atom to the ND1 atom of the distal histidine (‘usual’ ligands bind to the NE2 atom of the distal histidine). While it has been suggested (see, for example, Frauenfelder et al., 2001 ▶) that nitric oxide (NO) might be involved in some secondary functions of myoglobin, we considered it unusual compared with the ‘usual ligands’ with well studied roles and modes of binding to myoglobin.
2.7. Crystallographic validation
For crystallographic validation, we checked R factors, structure factors, Ramachandran outliers and MolProbity parameters (including clashscore) (Chen et al., 2010 ▶). For results, see Supporting Information §S2.
3. Results
3.1. PCA and DDM/RMSDD analysis of the myo291 and myo216 sets
3.1.1. Myo291
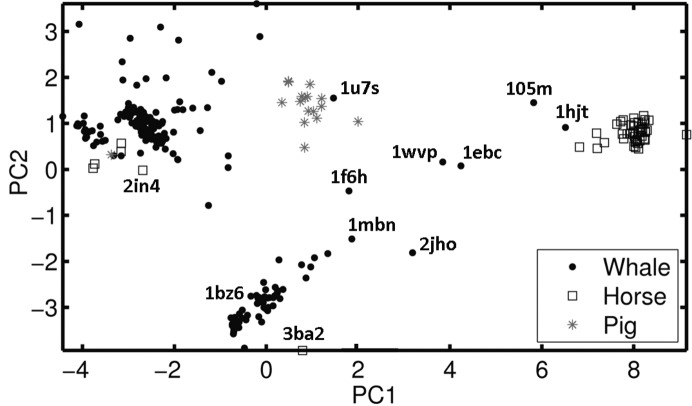
PCA and clustering of a large set of 291 myoglobin structures reveals distinct conformational clusters with high conformational consistency within each cluster. The clusters have a high correspondence with species-specific variations (Fig. 1 ▶). All but one of the pig myoglobin structures are in one cluster in the center of PC1/PC2 plane, and all but six of the horse myoglobin structures are in a well separated cluster on the extreme right of the PC1/PC2 plane. The whale myoglobin structures are mainly in two well separated groups: one at the bottom of the PC1/PC2 plane, containing most of the wild-type whale myoglobin structures, and another more diffuse group in the top left corner of the PC1/PC2 plane mostly containing whale myoglobin structures that are either mutants or contain unusual ligands (see §2.6). However, there are outliers of these two main clusters of sperm whale structures in many positions on the PC1/PC2 plane, including inside the pig and horse clusters. Five horse and one pig myoglobin outlier structures are in the upper cluster of whale structures. A more detailed presentation and analysis in three-dimensional PCA space are provided in Supporting Information §S3 (e.g. Supplementary Fig. S2). Interestingly, the horse myoglobin structures (PDB entries 3hc9, 3hep, 3hen and 3hed) that appear within the whale cluster are mutants that affect O2 and CO affinity, each containing the H64V mutation, while PDB entries 3hen and 3hed also contain a V67R mutation. Of the two other horse myoglobins showing departure from the main cluster, PDB entry 3ba2 (at the bottom of Fig. 1 ▶) has a strongly modified heme and has not been published in a refereed journal. Another outlier from the main horse cluster is PDB entry 2in4 (one of five horse myoglobins in the upper whale cluster), which contains a charge-neutralized heme. The only pig outlier (PDB entry 1mnh) contains both H64V and V67R mutations. Many of the structures that could be considered to be outliers contain distinct combinations of nonphysiological ligands or environmental conditions.
Figure 1.
Results of PCA and clustering of the myo291 set represented in two PC dimensions (see text). Some outliers are marked with their PDB codes.
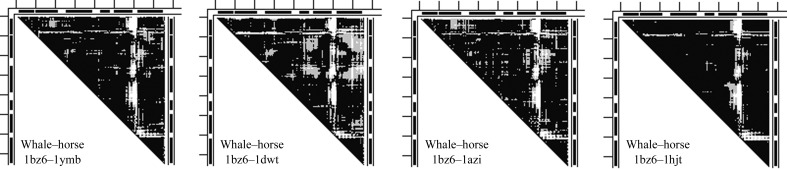
Closest to the horse cluster on the right of Fig. 1 ▶ is the whale outlier PDB entry 1hjt from the myo46 set (see Supplementary Table S3) and the next closest is the whale outlier PDB entry 1ebc from the same set. All structures in the PC1/PC2 plane (Fig. 1 ▶) were RMS fitted to the whale wild-type structure PDB entry 1bz6 (for its characteristics, see Supplementary Table S3). Fig. 2 ▶ compares the 1bz6–1ymb, 1bz6–1dwt and 1bz6–1azi DDMs of the whale structure 1bz6 with three structures from the horse cluster with the 1bz6–1hjt DDM of two whale structures. It is apparent that the main features of all of these DDMs are very similar. They are all dominated by a thick L-shaped white band with its corner at the GH fragment. Such a white L-shaped band identifies a difference of greater than 1 Å between the two molecules mapped in DDMs for all residues covered by the ends of the band (Rashin et al., 2009 ▶), i.e. all four DDMs indicate strong distortion of the GH region compared with the reference structure 1bz6. The possible causes of such a distortion are analyzed in §3.4.
Figure 2.
Comparison of DDMs between the reference whale structure 1bz6 and three horse myoglobin structures with the DDM of the same reference structure and the whale outlier 1hjt. The black bars at the top and sides represent helices; ticks are shown every 20 residues; black, DD < 0.5 Å; gray, DD < 1 Å; white, DD > 1 Å. A large white L-shaped strip indicates that the largest differences are located in the GH region.
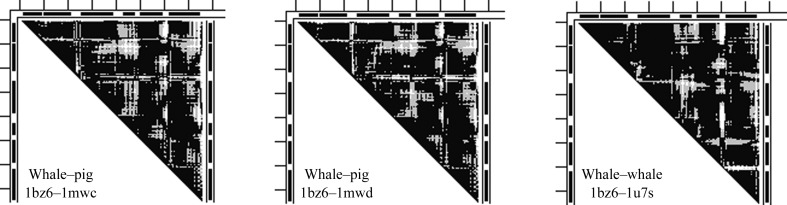
The whale structure 1u7s is in the pig cluster in the middle of Fig. 1 ▶. However, Supplementary Figs. S2(c) and S2(d) show that 1u7s is only close to the pig cluster in the PCA clustering projection on the PC1/PC2 plane in Supplementary Figs. S2(c); it is relatively far from this cluster in the projection of the PCA on the PC1/PC3 plane in Supplementary Fig. S2(d). Thus, 1u7s is not so close to the pig cluster in the three-dimensional PCA space. In contrast, 1hjt remains close to the horse cluster in both Supplementary Figs. S2(c) and S2(d), and thus they are close in the three-dimensional PCA space. Two of the three non-mutant structures in the pig cluster of Fig. 1 ▶ are compared with the reference 1bz6 in the 1bz6–1mwc and 1bz6–1mwd DDMs in Fig. 3 ▶ and clearly are not very similar to the 1bz6–1u7s DDM (RMSDD of 0.41–0.44 Å). Pig myoglobin has been described (Smerdon et al., 1990 ▶) as differing from the whale structure in the CD, EF and GH regions as well as in the HC region.
Figure 3.
Comparison of DDMs between the reference whale structure 1bz6 and two pig myoglobin structures with the DDM of the same reference structure and the whale outlier 1u7s. Notation is the same as in Fig. 2 ▶.
Across the myo291 set, 73% of the mutant structures are from crystals with P6 symmetry, while 87% of the whale mutants have P6 symmetry.
3.1.2. Myo216
PCA clustering of 216 whale myoglobin structures (the myo216 set) yielded two distinct dominant clusters, with multiple structures occupying a diffuse region (see Supplementary Fig. S3). One cluster contains most of the wild-type sequences and only one mutant 1a6g (D122N; see Supporting Information §S3). Two wild-type structures occupy the region of space between the major clusters: 2cmm (with a modified heme) and 1u7s (see Supplementary Fig. S3). The cluster with mutants contains the cryogenic 1u7r, which binds the unusual ligand imidazolium. Further subdivision of the myo291 set into smaller subsets and re-computation of the PCA on each elucidates additional relationships, as described in §3.2 below.
3.2. PCA and DDM/RMSDD analysis of the myo46 set
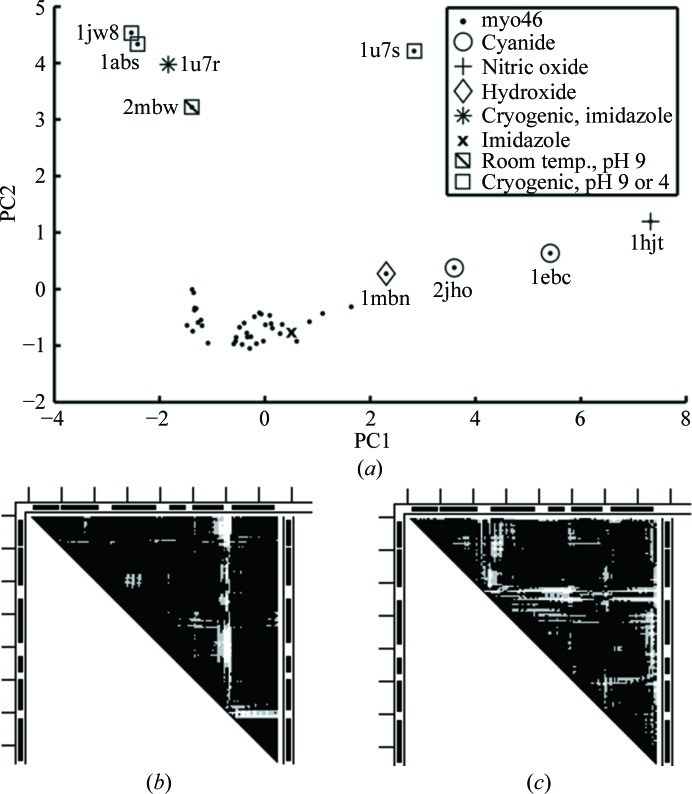
The PCA clustering of the myo46 set is shown in Fig. 4 ▶. In this simplest case, there is one dense cluster at the bottom with a protuberance of scattered outliers coming from it, and another well removed set of scattered outliers at the top of Fig. 4 ▶. The dense cluster contains room-temperature and cryogenic structures as well as one structure (PDB entry 1mbi) with the unusual ligand imidazole. At the beginning of the scattered protuberance, near the dense cluster, is a poorly solved first protein structure, 1mbn, with the unusual ligand hydroxide, followed to the right by three more outliers with unusual ligands (one of which, 2jh2, is cryogenic). It should be noted that 1swm with a bound azide ion is not among the outliers and that 1mbn might be an outlier not because of its unusual ligand (hydroxide) but because of the poor accuracy of its structure (see §3.2 above and §S2). The top outliers include four structures determined from crystals exposed to extreme pH conditions and one (1u7r) with an unusual ligand (imidazole). All but one (2mbw) of the structures in this scattered set are cryogenic. Thus, PCA clustering of the simplest myo46 set (except for 2mbw, 1mbi and 2jho) can be decently correlated with a few ‘identifiers’: unusual ligands and extreme pH with cryogenic temperature sends an outlier to one of two scattered sets of outliers. There are deviations from this correlation in each set of outliers: 2mbw in the upper set is at pH 9 but is not cryogenic, 2jho in the lower set has an unusual ligand (cyanide) and is cryogenic, and 1mbi liganded with imidazolium is not an outlier. (Note some similarity between the PCAs of myo216 and myo46 in Supplementary Figs. S3 and 4a.) PCA clustering, however, does not provide a quantitative description of conformational changes in the outliers and of their location, or allow a search for possible mechanisms of their formation. These can be obtained by applying the DDM/RMSDD-based method (Rashin et al., 2009 ▶). The results of this application as well as an analysis of possible mechanisms of conformational change are described below.
Figure 4.
(a) Results of PCA and clustering of the myo46 set represented in two PC dimensions. (b) A DDM (1bz6–1ebc) with an L-shaped white strip with its corner in the GH region, dominating the DDMs of outliers at the bottom of (a). (c) A DDM (1bz6–1u7r) with an L-shaped white strip with its corner in the CD region, dominating the DDMs of some outliers at the top of (a).
The PCA-based clustering of nine outliers is based on conformational differences between nine whale myoglobin structures RMS fitted to 1bz6, which reflect differences between these nine structures and 1bz6. Thus, directly related to these PCA-found outliers are fit-free calculations of the RMSDD (Rashin et al., 2009 ▶) between 1bz6 and all whale myoglobin structures from the myo46 set. The results of these RMSDD calculations are listed below (RMSDDs of less than 0.23 Å are not shown): 1bz6–1hjt, 0.47 Å; 1bz6–1u7s, 0.45 Å; 1bz6–1u7r, 0.43 Å; 1bz6–1ebc, 0.39 Å; 1bz6–1jw8, 0.38 Å; 1bz6–1mbn, 0.38 Å; 1abs–1bz6, 0.36 Å; 1bz6–2jho, 0.36 Å; 1bz6–1vxb, 0.35 Å; 1ajg–1bz6, 0.33 Å; 1ajh–1bz6, 0.32 Å; 1bz6–1jpb, 0.30 Å; 1bz6–1spe, 0.28 Å; 1bz6–1vxa, 0.27 Å; 1a6n–1bz6, 0.27 Å; 1bz6–1jp9, 0.27 Å; 1bz6–2mbw, 0.26 Å; 1bz6–2z6s, 0.24 Å; 1bz6–2z6t, 0.24 Å; 1a6m–1bz6, 0.23 Å. The first eight results in italics above correspond to eight of the nine PCA outliers. The RMSDDs for these eight pairs are the highest (0.36–0.47 Å) of the 20 listed. However, PCA outlier 2mbw has a much lower RMSDD of 0.26 Å. The difference in placing it, or not placing it, among the outliers might be owing to artifacts introduced by the RMS fitting (see §2.1).
3.3. DDM/RMSDD evaluation of all conformational differences in myo46
The coordinate differences between the reference 1bz6 and other myo46 structures are depicted by DDMs in Supplementary Fig. S4. Examination of these DDMs shows that the bottom set of outliers in the PCA in Fig. 4 ▶(a) all correspond to DDMs with a dominant white L-shaped line or strip with the corner of the L in the area of the GH loop, indicating a conformational change in this area exceeding 1 Å (Fig. 4 ▶ b). A dominating white L-shaped strip in the DDM of 1bz6–1u7r has its corner in the C–D (Fig. 4 ▶ c) area, indicating a conformational change in this area exceeding 1 Å (Rashin et al., 2009 ▶). Further examination of Supplementary Fig. S4 suggests that all outliers at the top of PCA in Fig. 4 ▶(a) correspond to DDMs which are not dominated by changes in the GH area.
RMSDD and Δ values were calculated for all 1035 pairs of 46 myoglobin structures (see §2).
Out of the 1035 pairs, only 85 have RMSDDs outside the coordinate uncertainty threshold and all Δs are within these thresholds; therefore, the Δs are not listed (see §2). Of these 85 RMSDD ‘outliers’ (Supporting Information §S5) one of the compared structures is either 1ebc or 1hjt in 48 pairs. If we exclude two N-terminal residues from the RMSDD comparisons (see Supporting Information §S5 and Supplementary Table S5), 17 more pairs would have RMSDDs below the uncertainty threshold. Without these 17 and 48 pairs, only 20 remaining pairs have ‘outlier’ RMSDDs and are given extra attention (room-temperature structures are in roman font, low-pH structures are in bold and cryogenic structures are in italics): 1abs–1mbn, 1abs–1spe, 1abs–1vxb, 1ajg–1u7r, 1ajg–1vxb, 1ajh–1u7r, 1ajh–1vxb, 1bzp–1u7s, 1jw8–1mbn, 1jw8–1spe, 1jw8–1vxb, 1mbn–1u7r, 1mbn–1vxb, 1spe–1u7s, 1spe–2jho, 1u7r–1u7s, 1u7r–1vxb, 1u7r–2jho, 1u7r–2mb5 and 1u7s–1vxb.
These pairs are made up of the 12 individual structures listed below with 1ebc and 1hjt added (mutants are underlined, 2mb5 was determined by neutron diffraction and 1u7r, 1mbn, 1ebc, 2jho and 1hjt have unusual ligands): 1abs, 1ajg, 1ajh, 1jw8, 2jho, 1u7r, 1u7s, 2mb5, 1bzp, 1mbn, 1spe, 1vxb, 1ebc and 1hjt.
It is clear from Fig. 5 ▶ that among the seven cryogenic structures only four pairs including 1u7r have an RMSDD outside the coordinate uncertainty threshold (Rashin et al., 2009 ▶). Among the seven room-temperature structures, only six pairs have an RMSDD outside the coordinate uncertainty threshold. 26 pairs, including one cryogenic and one room-temperature structure, have high RMSDDs beyond the uncertainty threshold, thus suggesting significant coordinate differences between the structures.
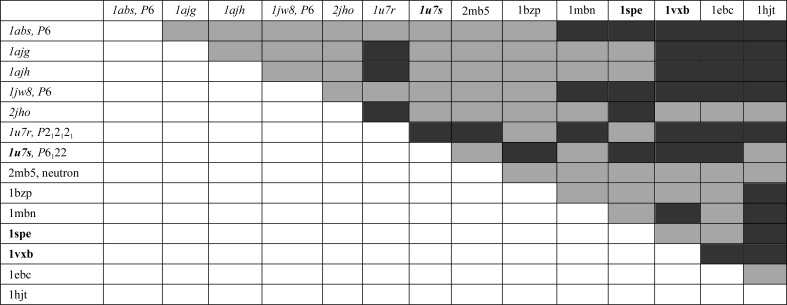
Figure 5.
RMSDDs in comparisons of 14 whale myoglobin structures. Black, RMSDD > 0.45 Å; gray, RMSDD < 0.46 Å. 1ebc and 1hjt are only compared with the structures of 20 outlier pairs (see text). Structures in italics are cryogenic, while structures in bold are at low pH.
Note that 1spe and 1vxb are structures at a low pH of 4 (see Fig. 5 ▶), which corresponds to pH denaturation, the degree of which should be larger in 1vxb. Experimental conditions were selected to maintain the integrity of the crystals (which were initially formed at neutral pH and then soaked in a lower pH buffer) and these structures probably contain unstable ‘trapped’ intermediates differing from the equilibrium states at pH 4 (Yang & Phillips, 1996 ▶). Uncoordinated movements represented by a spatter of white and gray spots in the DDMs 1bz6-1vxb and 1bz6-1spe (Supplementary Fig. S4) might agree with coordinate changes at intermediate stages of crystal-restrained denaturation.
DDMs of pairs of myoglobin structures with high RMSDD (with the exception of 1bz6–1u7s, which has an RMSDD below the uncertainty threshold) are shown in Supplementary Fig. S5. Analysis of the DDMs of pairs of structures with PDB codes A and B in Supplementary Fig. S5 supports the impression that all but a few DDMs A–B in Supplementary Fig. S5 are mainly superpositions of the reference DDMs 1bz6–A and 1bz6–B (Supplementary Fig. S4) in which structures A and B are compared with the reference structure 1bz6.
The DDMs in Supplementary Fig. S4 show a drastic difference between two myoglobins with the same P6 symmetry, a D122N mutation, a Met initiator and pH 9, but of which one (1jw8) was studied at cryogenic temperature and the other (2mbw) apparently at room temperature (the temperature is not listed in the PDB). The same is true for the imidazole-liganded cryogenic structure (1u7r) and that (1mbi) apparently at room temperature (the temperature is not listed in the PDB). This suggests a significant contribution of low temperature to coordinate differences between structures of the same protein. Such an effect had originally been discovered regarding myoglobin in 1987 (Frauenfelder et al., 1987 ▶). However, the myoglobin structures studied in this work have not been published (Rashin et al., 2009 ▶). While the temperature effect found here is apparently significant, we did not find any pair of native myoglobin structures with P21 symmetry (one at cryogenic and one at room temperature) at neutral pH in the deoxy-, oxy-, met- or CO-state and with RMSDDs above the uncertainty threshold. It may be that the extra contribution to the RMSDDs in the 1987 study came from C-terminal residues 152 and 153, which were excluded from comparison in our study (see also Supporting Information §S8).
3.4. Analysis of the roles of intermolecular bonds in crystals
Next, we check whether new or stronger intermolecular crystal (hydrogen) bonds form where the DDM indicates a significant conformational difference (a white strip or spot) in the GH area. In our reference structures 1bz6, 1bzp and 1bzr there is a weak intermolecular hydrogen bond to the GH area from Lys62 which is replaced or strengthened in other myoglobin structures (see Table 1 ▶)
Table 1. Intermolecular bonds with the GH region in myoglobin crystals.
| No. | PDB code | Bonds (length in Å) | Comments |
|---|---|---|---|
| 1 | 1bz6 | Arg118 O–Lys62 NZ (3.3) | Reference whale structure (P21 symmetry) |
| 2 | 1hjt | His119 ND–Lys62 NZ (3.0), Ser117 O–Lys63 NZ (3.3) | |
| 3 | 1ebc | Asp122 OD2–Lys62 NZ (3.2) | |
| 4 | 2jho | Arg118 O–Lys62 NZ (2.5) | Much stronger than reference and strong GH band in DDM |
| 5 | 1u7s | Gly121 O–Ala125 N (2.8), Ala125 N–Gly121 O (2.8) | Symmetric from second molecule |
| 6 | 1mwc | Asp126 OD2–Lys63 NZ (3.1) | Pig myoglobin (I1211 symmetry) |
| 7 | 1ymb | Asp122 OD1–Lys62 NZ (3.1), His119 ND1–Lys62 NZ (3.1), Ser117 O–Lys63 NZ (2.7) | Horse myoglobin (P21 symmetry) |
| 8 | 1dwt | Asp122 OD1–Lys62 NZ (2.8), His119 ND1–Lys62 NZ (3.2), Ser117 O–Lys63 NZ (2.8) | Horse myoglobin (P21 symmetry) |
| 9 | 1azi | Asp122 OD1–Lys62 NZ (3.1), Ser117 O–Lys63 NZ (2.8) | Horse myoglobin (P21 symmetry) |
| 10 | 1abs | His119 ND1–Glu18 O (3.3), Asn122 OD1–Glu18 OE1 (2.9), Gly121 O–Lys77 NZ (2.9), Phe123 O–Gln91 NE2 (2.9), Gly124 O–Gln91NE2 (3.3) | Whale; bonds are not to the vicinity of distal His64 (P6 symmetry) |
| 11 | 1jw8 | Same as in 1abs with bond lengths within 0.1 Å | Whale: as in 1abs |
In the cases of 1hjt, 1ebc and 2jho, which all have P21 symmetry and an unusual sixth ligand, there is a correlation between the significant change in the DDM involving GH and either strengthening of the reference bond (2jho) or the formation of new bonds (1hjt and 1ebc) compared with the reference intermolecular bond from near the distal His64 (which can be perturbed by unusual ligands) to the neighborhood of the GH region (however, see §3.6). Combinations of these intermolecular bonds are also found in most horse myoglobins (e.g. 1ymb, 1dwt and 1azi in Table 1 ▶).
A strong change in the GH region compared with the reference structures by the formation in 1u7s of two new strong intermolecular main-chain bonds (2.8 Å long) between the GH regions of neighboring monomers correlates with a low pH of 4.5 of the cryogenic structure 1u7s. Pig myoglobin 1mwc, which is close to the whale outlier 1u7s in Fig. 1 ▶, has only one intermolecular bond (3.1 Å in length) to the GH region, which is weaker than either of the two bonds in 1u7s. GH distortion in 1mwc is much weaker than in 1u7s, as indicated by the lack of a horizontal white band at GH of the 1bz6–1mwc DDM in Fig. 3 ▶.
1abs (at 20 K) and 1jw8 (at 100 K) have practically identical DDMs but a number of different intermolecular hydrogen bonds as calculated with CCP4.
However, Supplementary Fig. S7, which contains the alignment of bonds in 1bz6 and 2mbw, shows several new intermolecular bonds in 2mbw compared with the reference 1bz6, while the 1bz6–2mbw DDM in Supplementary Fig. S4 shows only very minimal differences between these two structures. Thus, as it has previously been shown that mere differences in intermolecular contacts do not usually indicate conformational differences (Kondrashov et al., 2008 ▶), here we find that differences in intermolecular bonding also do not necessarily indicate conformational differences.
Strong distortion of the distal region in the cryogenic 1u7r suggests that the known room-temperature intramolecular distortions (1mbi) of this region by the positively charged imidazole ligand in the sixth coordination site are significantly enhanced at low temperature (compare the 1bz6–1u7r and 1bz6–1mbi DDMs in Supplementary Fig. S4). This suggests a need for a study of the changes in intramolecular long-range hydrogen bonding between different structures (see §3.5).
3.5. Changes in long-range intramolecular and intermolecular hydrogen bonds
Even in the high-resolution reference room-temperature structures with P21 symmetry (1bz6, 1bzp and 1bzr), six out of a total of 34 long-range bonds involving side chains are not fully preserved. Only ten of the 34 long-range bonds in the reference structures are intermolecular and three out of these nine are not preserved among all reference structures. A high-resolution cryogenic structure (2z6s) retains 29 of the 34 long-range bonds observed in the reference room-temperature structures. Of the five unpreserved long-range bonds, four are intermolecular and only one is intramolecular (Supplementary Table S6). This might suggest that cryogenic temperature contracts molecules more than the intermolecular medium, because with a uniform contraction of cryogenic unit cells (see Supporting Information §S2) one might expect a shortening of the intermolecular distances and thus easier formation of intermolecular bonds.
Structures with unusual ligands, symmetries different from P21 or low pH lose between 12 (1hjt) and 19 (1u7r) of the 34 long-range bonds observed in the reference structures, but form many new contacts including new intermolecular bonds (see Supplementary Table S6 for a detailed albeit partial listing).
Interestingly, almost all of the myoglobin structures (see Supplementary Table S6) preserve six invariant (preserved) intramolecular long-range bonds: Lys42 NZ–Lys98 O, Lys77 NZ–Glu18 OE, His82 NE2–Asp141 OD2 and Ile99 O–Tyr146 OH. Additionally, Lys79 NZ–Glu4 OE1,2 (absent only in 2cmm) is practically invariant; 2cmm has a porphin instead of a heme and makes a Ser35 O–Glu4 N intermolecular bond instead of the invariant Lys79 NZ–Glu4 OE1,2. Similarly, His119 NE2–His24 NE2 is practically invariant and is only missing in the oldest and least accurate structure 1mbn. We are not aware of any previous discussions of the role of these invariant long-range bonds (except for Ile99 O–Tyr151 OH) in myoglobin structures. Supplementary Table S6 does not list all of the myo46 structures, but we have confirmed the preservation of the same six invariant intramolecular bonds in all of them. All non-mutant horse and pig structures preserve five of these bonds (they have Asp4 instead of Glu4, which cannot form the same longer bond as in the whale structures). We also checked the preservation of the residues involved in these six intramolecular bonds in the entire myo291 set. We found no substitutions except for that of Ile99 (involved in the Ile99 O–Tyr146 OH bond) by Ala99 and Val99 in 1cik and 1cio, respectively. However, this still allows the invariant Ile99 O–Tyr146 OH bond because only the main-chain O of residue 99 and not its side chain is involved in this bonding. As the myo291 set involves the most relevant mutants, it appears that nobody has attempted to mutate residues in the preserved intramolecular hydrogen bonds to investigate whether this could have a significant effect on the structures.
3.6. Factors correlating with crystallization in P6 versus P21 symmetry
The three myoglobin structures with P6 symmetry in Supplementary Table S1 all are characterized by three factors: (i) the presence of the N-terminal methionine in addition to the regular 153 residues in the sperm whale myoglobin sequence; (ii) the mutation D122N (if Met is given sequence number 0); and (iii) they were studied at pH 9.0. It is unknown whether these three factors determine crystallization with P6 symmetry. We did not intend to perform an exhaustive analysis here of many P6 myoglobin structures. It had been suggested (Phillips, 1990 ▶) that P6 symmetry is necessarily caused by the initiator Met residue at the N-terminus. Supplementary Fig. S6 (with the DDMs of six structures of interest compared with reference 1bz6) provides examples of other molecular characteristics that override some of these three factors.
The DDM of the first structure crystallized in space group P6, 2mbw, is characterized by three standard factors (see above). The mutation allows the new intermolecular hydrogen bond Asn122 ND2–Glu18 OE1, which is impossible for the native Asp122. The DDM of the second structure with P6 symmetry, 1tes, is almost indistinguishable from that of the first, having added an ‘unusual’ sixth coordination ligand. The DDM of the third structure with P6 symmetry, 1ch2, is practically identical to the first (2mbw), while having a second mutation L89F in addition to the three standard features of P6 crystals. The fourth P6 structure (1mti) has a different second mutation, F46L, and a somewhat more complex DDM than the previous three. 2blh is the fifth P6 structure and has a second mutation L29W in addition to the ‘standard’ D122N. Additionally, it has differences in two of the other features standard for P6 myoglobin crystals: it does not have an initiator methionine and was crystallized at pH 7.8. Its DDM is quite drastically different from all previous DDMs of P6 structures in Supplementary Figs. S4 and S6. The DDM of the sixth structure, 1dti, in Supplementary Fig. S6 characterizes a structure with all three features typical for P6 crystals but that contains a second mutation, H97D, and belongs to space group P212121. In 1dti the intermolecular bond Asn122–Glu18, which is present in all D122N-mutated P6 structures, is replaced by the intramolecular bond Asn122–Lys16. Two additional structures without the standard P6 features, but including heme substitutes, crystallize in P6 (1iop with 6,7-dicarboxyl hemin) and in P212121 (2cmm with porphin instead of heme). These two structures also have cyanide as their sixth ligand coordinated to iron and the distal histidine. Another cryogenic P6 structure (2w6w; DDM not shown) has Asp122 unmutated, the initiator Met crystallographically unresolved and was crystallized at pH 8.5 but contains four buried Xe atoms. All of these data indicate that the crystallization symmetry is associated with numerous parameter changes.
4. Discussion
Our study indicates that in smaller data sets (e.g. pig and horse myoglobins) it can be relatively easy to identify the causes of coordinate differences. Studying a combined mammalian data set allows the elucidation of possible causes of coordinate differences that are otherwise hidden in the large (216) whale set.
Our results show that among 46 sperm whale myoglobin structures, only 85 of over 1000 pairwise comparisons reveal coordinate differences beyond the coordinate uncertainty thresholds (Rashin et al., 2009 ▶). Most of these 85 outliers can be explained by (or correlated with) one or a few specific differences between the compared structures. There are almost always exceptions where these factors do not lead to coordinate differences beyond the uncertainty thresholds.
One such factor correlated to significant coordinate differences in the range commonly interpreted as biologically important are the unusual ligands in the sixth coordination site forming strong hydrogen bonds with the heme iron and the distal histidine (His64) or pushing it out of its usual position. These unusual ligands are cyanide (1ebc and 2jho), nitric oxide (1hjt), imidazole (1u7r) and hydroxyl ion (1mbn). Neither azide ion (1swm) nor ethyl isocyanide (1tes) show any significant effects. However, as Fig. 5 ▶ shows, even 1ebc or 1hjt do not always differ beyond the coordinate uncertainty threshold for all structures. While the reasons for the presence or absence of such strong effects in whale myoglobins (i.e. distortions of the GH region with the formation of specific crystal bonds) are not clear, the same intermolecular crystal bonds, leading to significant distortions of the GH region, are a repeating feature in practically all horse myoglobins with or without unusual ligands. A crystallographic study of mutants eliminating these intermolecular hydrogen bonds could significantly clarify the dispute on the difference between the crystal and solution structures. Structural studies of mutants of conserved intramolecular hydrogen bonds might also be illuminating.
However, the distal histidine is a kind of ‘lid’ (often found with biologically significant coordinate differences) over the ligand-binding site of myoglobin (Scott et al., 2001 ▶). Therefore, the introduction of unusual ligands strongly interacting with this ‘lid’ and consequent coordinate changes beyond the uncertainty threshold can be classified as biologically important. This might suggest that the bound substrate analogs often used in crystallographic studies of protein mechanisms can, like unusual distal ligands in myoglobin, cause rather large loop movements which are seemingly biologically relevant but are in fact caused by intermolecular crystal contacts/bonds and/or different protein–substrate contacts/bonds, and have nothing to do with protein mechanism in solution. Unfortunately, contrary to what is typically observed in the active-site lid movements of most proteins (Rashin et al., 2009 ▶, 2010 ▶), lid movements of the distal histidine in myoglobin seem to be limited to its side chain and do not involve any large repositioning of its main chain. Owing to this limitation, functional movements, when considering Cα solely, are undetectable by any method, including PCA (see Supporting Information §S4).
It should be noted that in some cases no new (compared with the reference structures) intermolecular hydrogen bonds form or such bonds existing in the reference structures do not contract, and the observed conformational change proceeds intramolecularly (e.g. as in the case of the imidazole ligand).
The low-pH structures 1spe, 1vxb (pH 4.0, room temperature) and 1u7s (pH 4.5, T = 100 K) are of particular interest. At room temperature, myoglobin undergoes pH denaturation around pH 4.0, and 1spe and 1vxb are partially denatured structures/molecules trapped in the folded form by the crystal. Main-chain intermolecular hydrogen bonds, resembling β-structure, form between residues 121 and 125 in the cryogenic 1u7s. This leads to further questions: does myoglobin start to denature at pH 4.5 and cryogenic temperature? Are the thermodynamic characteristics of the various interactions and their magnitudes the same as those at physiological temperatures? Thus, for the structure 1u7s there are two factors that may cause coordinate changes: cryogenic temperature and low pH. Since in myoglobin cryogenic temperature (which is biologically irrelevant) amplifies coordinate changes through mechanisms that are not yet understood, the wide use of structures determined at cryogenic temperatures in protein-function studies may also introduce biologically irrelevant conformational changes that can easily lead to misinterpretations (see Fraser et al., 2011 ▶).
It might also be worth noting a recent suggestion to test, by comparing the set of four crystal structures with an NMR myoglobin ensemble (Kondrashov et al., 2008 ▶; see also Fraser et al., 2011 ▶), the hypothesis that myoglobin structures in different crystalline forms are conformers selected out of the solution state by comparing the set of four crystal structures with an NMR myoglobin ensemble (Kondrashov et al., 2008 ▶; see also Fraser et al., 2011 ▶). This hypothesis does not seem to have a strong physical justification, even if one disregards the uncertainties in a molecular-dynamics simulation of the NMR solution ensemble. Four myoglobin crystal structures with different symmetries do not belong to the same thermodynamic ensemble because they have different ligands, have mutations (i.e. in P6 molecules) and represent very different solution conditions. An NMR ensemble is supposed to represent identical molecules under the same conditions. Furthermore, with three terminal residues not included in the structural comparison (Kondrashov et al., 2008 ▶), the agreement between the ‘ensemble’ of four crystal structures and the NMR ensemble has a correlation coefficient of only 0.56 (see, however, Csermely et al., 2010 ▶). Our results rather support the view that ‘the crystal state may correspond to a global minimum of free energy where biologically relevant interactions are sacrificed in favor of (biologically) unspecific contacts’ (Krissinel, 2010 ▶).
Pairwise coordinate differences, as observed in this work, usually correlate with more than one factor, and more factors can be found later. Simplistic ideas of intermolecular contacts or more specific intermolecular hydrogen bonds as a universal cause of coordinate differences are found to be inadequate and to provide explanations only in some cases. While such differences in myoglobins are amplified when one of the compared structures is cryogenic, the temperature factor remains cryptic (see §3.3 and Supporting Information §S8). What other factors will emerge from further studies cannot be foreseen, but the methodology of protein structure validation does need to be further developed and broadly applied. We hope that the study presented here can help to move towards this goal.
Supplementary Material
Supporting Information.. DOI: 10.1107/S1399004713028812/dz5300sup1.pdf
Acknowledgments
We thank Dr Lesa Offermann for valuable comments. We also thank Professor Jane Richardson for a stimulating and productive discussion. This work was supported by NIH Grants R01GM072014 and R01GM053163.
Footnotes
Supporting information has been deposited in the IUCr electronic archive (Reference: DZ5300).
References
- Bahadur, R. P., Chakrabarti, P., Rodier, F. & Janin, J. (2004). J. Mol. Biol. 336, 943–955. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Carugo, O. & Argos, P. (1997). Protein Sci. 6, 2261–2263. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Csermely, P., Palotai, R. & Nussinov, R. (2010). Trends Biochem. Sci. 10, 539–546. [DOI] [PMC free article] [PubMed]
- Dasgupta, S., Iyer, G. H., Bryant, S. H., Lawrence, C. E. & Bell, J. A. (1997). Proteins, 28, 494–514. [DOI] [PubMed]
- DePristo, M. A., de Bakker, P. I. W. & Blundell, T. L. (2004). Structure, 12, 831–838. [DOI] [PubMed]
- Fraser, J. S., van den Bedem, H., Samelson, A. J., Lang, P. T., Holton, J. M., Echols, N. & Alber, T. (2011). Proc. Natl. Acad. Sci. USA, 108, 16247–16252. [DOI] [PMC free article] [PubMed]
- Frauenfelder, H., Hartmann, H., Karplus, M., Kuntz, I. D., Kuriyan, J., Parak, F., Petsko, G. A., Ringe, D., Tilton, R. F., Connolly, M. L. & Max, N. (1987). Biochemistry, 26, 254–261. [DOI] [PubMed]
- Frauenfelder, H., McMahon, B. H., Austin, R. H., Chu, K. & Groves, J. T. (2001). Proc. Natl. Acad. Sci. USA, 98, 2370–2374. [DOI] [PMC free article] [PubMed]
- Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J. & Lopez, R. (2010). Nucleic Acids Res. 38, W695–W699. [DOI] [PMC free article] [PubMed]
- Henrick, K. & Thornton, J. M. (1998). Trends Biochem. Sci. 23, 358–361. [DOI] [PubMed]
- Horimoto, K. & Toh, H. (2001). Bioinformatics, 17, 1143–1151. [DOI] [PubMed]
- Janin, J. & Rodier, F. (1995). Proteins, 23, 580–587. [DOI] [PubMed]
- Janin, J. & Wodak, S. J. (1983). Prog. Biophys. Mol. Biol. 42, 21–78. [DOI] [PubMed]
- Jaskolski, M., Gilski, M., Dauter, Z. & Wlodawer, A. (2007). Acta Cryst. D63, 611–620. [DOI] [PubMed]
- Jolliffe, I. T. (2002). Principal Component Analysis New York: Springer-Verlag.
- Kleywegt, G. J. (2009). Acta Cryst. D65, 134–139. [DOI] [PMC free article] [PubMed]
- Kondrashov, D. A., Zhang, W., Aranda, R., Stec, B. & Phillips, G. N. Jr (2008). Proteins, 70, 353–362. [DOI] [PubMed]
- Krebs, W. G., Tsai, J., Alexandrov, V., Junker, J., Jansen, R. & Gerstein, M. (2003). Methods Enzymol. 374, 544–584. [DOI] [PubMed]
- Krissinel, E. (2010). J. Comput. Chem. 31, 133–143. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Lionetti, C., Guanziroli, M. G., Frigerio, F., Ascenzi, P. & Bolognesi, M. (1991). J. Mol. Biol. 217, 409–412. [DOI] [PubMed]
- Phillips, G. N. Jr (1990). Biophys. J. 57, 381–383. [DOI] [PMC free article] [PubMed]
- Rashin, A. A., Rashin, A. H. L. & Jernigan, R. L. (2009). Acta Cryst. D65, 1140–1161. [DOI] [PMC free article] [PubMed]
- Rashin, A. A., Rashin, A. H. L. & Jernigan, R. L. (2010). Biochemistry, 49, 5683–5704. [DOI] [PMC free article] [PubMed]
- Rashin, A. A., Rashin, B. H., Rashin, A. & Abagyan, R. (1997). Protein Sci. 6, 2143–2158. [DOI] [PMC free article] [PubMed]
- Scott, E. E., Gibson, Q. H. & Olson, J. S. (2001). J. Biol. Chem. 276, 5177–5188. [DOI] [PubMed]
- Sevcik, J., Dodson, E. J. & Dodson, G. G. (1991). Acta Cryst. B47, 240–253. [PubMed]
- Shindyalov, I. N. & Bourne, P. E. (1998). Protein Eng. 11, 739–747. [DOI] [PubMed]
- Smerdon, S. J., Oldfield, T. J., Dodson, E. J., Dodson, G. G., Hubbard, R. E. & Wilkinson, A. J. (1990). Acta Cryst. B46, 370–377. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, F. & Phillips, G. N. Jr (1996). J. Mol. Biol. 256, 762–774. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S1399004713028812/dz5300sup1.pdf