Abstract
The Src family of protein kinases (SFKs) plays key roles in regulating fundamental cellular processes, including cell growth, differentiation, cell shape, migration and survival, and specialized cell signals in various malignancies. The pleotropic functions of SFKs in cancer make them promising targets for intervention. Here we sought to investigate the role of miR-205 in inhibition of Src-mediated oncogenic pathways in renal cancer. We report that expression of miR-205 was significantly suppressed in renal cancer cell lines and tumors when compared with normal tissues and a non-malignant cell line, and is correlated inversely with the expression of SFKs. miR-205 significantly suppressed the luciferase activity of reporter plasmids containing the 3’UTR sequences complementary to either Src, Lyn or Yes, which was abolished by mutations in these 3’UTR regions. Over-expression of miR-205 in A498 cells reduced Src, Lyn and Yes expression both at mRNA and protein levels. Proliferation of renal cancer cells was suppressed by miR-205, mediated by the phosphoSrc-regulated ERK1/2 pathway. Cell motility factor- FAK and STAT3 activation was also inhibited by miR-205. Transient as well as stable over-expression of miR-205 in A498 cells resulted in induction of G0/G1 cell cycle arrest and apoptosis as indicated by decreased levels of cyclin D1 and cMyc, suppressed cell proliferation, colony formation, migration, and invasion in renal cancer cells. miR-205 also inhibited tumor cell growth in vivo. This is the first study demonstrating that miRNA-205 inhibits protooncogenic Src family of kinases indicating a therapeutic potential of miR-205 in the treatment of renal cancer.
Keywords: MicroRNA-205, Src, Lyn, Yes, renal cancer
Introduction
Src family kinases (SFKs) are prototypical modular signaling proteins and the largest family of non-receptor protein tyrosine kinases (1-3). SFKs have been shown to be up-regulated in multiple types of human tumors, with Src activity increasing proportionally to the progressive stages of the disease (1, 4). Of the 9 family members, c-Src, Fyn and Yes are widely expressed in tissues and appear to play an important role in the regulation of cell adhesion, cell growth, and differentiation (5). Among the SFKs, Src itself is most frequently implicated in human cancer, and previous studies have shown that, in mouse models, Src activation is associated with progression and metastasis in pancreatic (6) and colorectal (7) carcinomas. In prostate cancer cells in vitro, inhibition of SFKs decreases proliferation (8) and, more profoundly, invasion and migration (9); the latter through selective inhibition of phosphorylation of Src substrates, such as focal adhesion kinase (FAK) and Crk associated substrate (10). In renal cancer Src has been shown to contribute to the appearance of malignant phenotypes particularly due to the resistance against apoptosis by Bcl-xL and angiogenesis stimulated by Src-Stat3-VEGF signaling (11). The pleiotropic effects of Src activity are due to the multiple signal pathways engaged by Src and its accompanying kinases. Src is able to channel phophorylation signals through Ras/Raf/extracellular signal-regulated kinase (ERK) 1/2 and in certain cells, phophatidylinositol 3-kinase (PI3K)/AKT pathways. Somewhat selective to SFKs is their ability to activate signal and transducer of transcription (STAT) 3 and β-catenin, which leads to the activation of c-Myc (12, 13), and consequently cyclin D1 (14, 15). Overall, these studies suggest that Src plays pleiotropic roles in cancer, often in a cell dependent manner and that Src is a promising target for intervention. Here we provide the first demonstration that inhibition of SFKs can be effectively achieved by microRNA-205 (miR-205) in renal cancer.
Renal cell carcinoma (RCC) is the seventh most common cancer in the United States and was predicted to result in nearly 13 000 deaths in 2009 (16). Surgery is the first line of treatment with successful resection, often resulting in long-term disease-free status. Although the overall survival rate is more than 60% over 5 years (16), ~30% of patients who are diagnosed with localized RCC develop metastatic recurrence (17). These patients have a very poor prognosis because of the refractory nature of RCC to current treatment regimens. Therefore, there has been much interest in the identification of biomarkers for RCC to better predict cancer development and prognosis.
MicroRNAs (miRNAs) are small, non-coding RNAs that have been found to regulate expression through targeted repression of gene transcription and translation. These endogenous, silencing RNAs have been shown to play important roles in development and differentiation (18, 19), cellular stress responses (20), and cancer (21). Specific subsets of miRNAs have also been shown to be dysregulated in various solid tumors (22, 23). Due to their tremendous regulatory potential and tissue-specific and disease-specific expression patterns (24, 25), there is increasing evidence that miRNA expression profiles could be indicative of disease risk and burden. Thus, miRNAs are being assessed as possible biomarkers to aid in the diagnosis and prognosis of different cancers (26, 27). Here we report that miR-205 is significantly down-regulated in renal cancer tissue samples and cell lines. In addition, we examined the consequences of miR-205 over-expression and identified the Src family of kinases as direct targets of miR-205 in renal cancer.
Materials and Methods
Cell culture, plasmids and transfection
Human renal cell carcinoma cell lines A498, ACHN, Caki1, 769-P and a non-malignant renal cell line HK-2 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and grown according to ATCC protocol (28). Plasmids pEZX-MT01 miRNA 3’UTR target expression clones for Src (HmiT017696-MT01), Lyn (HmiT010935-MT01), Yes (HmiT018569-MT01), LCK (CS-HmiT010565-MT01) and miRNA Target clone control vector for pEZXMT01 (CmiT000001-MT01) (GeneCopoeia, Rockville, MD), miRNASelect™ pEP-miR Null control vector (pEP Null), miRNASelect™ pEP-hsa-mir-205 expression vector (pEP miR-205) (Cell Biolabs Inc, San Diego CA) were purchased. TaqMan probes for hsa-miR-205 (miR-205), anti-miR-205 and negative controls pre-miR and anti-miR-Control (cont-miR) were purchased from Applied Biosystems (Foster City, CA). siRNA duplexes ((Src (Human)-3 unique 27mer siRNA duplexes (SR304574)) were purchased from Origene (Origene Technologies, Inc., Rockville, MD).
Quantitative real-time PCR
Tissue samples from radical nephrectomy were obtained from the Veterans Affairs Medical Center, San Francisco, CA, USA. Total RNA was extracted and assayed for mature miRNAs and mRNAs using the TaqMan MicroRNA Assays and Gene Expression Assays, respectively, in accordance with the manufacturer's instructions (Applied Biosystems). All RT reactions were run in a 7500 Fast Real Time PCR System (Applied Biosystems). Relative expression was calculated using the comparative Ct.
Flow cytometry, cell viability, migratory, clonability and invasion assays
FACS analysis for cell cycle and apoptosis was done 72 hours post-transfection using nuclear stain DAPI for cell cycle analysis or ANNEXIN V-FITC /7-AAD KIT (Beckman Coulter, Inc. Fullerton, CA) for apoptosis analysis according to the manufacturer's protocol. Cell viability was determined at 24, 48 and 72 h by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer's protocol. For colony formation assay, cells were seeded at low density (1000 cells/plate or 200 cells/plate) and allowed to grow till visible colonies appeared. Then, cells were stained with Giemsa and colonies were counted. An artificial ‘‘wound’’ was created on a confluent cell monolayer and photographs were taken after 24 hrs for migration assay. Also a cytoselect 24-well cell migration and invasion assay kit (Cell Biolabs, Inc) was used for migration and invasion assays according to manufacturer's protocol.
Immunoblotting
Immunoblotting was performed as described previously (29). Briefly protein was isolated from 70-80% confluent cultured cells using the M-PER Mammalian Protein Extraction Reagent (Pierce Biotechnology, Rockfield, IL) following the manufacturer's directions. Equal amounts of protein were resolved on 4-20% SDS polyacrylamide gels and transferred to nitrocellulose membrane. The resulting blots were blocked with 5% non-fat dry milk and probed with antibodies. All antibodies were obtained from Cell Signalling Technology Inc. Denver, MA except c-Myc and cyclin D1 which were purchased from BD Pharmingen (BD Biosciences). Blots were visualized using enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL).
Luciferase Assays
The Src, Lyn, Yes, Lck and Control vectors were purchased from GeneCopoeia and named as Src-3’UTR, Lyn-3’UTR, Yes-3’UTR, Lck-3’UTR and Empty-Vector, respectively. Mutated 3’UTR sequences of Src, Lyn and Yes complementary to miR-205 were cloned and named Src-Mut, Lyn-Mut and Yes-Mut. For reporter assays, cells were transiently transfected with wild-type or mutant reporter plasmid and miR-205 or control-miR. Firefly luciferase activities were measured by using the Dual Luciferase Assay (Promega, Madison, WI) 24 hr after transfection and the results were normalized with Renilla luciferase. Each reporter plasmid was transfected at least three times (on different days) and each sample was assayed in triplicate.
Stable cell generation and in vivo study
A498 cells were transfected with pEP Null vector and pEP miR-205 vector (Cellbiolabs, San Diego CA) and selected with puromycin (1μg/mL). pEP miR-205 vector was labeled with RFP. After transfection cells were observed under a microscope to check for red fluorescence and then selected with a cell sorter (BD FACSAria II (BD Biosciences, San Jose, CA). The sorted cells were grown in puromycin and real time quantitative PCR was performed to check the expression of miR-205. For in vivo studies, 5×106 cells were injected into nude mice subcutaneously and tumor growth was followed for 28 days. We also looked at the antitumor effects of miR-205 by local administration in established tumors. Each mouse was injected with 7.5×106 cancer cells. Once palpable tumors developed (average volume 80mm3), 6.25 μg synthetic miRNA complexed with 1.6 μl siPORT Amine transfection reagent (Ambion, Austin, TX) in 50 μl PBS was delivered seven times intratumorally in 3-day intervals. Tumor growth was followed for 21 days from first injection. All animal care was in accordance with the institutional guidelines.
Statistical analysis
All quantified data represents an average of at least triplicate samples or as indicated. Error bars represent standard deviation of the mean. Statistical significance was determined by the Student's t-test and two-tailed p values less than 0.05 were considered significant.
Results
miR-205 is downregulated in renal carcinoma, and its expression is inversely correlated with that of Src family of kinases
Preliminary microarray data revealed that miR-205 was highly downregulated in renal cancer cell lines compared to the non-malignant HK-2 cell line (data not shown). We validated the microarray data by miRNA-quantitative RT-PCR (miR qRT-PCR) analysis and results confirmed that miR-205 was downregulated in all the cancer cell lines (Figure 1A). To examine the clinical relevance of miR-205, its expression was analyzed in carcinoma and normal renal tissue samples. Patients and tumor characteristics are summarized in Supplemental Table 1. Almost all carcinoma samples showed significant down regulation of miR-205 expression with respect to the normal samples and an overall lower relative average expression was observed in carcinoma compared to normal samples (Figure 1B). These results suggest a potential tumor suppressor role for miR-205 in renal carcinoma. To identify the potential targets of miR-205, we used different algorithms that predict the mRNA targets of a miRNA: miRanda (30) microRNA target predictions (31), TargetScan (32), and PicTar (33). Among the list of potential targets of miR-205 were mRNAs encoding the Src family of kinases, Src, Lyn, Yes and LCK. The seed sequence of miR-205 was complementary to the 3’UTR of these genes (Figure 1C). To investigate the correlation between expression of miR-205 and that of Src, Lyn and Yes, we measured expression of Src, Lyn and Yes at the mRNA and protein levels in the same panel of cell lines and in individual sets of 12 pairs of tissue samples (Supplemental Figure 1). The mRNA and protein expression levels of these genes were higher when compared to the non-malignant cell line (Figure 1D-E), although the absolute level of expression varied among different cell lines. The relative miR-205 expression was higher in the normal tissue samples compared to paired tumor samples, whereas the mRNA expression of Src, Lyn and Yes was high in tumors compared to their normal samples (Supplemental Fig. 1). This data clearly demonstrated an inverse correlation between the expression of miR-205 and that of Src, Lyn and Yes in renal cancer, suggesting that these genes are targets of miR-205.
Figure 1. miR-205 expression is downregulated in renal cancer and inversely correlated with expression of Src, Lyn and Yes.
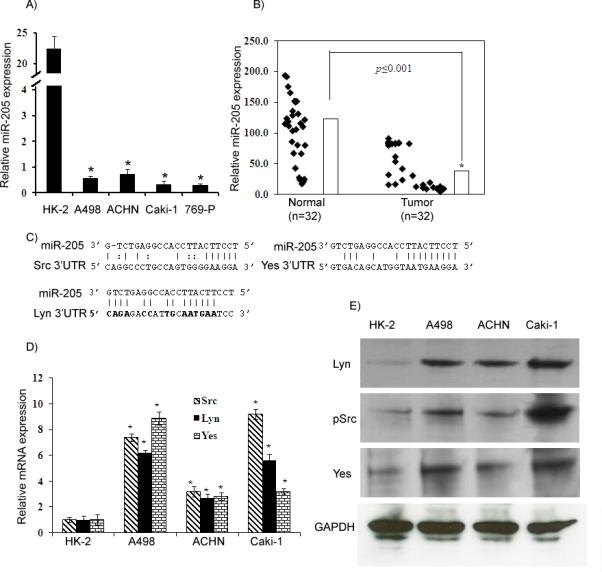
A) Quantitative RT-PCR analysis of miR-205 expression levels in renal cancer and nonmalignant cell lines. B) miR-205 expression in a cohort of renal cancer and normal tissue samples. C) The miR-205 seed sequence is complementary to the 3’UTR of Src, Lyn and Yes. D-E) Src, Lyn and Yes mRNA and protein expression in human renal cancer and nonmalignant cell lines. *p<0.05
The Src family members are direct targets of miR-205
We investigated whether the 3’UTR of Src, Lyn, Yes and LCK are functional targets of miR-205 in renal cancer. Transient transfection of human A498 cancer cells with Src, Lyn, and Yes 3’UTR plasmids along with miR-205 led to a significant decrease in relative luciferase units when compared with empty vector and cont-miR or empty vector and miR-205 (Figure 2A-D). No significant difference was observed in the case of LCK-3’UTR (data not shown). The luciferase activity of the reporter vectors containing a mutated 3’UTR of the respective genes was unaffected by miR-205 (Figure 2A-D). These results indicate that members of the Src family of kinases, Src, Lyn and Yes (but not LCK) are direct targets of miR-205 in renal cancer.
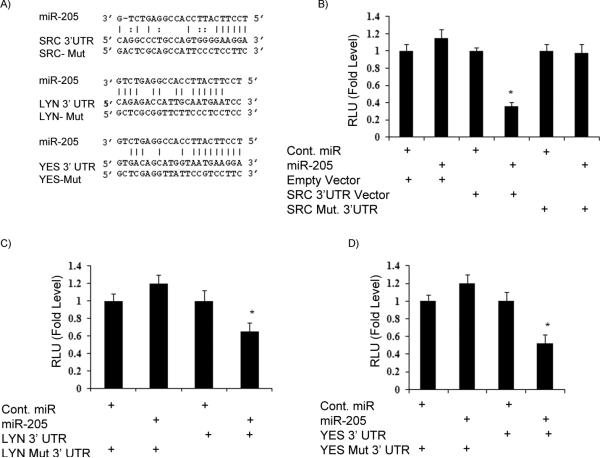
Figure 2. Src, Lyn and Yes 3’UTRs are targets of miR-205.
A) The 3’UTR sequences of Src, Lyn and Yes and mutant sequences that abolished binding. B-D) Luciferase assays showing decreased reporter activity after co-transfection of either Src-3’UTR, Lyn-3’UTR or Yes-3’UTR with miR-205 in A498 cells. The mutant 3’ UTRs of either gene had no effect on reporter activity. *p<0.05
miR-205 suppresses Src family members and negatively regulates the Ras/Raf/ERK1/2 pathway in renal carcinoma
We then determined whether the overexpression of miR-205 could regulate the levels of Src, Lyn and Yes mRNA or protein and alter downstream signaling events. A498 cells were transfected with miR-205, resulting in miR-205 overexpression as determined by miR qRT-PCR analysis (Figure 3A). miR-205 transient transfection significantly downregulated Src, Lyn and Yes at the mRNA level (Figure 3B). Western blot analysis also confirmed the downregulation of these genes at the protein level with miR-205 overexpression (Figure 3C). These results support the notion that miR-205 binds to the 3’UTR of these genes and regulates their expression. Src family kinases have been shown to be up-regulated in multiple types of human tumors. c-Src itself is widely expressed in tissues and plays an important role in the regulation of cell adhesion, cell growth, and differentiation (5). It is frequently implicated in human cancer, and previous studies have shown that, in mouse models, Src activation is associated with pancreatic cancer progression and metastasis (6, 34). Therefore we analyzed its role in response to miR-205 overexpression. Src has been reported to be able to channel phosphorylation signals through the Ras/Raf/extracellular signal-regulated kinase (ERK)1/2 (35). Src also activates signal and transducer of transcription (STAT) 3, a Src target and key transcriptional factors of c-Myc and cycling D1 (36, 37) which leads to their activation(12, 13, 38). Inhibition of Src has been found to inhibit cancer cell proliferation (8), invasion and migration (9); the later through selective inhibition of phophorylation of Src substrates, such as FAK and Crk-associated substrate (10). To determine whether the above mentioned effectors are affected by miR-205-mediated suppression of Src, A498 cells were transfected with miR-205 or cont-miR. Western blot analysis showed reduced levels of the members of the phopho-Erk 1/2 pathway, phospho-STAT3, phosphor-FAK, c-Myc and cyclin D1 (Figure 3D) in cells with suppressed phospho-Src expression following miR-205 overexpression. We next inhibited the endogenous expression of miR-205 in A498 cells by transfecting anti-miR-205 (Figure 3E), an inhibitory oligonucleotide designed specifically to bind and sequester the mature miR-205 sequence to see if the expression of target genes is rescued by inhibiting miR-205. Indeed the expression of all the three genes was restored at both the protein and mRNA levels (Figure 3F) in anti-miR-205 transfected cells. These data indicate that miR-205 targets Src, which in turn results in suppression of the ERK pathway and the genes involved in migration/invasion and proliferation.
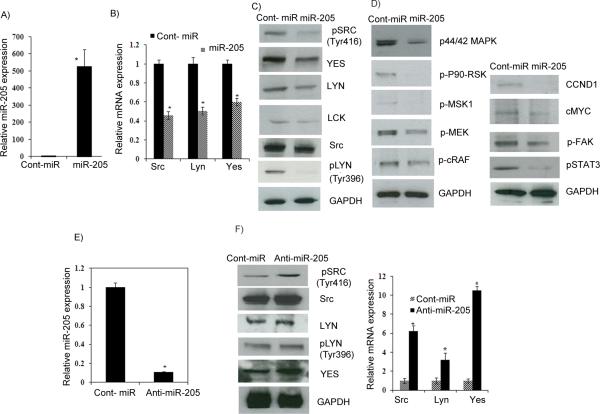
Figure 3. miR-205 suppresses Src, Lyn and Yes expression and regulates the ERK1/2 pathway.
A) Relative miR-205 expression level. B-C) Quantitative RT-PCR and Western blot analysis showing decreased Src, Lyn and Yes expression. D) Western analysis showing a decrease in the ERK1/2 pathway, cyclin D1, cMyc, a decrease in phosphoSTAT3, and FAK in miR-205 transfected A498 cells. E) Relative miR-205 expression in anti-miR-205 transiently transfected A498 cells F) Western blot and qRT-PCR analysis showing that Src, Lyn and Yes expression was rescued in cells transfected with anti-miR-205.
MicroRNA-205 induces apoptosis, cell cycle arrest, impairs cell viability, migratory, clonability and invasive properties of renal cancer cells
Since Src has been reported to be involved in cancer cell proliferation (8) and invasion and migration (9), we sought to determine if downregulation of Src by miR-205 has effect on the cell cycle, viability, migratory or invasion properties of A498 cancer cells. A significant decrease in cell proliferation was observed over time in A498 cells expressing miR-205 (Figure 4A) as compared to cells expressing cont-miR. The miR-205-transfected cells also had low colony formation ability, as both size and number of foci in miR-205 expressing cells were suppressed when compared to cont-miR expressing cells (Figure 4B). To determine if miR-205 affects renal cancer cell migration or invasiveness, wound healing, migration and invasion assays were performed. miR-205 over-expressing A498 cells were less proficient than cont-miR transfected cells in closing an artificial wound created over a confluent monolayer (Figure 4C). Less absorbance was observed at 560nm with miR-205 transfected cells compared to cont-miR in the migration assay (Fig. 4C). miR-205 overexpression also significantly reduced the invasiveness of A498 cells (Figure 4D). FACS (fluorescence activated cell sorting) analysis revealed that re-expression of miR-205 leads to a significant increase (10%±3%) in the number of cells in the G0-G1 phase of the cell cycle while the S-phase population decreased from 15%±4% to 5%±3% suggesting that miR-205 causes a G0-G1 arrest in miR-205 transfected A498 cells compared to a nonspecific microRNA control (Cont-miR) (Figure 4E). FACS analysis for apoptosis was performed using Annexin-V-FITC-7-AAD dye. The percentage of total apoptotic cells (early apoptotic + apoptotic) was significantly increased (14%±3%) in response to miR-205 transfection compared to cont-miR (4%±2%) with a corresponding 10%±4 decrease in the viable cell population (Figure 4F). All the functional assays were confirmed in the 769-P cell line which is from the same tumor type as A498 cells and the results were consistent (Supplemental Fig. 2). These results indicate that suppression of phopho-Src by miR-205 inhibits renal cell proliferation, invasion and migration by inhibiting phophorylation of focal adhesion kinase (FAK), a Src substrate, c-Myc, a Src target gene (39) and cyclin D1, the rate limiting factor for cellular proliferation (40, 41).
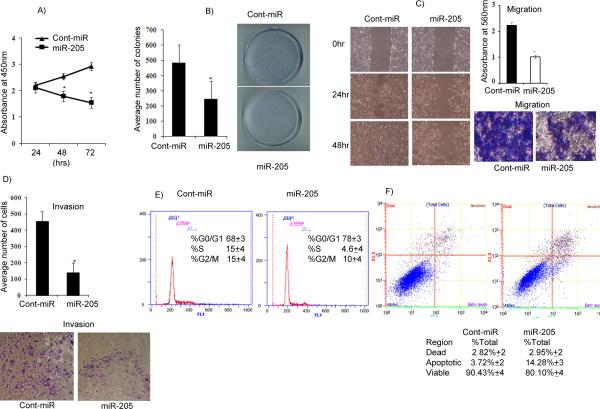
Figure 4. Transient transfection of miR-205 inhibits renal cancer cell proliferation, colony formation, migration, invasion, and induces apoptosis and cell cycle arrest in A498 cells.
A) Proliferation of A498 cells after miR-205 transfection was significantly reduced compared to cont-miR. B) miR-205 overexpression significantly inhibits colony formation of A498 cells. C) Wound healing and migration assays of A498 cells transfected with miR-205. D) Invasion assay shows a significant decrease in the number of invading A498 cells transfected with miR-205. E) Cell cycle analysis showing an increase in the G0/G1 phase of A498 cells overexpressing miR-205. F) Apoptosis assay showing induction of apoptosis by miR-205. *p<0.05.
Src inhibition by siRNA mimics miR-205 reconstitution in renal cancer and attenuation of miR-205 in non-malignant cells increases proliferation, migration and invasion
Phenocopy experiments inhibiting Src expression by siRNA were also performed (Figure 5). We initially tested three siRNAs to achieve 80-90% Src gene knockdown and confirmed the results at the mRNA and protein levels (Figure 5A). Then we selected one siRNA (S-1) for further experiments. Our results showed that siRNA inhibition of Src caused decreased cell viability (Figure 5B), migratory and invasive (Figure 5C) capability of A498 cancer cells. We also observed a G0/G1 cell cycle arrest (14%), whereas there was a decrease of 11% in S-phase cell population (Figure 5D). Almost 5% of the cells were in the apoptotic fraction in Src siRNA transfected cells compared to non-specific control (Figure 5D). These results provide evidence that inhibition of Src by miR-205 reconstitution is responsible for the observed phenotype in renal cancer cells. We also knocked down the expression of miR-205 in HK-2 cells that expressed high levels of miR-205 (Figure 6A) and determined its effect on cell growth, migration and invasion. Our results showed that cells transfected with anti-miR-205 showed increased proliferation (Figure 6B), migration (Figure 6C) and invasion (Figure 6D) compared to control anti-microRNA. These results show that miR-205 is an important tumor suppressor microRNA in renal cancer and attenuation of this microRNA in over expressing non-malignant renal cells increases their proliferative, migratory and invasive capability.
Figure 5. Knock-down of Src by small interfering RNA (siRNA).
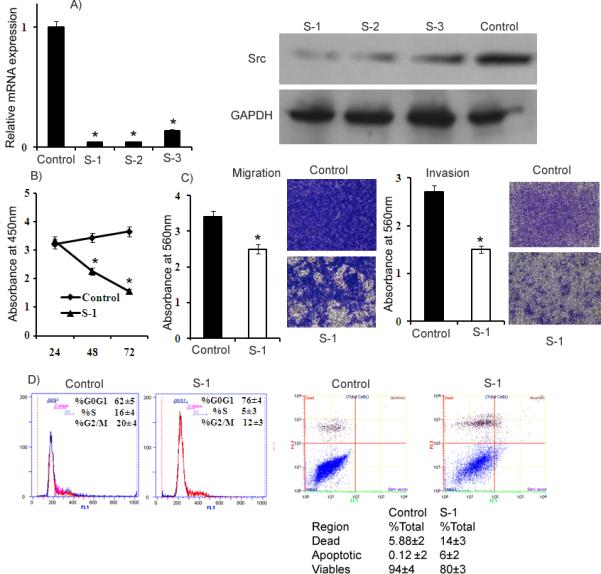
A) Relative Src mRNA levels assessed by qRT-PCR in A498 cells transfected with 50nM siRNA duplexes (S-1, S-2 and S-3) and a nonsilencing siRNA duplex (Control). Src protein levels were assessed by Western blot in A498 cells transfected with 50nM siRNA duplexes and a nonsilencing siRNA duplex. B) Proliferation of A498 cells after S-1 transfection was significantly reduced compared to control. C) A significant decrease was observed in the migratory capability of A498 cells after siRNA (S-1) transfection compared to Control. Invasion assay shows a significant decrease in the number of invading A498 cells transfected with S-1. D) Cell cycle analysis showing an increase in the G0/G1 phase of A498 cells transfected with S-1. Apoptosis assay showing induction of apoptosis after Src knockdown by S-1. *p<0.05.
Figure 6. Attenuation of miR-205 expression by anti-miR-205 in HK-2 cells.
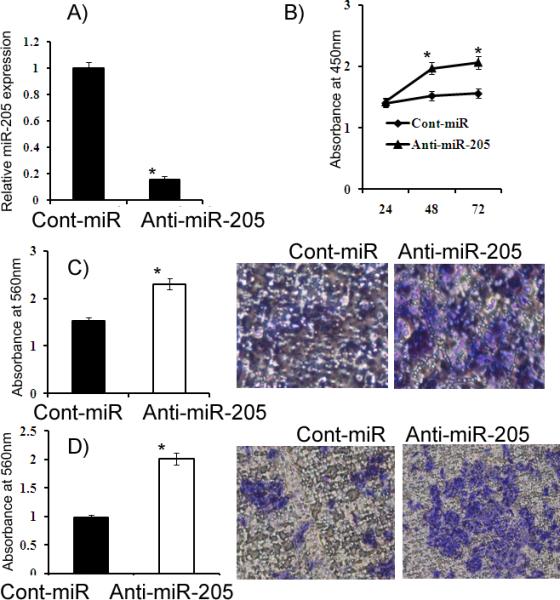
A) Relative miR-205 expression. B) HK-2 cells had increased proliferation after anti-miR-205 transfection compared to the anti-miR-Control (Cont-miR). C-D) Migration and invasion assays. *p<0.05
miR-205 inhibits tumor growth in-vivo
The antitumor effect of miR-205 stably transfected in A498 cells was determined by performing phenocopy experiments in-vitro (Supplemental Figure 3) and confirmed by in vivo experiments. Stable overexpression of miR-205 dramatically suppressed tumor growth in vivo upon subcutaneous injection into nude mice when compared to cells expressing control vector (Figure 7A). We further checked the expression of miR-205 or Src, Lyn and Yes in 8 harvested tumors, four from pEP Null control group and four from pEP miR-205 group. Our results showed that miR-205 expression was significantly high with a corresponding significant decrease in the target gene expression in tumors that had pEP miR-205 compared to the pEP Null control (Supplemental Figure 4AB). Since overexpression of miR-205 inhibited cell growth in-vitro, we performed an additional experiment to check the antitumor effect of miR-205 after local administration in established A498 tumors. Indeed, the tumor volume regressed from 81mm3 to 5mm3 with miR-205 compared to cont-miR in which tumor volume increased from 80mm3 to 306mm3 (Figure 6B). These results show that miR-205 suppressed cancer growth both in-vitro and in-vivo.
Figure 7. miR-205 inhibits tumor growth in vivo. *p<0.05.
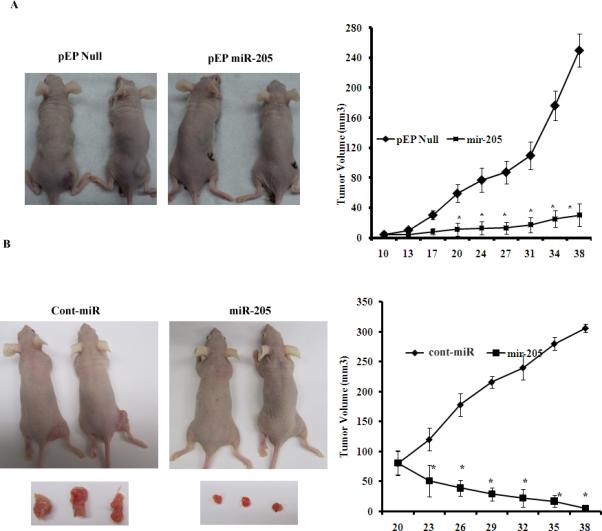
A) Tumor volume following subcutaneous injection of stable A498 cells expressing miR-205 was significantly reduced. B) Tumor volume following intratumoral injection of Cont-miR or miR-205 precursor into established tumors. *p<0.05.
Discussion
In this study we provide evidence that miR-205 interdicts SFK pathways by inhibiting their expression at both the mRNA and protein level. Our results show that Src inhibition by miR-205 leads to growth suppression and cell cycle arrest in renal cancer and are accompanied by inactivation of ERK1/2 and downregulation of c-Myc and cyclin D1. Focal adhesion kinase and STAT3 phophorylation were also decreased by diminished Src activity, leading to significantly reduced cell migration and invasion.
Expression of miR-205 in cancer is controversial because it has been found to be either up-regulated (42) or downregulated (43) in tumors. In the present study, we examined the expression pattern and functional significance of miR-205 in renal cancer. We found miR-205 to be significantly downregulated in tumor samples when compared to adjacent normal samples. The downregulation of miR-205 expression was also observed in RCC cell lines when compared to a non-malignant cell line. This is consistent with a previous microarray analysis of 27 samples of kidney cancer tissues that showed downregulation of miR-205 (42). The significant suppression of miR-205 expression in tumors and cancer cell lines suggests a tumor suppressor role in renal carcinoma. However, neither the functional role nor the targets of miR-205 in renal cancer have been previously defined.
An obstacle to understanding miRNA function has been the relative lack of experimentally validated targets. To determine potential targets of miR-205 action, several in silico algorithms were utilized to identify SFKs as putative targets of miR-205. The SFKs plays an important role in the regulation of cellular proliferation and cell cycle progression (5). Our results indicate an inverse correlation between expression of miR-205 and that of phosphoSrc, lyn and Yes in cell lines and tissues samples. We demonstrated that miR-205 directly targets the 3’UTR of phosphoSrc, lyn and Yes, as its overexpression were associated with suppression of luciferase activity. In addition, a significant downregulation of phosphoSrc, lyn and Yes protein and mRNA levels was observed after miR-205 overexpression indicating that phosphoSrc, lyn and Yes mRNAs are targets of miR-205.
Due to the reported importance of phosphoSrc in renal cancer (11), we further characterized its role in response to miR-205. It has been reported that Src is involved in multiple signaling pathways including Ras/Raf/ERK1/2, PI3K/AKT, β-catenin/c-Myc/cyclin D1 and FAK/p130CAS/MMP-9 that induce growth, survival and migration in various types of cancer cells (35). We observed that inhibition of phosphoSrc by miR-205 overexpression reduced signaling via the ERK1/2 pathway. A previous study by Chang et al. (35) and others (44) have shown that Src inhibition by small molecule inhibitors induced apoptosis and cell cycle arrest at the G0/G1 phase of the cell cycle in prostate cancer cell lines. Our results revealed that inhibition of phosphoSrc by miR-205 overexpression induced apoptosis and G0/G1 arrest in renal cancer cells. This effect on the cell cycle prompted us to study the effect on c-Myc, a Src target gene (39), cyclin D1, the rate limiting factor for cellular proliferation (41) and phosphoSTAT3, a Src target and key transcriptional factor for c-Myc and cyclin D1 (37). We found that all these genes were downregulated at the protein level. Our results indicate that miR-205 inhibited renal cell migration and invasion and also downregulated phosphoFAK, a Src substrate in renal cancer cells. Inhibition of Src has been found to decrease the invasion and migration of prostate cancer cells (9) through selective inhibition of phosphorylation of Src substrates, such as focal adhesion kinase (FAK). To determine whether Src inhibition is responsible for the phenotype observed after miR-205 reconstitution, we performed phenocopy experiments inhibiting Src expression by siRNA. Our results showed that inhibition of Src was responsible for decreased cell viability, migratory and invasive capability of A498 cancer cells. We also observed a G0/G1 cell cycle arrest (14%), whereas there was a decrease of 11% in S-phase cell population. Almost 5% apoptotic cells were observed in Src siRNA transfected cells compared to non-specific control. These results prove that tumor suppressive effect of miR-205 is mediated by Src inhibition in renal caner. We further attenuated miR-205 expression in non-malignant HK-2 cells that expressed higher levels of miR-205 and determined its effect on cell growth, migration and invasion. Our results showed that cells transfected with anit-miR-205 showed more proliferation, migration and invasion compared to control anti-microRNA. These results indicate that miR-205 is important tumor suppressor microRNA in renal cancer and attenuation of this microRNA in over expressing non-malignant renal cells increases their proliferative, migratory and invasive capability.
The anti-proliferative effects of miR-205 observed in this study, mediated by suppression of phosphoSrc and downstream target genes, were confirmed following stable overexpression of miR-205 in A498 cells. In vivo studies demonstrated a striking reduction in subcutaneous tumor cell growth in mice injected with stable A498 cancer cells overexpressing miR-205. Furthermore, results from local administration of miR-205 in established tumors revealed a dramatic regression of tumor growth compared to the cont-miR. In conclusion, our study demonstrates an important tumor suppressor role for miR-205 in renal cancer.
The Src family of kinases (SFKs) is essential for many important tumorigenic phenotypes including proliferation, invasion, migration, epithelial-mesenchymal transition (5, 45), apoptosis, survival, angiogenesis, etc. Thus, the activity of SFKs increase in progressive stages of tumors, with the highest activity observed in metastatic lesions (46). Increasing evidence from molecular and pharmacologic studies suggests that inhibition of Src, the prototype SFK member, inhibits tumor function associated with metastasis, including migration, invasion and expression of the proangiogenic molecules, such as interleukin-8 and vascular endothelial growth factor (47). In addition, recent studies indicate that Src plays critical roles in host cells in the tumor microenvironment, as well as in the tumor cells that contribute to metastasis (4). Several studies have shown that Src-mediated phophorylation of VE-cadherin, a cell adhesion molecule that is essential to vascular cell-to-cell junctional integrity, directly leads to increased vascular permeability, thus facilitating intravasation and extravasation of migratory tumor cells (48). Thus Src plays pleiotropic roles in cancer making it a promising therapeutic target for intervention. Our study is the first report demonstrating that miR-205 inhibits the protooncogenic Src family of kinases indicating the therapeutic potential of miR-205 in the treatment of renal cancer.
Supplementary Material
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by Grants RO1CA138642, RO1CA154374, T32DK007790 (NIH), VA Research Enhancement Award Program (REAP) and Merit Review grants.
References
- 1.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–58. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 2.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 3.Martin GS. The hunting of the Src. Nat Rev Mol Cell Biol. 2001;2:467–75. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 4.Park SI, Shah AN, Zhang J, Gallick GE. Regulation of angiogenesis and vascular permeability by Src family kinases: opportunities for therapeutic treatment of solid tumors. Expert Opin Ther Targets. 2007;11:1207–17. doi: 10.1517/14728222.11.9.1207. [DOI] [PubMed] [Google Scholar]
- 5.Stein PL, Vogel H, Soriano P. Combined deficiencies of Src, Fyn, and Yes tyrosine kinases in mutant mice. Genes Dev. 1994;8:1999–2007. doi: 10.1101/gad.8.17.1999. [DOI] [PubMed] [Google Scholar]
- 6.Ito H, Gardner-Thorpe J, Zinner MJ, Ashley SW, Whang EE. Inhibition of tyrosine kinase Src suppresses pancreatic cancer invasiveness. Surgery. 2003;134:221–6. doi: 10.1067/msy.2003.224. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright CA, Meisler AI, Eckhart W. Activation of the pp60c-src protein kinase is an early event in colonic carcinogenesis. Proc Natl Acad Sci U S A. 1990;87:558–62. doi: 10.1073/pnas.87.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardo LJ, Lee FY, Chen P, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–61. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 9.Nam S, Kim D, Cheng JQ, et al. Action of the Src family kinase inhibitor, dasatinib (BMS-354825), on human prostate cancer cells. Cancer Res. 2005;65:9185–9. doi: 10.1158/0008-5472.CAN-05-1731. [DOI] [PubMed] [Google Scholar]
- 10.Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–16. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 11.Yonezawa Y, Nagashima Y, Sato H, et al. Contribution of the Src family of kinases to the appearance of malignant phenotypes in renal cancer cells. Mol Carcinog. 2005;43:188–97. doi: 10.1002/mc.20109. [DOI] [PubMed] [Google Scholar]
- 12.Furstoss O, Dorey K, Simon V, Barila D, Superti-Furga G, Roche S. c-Abl is an effector of Src for growth factor-induced c-myc expression and DNA synthesis. EMBO J. 2002;21:514–24. doi: 10.1093/emboj/21.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas A, Szatmari E, Orbok A, et al. Hyperosmotic mannitol induces Src kinase-dependent phosphorylation of beta-catenin in cerebral endothelial cells. J Neurosci Res. 2005;80:855–61. doi: 10.1002/jnr.20521. [DOI] [PubMed] [Google Scholar]
- 14.Taj MM, Tawil RJ, Engstrom LD, et al. Mxi1, a Myc antagonist, suppresses proliferation of DU145 human prostate cells. Prostate. 2001;47:194–204. doi: 10.1002/pros.1063. [DOI] [PubMed] [Google Scholar]
- 15.Devi GR, Oldenkamp JR, London CA, Iversen PL. Inhibition of human chorionic gonadotropin beta-subunit modulates the mitogenic effect of c-myc in human prostate cancer cells. Prostate. 2002;53:200–10. doi: 10.1002/pros.10151. [DOI] [PubMed] [Google Scholar]
- 16.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 17.Pantuck AJ, Zisman A, Belldegrun AS. The changing natural history of renal cell carcinoma. J Urol. 2001;166:1611–23. [PubMed] [Google Scholar]
- 18.Hornstein E, Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 19.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 20.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–5. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 22.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 23.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Saunders MA, Lim LP. (micro)Genomic medicine: microRNAs as therapeutics and biomarkers. RNA Biol. 2009;6:324–8. doi: 10.4161/rna.6.3.8871. [DOI] [PubMed] [Google Scholar]
- 25.Bargaje R, Hariharan M, Scaria V, Pillai B. Consensus miRNA expression profiles derived from interplatform normalization of microarray data. RNA. 16:16–25. doi: 10.1261/rna.1688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi R, Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 17:229–35. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–31. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 28.Majid S, Dar AA, Ahmad AE, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–70. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majid S, Dar AA, Saini S, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Trevino JG, Summy JM, Lesslie DP, et al. Inhibition of SRC expression and activity inhibits tumor progression and metastasis of human pancreatic adenocarcinoma cells in an orthotopic nude mouse model. Am J Pathol. 2006;168:962–72. doi: 10.2353/ajpath.2006.050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang YM, Bai L, Liu S, Yang JC, Kung HJ, Evans CP. Src family kinase oncogenic potential and pathways in prostate cancer as revealed by AZD0530. Oncogene. 2008;27:6365–75. doi: 10.1038/onc.2008.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–30. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 37.Prathapam T, Tegen S, Oskarsson T, Trumpp A, Martin GS. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci U S A. 2006;103:2695–700. doi: 10.1073/pnas.0511186103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowman T, Broome MA, Sinibaldi D, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A. 2001;98:7319–24. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barone MV, Courtneidge SA. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature. 1995;378:509–12. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 40.Albanese C, Johnson J, Watanabe G, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–97. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe G, Howe A, Lee RJ, et al. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci U S A. 1996;93:12861–6. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 44.Nam S, Buettner R, Turkson J, et al. Indirubin derivatives inhibit Stat3 signaling and induce apoptosis in human cancer cells. Proc Natl Acad Sci U S A. 2005;102:5998–6003. doi: 10.1073/pnas.0409467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah AN, Gallick GE. Src, chemoresistance and epithelial to mesenchymal transition: are they related? Anticancer Drugs. 2007;18:371–5. doi: 10.1097/CAD.0b013e32801265d7. [DOI] [PubMed] [Google Scholar]
- 46.Heidenreich A, Varga Z, Von Knobloch R. Extended pelvic lymphadenectomy in patients undergoing radical prostatectomy: high incidence of lymph node metastasis. J Urol. 2002;167:1681–6. [PubMed] [Google Scholar]
- 47.Gray MJ, Zhang J, Ellis LM, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–20. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 48.Criscuoli ML, Nguyen M, Eliceiri BP. Tumor metastasis but not tumor growth is dependent on Src-mediated vascular permeability. Blood. 2005;105:1508–14. doi: 10.1182/blood-2004-06-2246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.