Abstract
Background
MicroRNAs (miRNAs) are small noncoding RNAs that regulate the expression of approximately 30% of all human genes. They play important roles in numerous cellular processes including development, proliferation, and apoptosis. It is currently believed that miRNAs elicit their effect by silencing the expression of target genes. Here we show that microRNA-205 (miR-205) induces the expression of IL24 and IL32 tumor suppressor genes by targeting specific sites in their promoters.
Methods
Methods used in this study include transfection of small RNAs, quantitative-real-time-PCR, in-situ hybridization, fluorescence labeled in-situ hybridization, cell cycle, apoptosis, cell viability, migratory, clonability and invasion assays, immunoblotting, luciferase reporter, nuclear run-on and chromatin immunoprecipitation assays.
Results
Our results revealed that miR-205 is silenced in prostate cancer. Its re-expression induced apoptosis and cell cycle arrest. It also impaired cell growth, migration, clonability and invasiveness of prostate cancer cells. MicroRNA-205 induced tumor suppressor genes IL24 and IL32 at both mRNA and protein levels. Induction of in-vitro transcription and enrichment of markers for transcriptionally active promoters in IL24 and IL32 genes was observed in response to miR-205.
Conclusion
In this study we identify a new function for miR-205 to specifically activate tumor suppressor genes by targeting specific sites in their promoters. These results corroborate a new function that miRNAs have in regulating gene expression at the transcriptional level. The specific activation of tumor suppressor genes (e.g., IL24, IL32) or other dysregulated genes by microRNAs may contribute to the novel therapeutic approach in the treatment of prostate cancer.
Keywords: MicroRNA-205, IL24, IL32, Transcriptional activation, Prostate cancer
Introduction
MicroRNAs (miRNAs) are non-protein-coding sequences thought to regulate the expression of up to 60% of human genes, either by inhibiting mRNA translation or inducing its degradation 1. These small RNAs play important roles in numerous cellular processes including development, proliferation, and apoptosis 2. In addition to their crucial role in cellular differentiation and organism development 3, miRNAs are frequently misregulated in human cancers 4, 5, and can act as either potent oncogenes or tumor suppressor genes 6.
Prostate cancer (PCa) is the most frequently diagnosed malignant tumor and the second-leading cause of cancer deaths in American men. It is estimated that in 2009 there will have been more than 192, 280 newly diagnosed prostate cancer cases and more than 27, 360 attributed deaths 7. Although surgery and radiotherapy are generally effective against clinically localized PCa, androgen ablation, the treatment of choice for advanced disease, only leads to temporary tumor regression 8. The lack of available treatment options for effectively eradicating advanced PCa makes the development of alternative approaches urgent. Understanding the molecular alterations that distinguish progressive from non-progressive disease will help identify novel prognostic markers or therapeutic targets. Some aberrantly produced miRNAs have been discovered in PCa cell lines, xenografts, and clinical samples 9. These miRNAs may play critical roles in the pathogenesis of prostate cancer.
MicroRNA-205 (miR-205) expression in cancer is controversial because it has been found to be either up-regulated 10, 11 or down-regulated 12, 13 in tumor compared with normal tissues. It is currently believed that miRNAs elicit their effect by silencing the expression of target genes 14. Given the functional complexity of miRNA mediated gene regulation, it is unlikely that the effects of these molecules are limited to gene silencing. This study was undertaken to investigate the potential involvement of miR-205 in positively regulating the expression of tumor suppressor genes IL24 and IL32.
IL24 has been reported to be a novel tumor-suppressor gene whose expression is lost during tumorigenesis 15. Gene transfer of an adenovirus-expressed mda-7/IL24 into numerous histologic types of human tumor cell lines, including melanoma, gliobastoma, breast, lung, pancreatic, and others, resulted in tumor-specific growth arrest 16–19. In addition to its direct apoptosis-inducing properties, IL24 shows antiangiogenic, radiosensitizing, immunostimulatory, and potent “bystander” antitumor activity 20–22. These divergent anticancer properties of mda-7/IL24 make it an ideal candidate for anticancer therapy. A recent Phase 1 clinical trial using a replication-incompetent adenovirus expressing IL24, showed evidence of clinical activity with limited toxicity. Thus IL24 is being hailed as a ‘magic bullet’ for cancer gene therapy. IL32 is a novel cytokine implicated in inflammation 23, and cell death 24. IL32 is known to associate specifically with apoptotic T cells and ectopic expression of IL32 in HeLa cells causes apoptosis 24. Our data showed that there was strong induction of IL24 and IL32 tumor suppressor genes in response to miR-205. This study identifies a new function for miR-205 in regulating tumor suppressor genes.
Experimental Procedures
Cell culture and micro-RNA transfection
Human prostate carcinoma cell lines (LNCaP, PC3 and Du145) and a non-malignant epithelial prostate cell line (RWPE-1) were obtained from the American Type Culture Collection (Manassas, VA). All cell lines were cultured as described earlier 25. Transfection of miRNA was carried out by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufactures’s protocol. All miR-RNA treatments proceeded for 72 h.
Quantitative real-time PCR
Mature miRNAs and other mRNAs were assayed using the TaqMan MicroRNA Assays and the Gene Expression Assays, respectively, in accordance with manufacturer's instructions (Applied Biosystems, Foster City, CA). Samples were normalized to RNU48 or GAPDH as indicated. Comparative real-time PCR was performed in triplicate, including no-template controls.
In Situ Hybridization
For cell lines, cells were grown on sterile slides in 100-mm Petri dishes for 48 h. These cells were then stained by using DIG-labeled locked nucleid acid (LNA)-based probes specific for mir-205 following the manufacturer's protocol (www.exiqon.com) and finally detected by using anti-DIG-Fluorescein, Fab Fragments (Roche Applied Science, Indianapolis, IN). Nuclei were routinely stained by using DAPI. Tissue array slides contained human normal adjacent prostate tissue samples (n =15), second stage (n =14), third stage (n=52), fourth stage (n=6) and metastatic (n =8) carcinomas. Hybridizations were performed overnight at 52°C after the addition of 50 nM DIG-labeled locked nucleid acid (LNA)-based probes specific for miR-205 and U6 (Exiqon,Inc Woburn, MA). Alkaline phosphatase activity was detected using BM Purple AP Substrate (Roche Applied Science, Indianapolis, IN). ISH results were semiquantitatively graded according to the intensity of staining and scored from 1 to 4, and then normalized to U6 levels.
Immunoblotting
Immunoblotting was performed as described earlier 26. Antibodies specific for p21WAF1 (05-345; Upstate), p27 (2552; Cell Signaling), IL24 (ab56811; abcam), IL32 (ab62580), p38 (9212; Cell Signaling), BAK (53251; AnaSpec), FLIP (3210; Cell Signalling), BAX (2774; Cell Signalling), BID (sc-11423), SAP-JNK (9252; Cell Signalling) and GAPDH (2118; Cell Signalling) were used.
Flow cytometry, cell viability, migratory, clonability and invasion assays
FACS analysis was done 72 hours post-transfection using nuclear stain DAPI for cell cycle analysis or ANNEXIN V-FITC /7-AAD KIT (Beckman Coulter, Inc. Fullerton, CA) for apoptosis analysis according to the manufacturer’s protocol. Cell viability was determined at 24, 48 and 72 h by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. For colony formation assay, cells were seeded at low density (1000 cells/plate or 200 cells/plate) and allowed to grow for three weeks. Then, cells were stained with crystal violet and colonies were counted. An artificial “wound” was created on a confluent cell monolayer and photographs were taken after 24 hrs for migration assay. The invasiveness of PC3 cells was assessed by an invasion assay that was performed in modified Boyden chambers as described elsewhere 27.
Luciferase Assays
A 2.2kb promoter reporter vector of IL24 (ID: 114318_CHR1_P1626_R1(IL24)) was obtained from SwitchGear Genomics, along with the positive (ID: RPL10_PROM) and negative (ID: EMPTY_PROM) promoter control vectors. The assay was performed according to the manufacturer’s protocol (www.switchgeargenomics.com).
Nuclear Run-On Assays
Nuclear run-on assays were performed as described by Kim et al. 28. Metastatic prostate cancer PC3 cells were transfected with control or miR-205 duplexes (Ambion) at 50 nM final concentration, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Seventy two hour after transfection, 2 × 107 cells were washed with cold PBS, harvested, and lysed on ice in 0.5% Nonidet P-40 lysis buffer [10 mM Tris·HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2], and centrifuged at 500 × g for 10 min. Supernatants were removed and nuclei were incubated in reaction buffer [10 mM Tris·HCl (pH 8.0), 5 mM MgCl2, 0.3 mM KCl] and 2.5 mM NTP plus Biotin-16-UTP mix (Roche Applied Science, Indianapolis, IN) for 45 min at 30 °C. The transcription reaction was stopped by adding RNA STAT-60 Reagent (Tel-Test) to isolate total nuclear RNA, according to manufacturer recommended protocols. Biotinylated nascent RNA transcripts were isolated by incubation with streptavidin beads (Active Motif) for 2 h at room temperature on a rocking platform. Beads were collected by centrifugation and washed once with 2× SSC-15% formamide for 10 min and twice with 2× SSC for 5 min on a rocking platform. Biotinylated RNA was eluted from streptavidin beads in H2O or 10 mM EDTA pH 8.2 by incubation at 90 °C for 10 min and analyzed by qRT-PCR.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using EZ-ChIP Kit (Upstate Biotechnology, Billerica, MA) as described previously 26. Immunoprecipitations were performed using antibodies purchased from Upstate Biotechnology and recognized acetyl histone H3 (06-599), acetyl histone H4 (06-866), dimethyl-histone H3 lysine 4 (07-030). Power SYBR Green PCR Mastermix (Applied Biosystems, Foster city, CA) was used to perform real-time PCR on 7500 Fast Real Time PCR System (Applied Biosystems, Foster city, CA). Signals were also confirmed by conventional PCR and gel analyses.
Statistical analysis
Statistical analysis was performed using StatView version 5.0 for Windows as needed. Student’s t-test was used to compare the different groups. p values of less than 0.05 were regarded as statistically significant and are represented by * in the figures.
Results
MicroRNA-205 is down-regulated in prostate cancer cell lines and tissue specimens
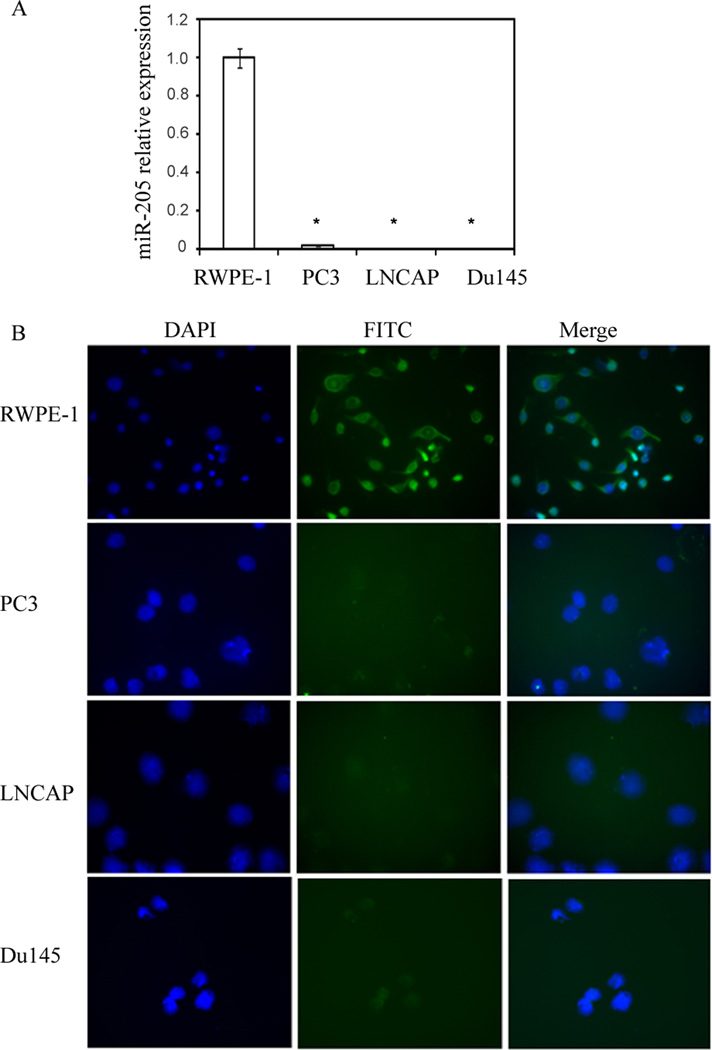
Quantitative real time PCR showed that miR-205 was either undetectable or highly repressed in both androgen-dependent (LNCaP) and androgen-independent (PC3, Du145) prostate cancer cells compared to non-malignant RWPE-1 cells (Figure 1A).
Figure 1.
miR-205 is silenced in human prostate cancer cell lines. (A) Relative miR-205 expression levels in cell lines, assessed by qRT-PCR. (B) In-situ hybridization (ISH) analysis of miR-205 expression in prostatic cell lines. The fluorescent signals are mostly cytoplasmic in location (green) and are present only in non-malignant RWPE-1 cells indicating the expression of miR-205 compared to cancer cell lines. The nuclei were stained with DAPI (blue).
Since, locked nucleic acid (LNA)-modified miRNA probes have been reported to markedly increase hybridization affinity to miRNAs compared with traditional RNA- or DNA-based probes, a digoxigenin (DIG)-labeled LNA-miR-205 probe was used to detect miR-205 abundance in human prostate cancer cell lines using in situ hybridization (ISH) and detected with a FITC-labelled anti-DIG antibody. Consistent with the above results, fluorescent signal was detected in RWPE-1 cells representing the abundance of miR-205 in this cell line where as it was absent in cancer cell lines (LNCaP, PC3 and Du145) (Figure 1B).
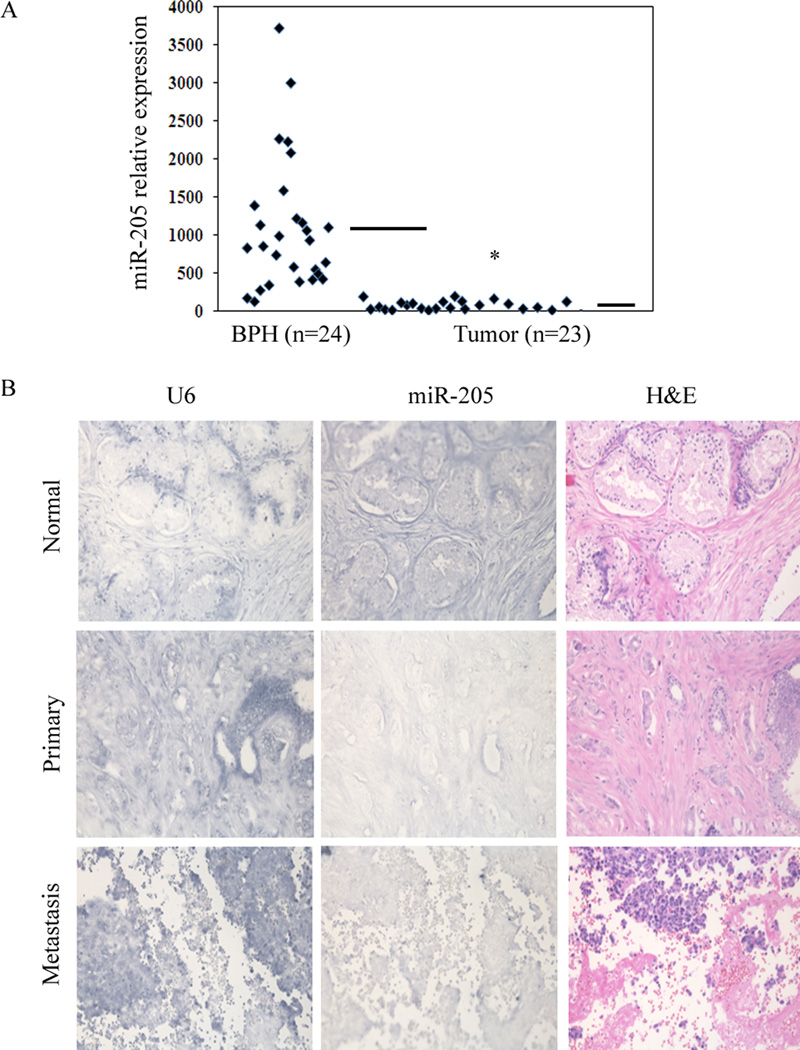
To examine the clinical relevance of miR-205, its expression was analyzed in carcinoma and benign prostate hyperplasia (BPH) tissue samples. Almost all the carcinoma samples showed significantly down regualted miR-205 expression with respect to the BPH samples and an overall lower relative average expression was observed in carcinoma compared to BPH samples (Figure 2A). ISH analysis of miR-205 expression was also performed on tissue arrays containing human normal adjacent prostate tissue samples (n =15), second stage (n =14), third stage (n=52), fourth stage (n=6) and metastatic (n =8) carcinomas. Normal adjacent prostate tissue samples highly expressed miR-205 indicated by the BM-Purple stain. Figure 2B depicts representative ISH results showing that normal adjacent sample expressed markedly increased miR-205 expression levels where as it is almost absent in primary and metastatic prostate cancer samples. These ISH results suggest that expression of miR-205 is significantly high in normal tissue samples compared to prostate cancer.
Figure 2.
miR-205 is silenced in human prostate tumors. (A) Relative miR-205 RNA expression levels in BPH (n=24) and tumor (n=23) tissues, assessed by qRT-PCR indicating significant expression of miR-205 in BPH samples as compared to tumor samples. (—) represents average value. (B) miR-205 in-situ hybridiztion (ISH) on prostate tissue microarrays. Representative pictures of miR-205 expression in normal, primary and metastatic prostate tissues (Middle), U6 staining confirming the preservation of intact small RNAs in the same cases (Left); H & E stained sections allowed the identification of tumors (Right).
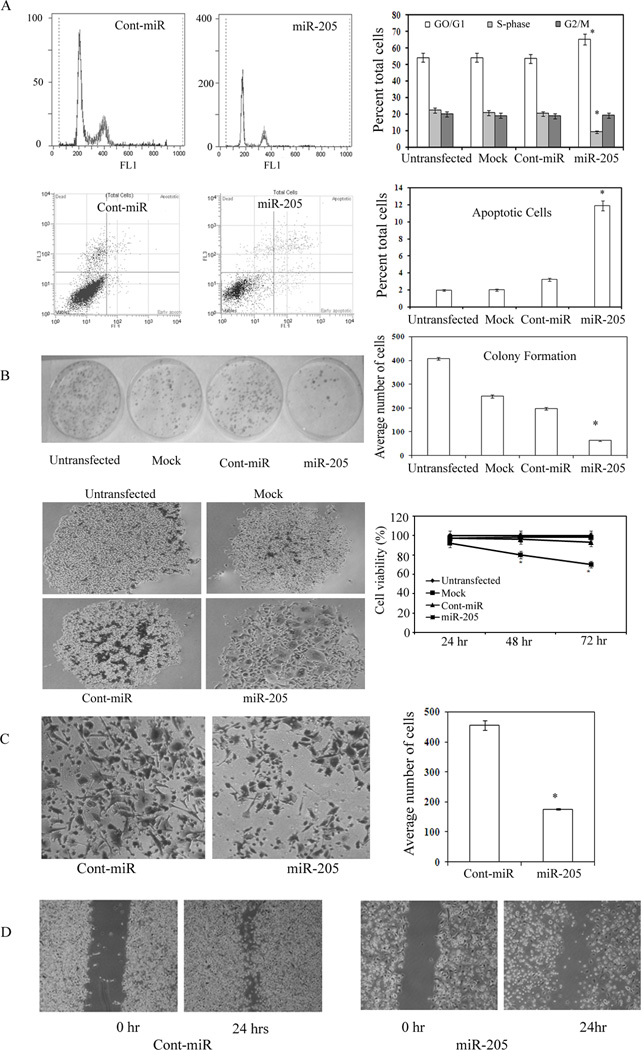
MicroRNA-205 induces apoptosis, cell cycle arrest, impairs cell viability, migratory, clonability and invasive properties of prostate cancer cells
FACS (fluorescence activated cell sorting) analysis revealed that re-expression of miR-205 leads to a significant increase (10%±3%) in the number of cells in the G0-G1 phase of the cell cycle while the S-phase population decreased from 21%±3% to 10%±2% suggesting that miR-205 causes a G0-G1 arrest in miR-205 transfected PC3 cells compared to a nonspecific microRNA control (Cont-miR) (Figure 3A). FACS analysis for apoptosis was performed using Annexin-V-FITC-7-AAD dye. The percentage of total apoptotic cells (early apoptotic + apoptotic) were significantly increased (11%±3%) in response to miR-205 transfection compared to cont-miR (3%±2%) with a corresponding 8%±2 decrease in the viable cell population (Figure 3A). Colony formation assay showed that over-expression of miR-205 significantly decreased the ability of metastatic PC3 cells to form colonies compared to the cont-miR expressing cells (Figure 3B). MicroRNA-205 caused drastic changes in cell morphology associated with growth arrest including increased size and a broad, flattened shape three days (72hrs) following transfection (Figure 3B). Cell viability after miR-205 over-expression was significantly decreased (8% to 30%) in a time-dependent manner (0 to 72hrs) compared to the cont-miR (Figure 3B) suggesting that miR-205 has an anti-proliferative effect in prostate cancer. Wound and transwell invasion assays were carried out to evaluate whether miR-205 affects the migratory and invasive capabilities of prostate cancer cells. The average number of invading cells transfected with miR-205 were significantly (190±11) lower than that of cont-miR (500±16) transfected cells (Figure 3C). We also found that miR-205 over-expressing cells were less proficient than equivalent cont-miR transfected cells at closing an artificial wound created over a confluent monolayer (Figure 3D) suggesting that miR-205 impairs the migratory capability of the metastatic PC3 prostate cancer cells.
Figure 3.
Re-expression of miR-205 induces apoptosis, cell cycle arrest, impairs cell viability, migration, clonability and invasive properties in PC3 cells. PC3 cells were transfected with a control miRNA or miR-205. Untransfected and mock transfected controls were included. (A) Representative histograms and graphical summary of cell-cycle profile changes in response to miR-205. Representative quadrants and graphical summary of apoptosis induced by miR-205. (B) Colony formation assay of PC3 cells transfected with miR-205 or control molecules. Graph is representative of 3 independent experiments. Over expression of miR-205 promotes an arrested phenotype in PC3 cells. The percent viability of metastatic PC3 cell lines decreased in response to miR-205 in a time dependent manner. (C) miR-205 significantly decreased invasive behavior of PC3 cells as compared to cont-miR transfected PC3 cells. Graph represents summary of three independent experiments. (D) PC3 cells transfected with miR-205 exhibited less migratory behavior than cont-miR transfected cells.
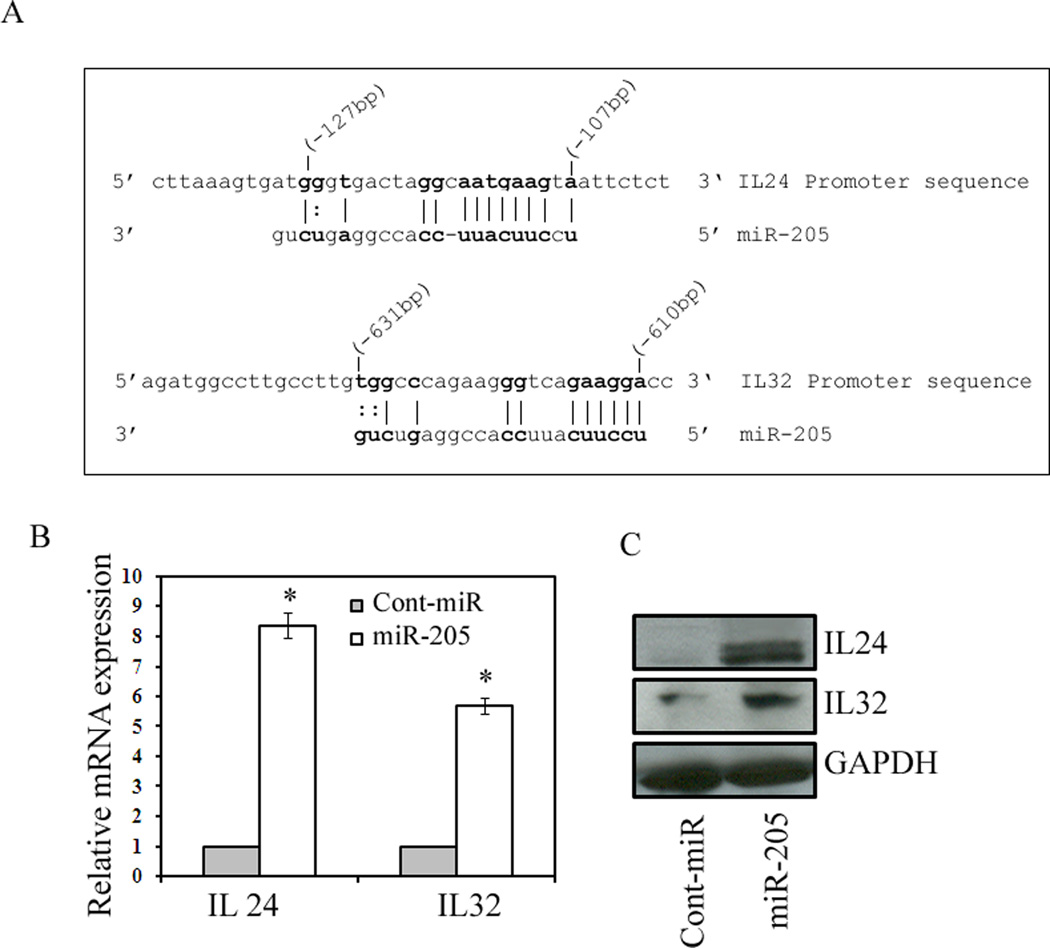
MicroRNA-205 targets the IL24 and IL32 promoters to induce gene expression
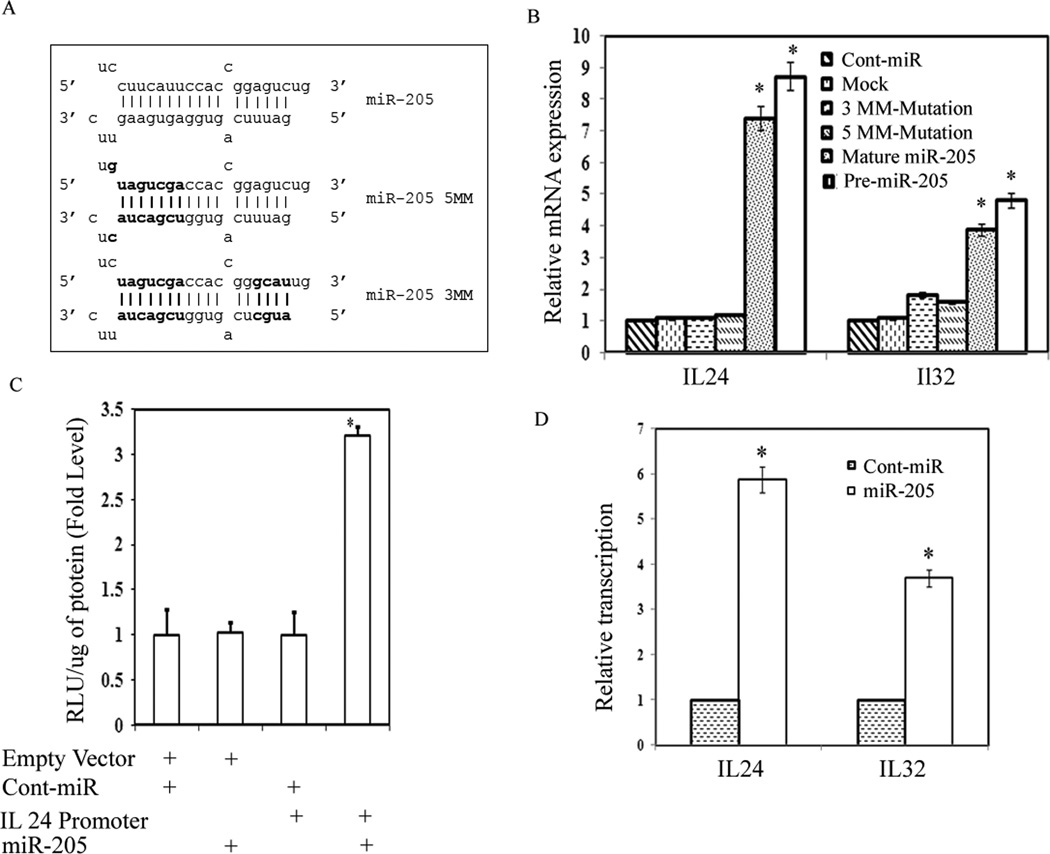
Our preliminary microarrayunpublished data showed that there was a strong induction of IL24 and IL32 genes in response to miR-205 transfection. To determine whether these genes have target sites for miR-205, we scanned 1kb promoters of both genes for sequences complementary to miR-205 using the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/). Scanning analysis revealed a potential binding sequence located at position -107 (IL24) and -610 (IL32) relative to the transcription start site in the promoters of both genes (Figure 4A). We transfected pre-miR-205 precursor (Ambion) and a control miR precursor (Ambion) into metastatic PC3 prostate cancer cells and analyzed IL24 and IL32 expression 72hrs post transfection. Analysis of mRNA expression revealed a profound induction in IL24 (~8 fold) and IL32 (~5 fold) levels after miR-205 transfection compared to cont-miR (Figure 4B). Induction of these genes was further confirmed by immunoblot analysis. As shown in Figure 4C, elevated levels of IL24 and IL32 protein levels strongly correlate to the increase in mRNA expression levels. These results suggest that miR-205 induces gene expression by targeting the promoters of these genes.
Figure 4.
miR-205 induces expression of tumor suppressor genes IL24 and IL32. (A) Sequence of the miR-205 target sites located at -107 (IL24) and -610 (IL32) relative to the transcription start sites. Bases in bold face correspond to those bases in miR-205 that are complementary to the target sites, including G:U/T wobble base-pairing. (B) Relative IL24 and IL32 mRNA expression levels in PC3 cells transfected with 50nM concentrations of the miR-205 or cont-miR for 72 h, assessed by qRT-PCR. (C) Induction of IL24 and IL32 proteins in response to miR-205 assessed by Western blot.
Sequence specificity for gene induction by miR-205 and direct interaction between miR-205 and promoter target sequence
To determine whether induction of IL24 and IL32 was specific to the sequence of miR-205, we synthesized two miR-205 mutants to create mismatches with the target sites. Mutation to 8 or 11 bases in the miR-205 sequence resulted in mutant derivatives miR-205-5MM and miR-205-3MM, respectively (Figure 5A). As shown in Figure 5B neither miR-205-5MM nor miR-205-3MM were capable of inducing IL24 or IL32 expression suggesting that the induction of these genes was specific to the sequence of miR-205.
Figure 5.
Sequence specificity for miR-205. (A) Mutations to 8 or 11 base pairs in miR-205 resulted in miR-205-5MM and miR-205-3MM, respectively. The mutated bases are shown in bold letters. (B) Relative IL24 and IL32 mRNA expression levels in PC3 cells transfected with 50nM concentrations of each indicated miRNA duplex for 72 h, assessed by qRT-PCR. (C) IL24 is a direct target of miR-205. IL24 promoter vector containing the binding site for miR-205 was purchased from SwitchGear Genomics and cotransfected with miR-205 or cont-miR and luciferase activity was measured after 48 hours. Luciferase activity was normalized to total protein and relative expression was determined for cont-miR and miR-205. RLU (Relative Luciferase units). (D) Activation of in-vitro transcription by miR-205. Nuclear run-on experiments for nascent IL24 and IL32 mRNA transcription in PC3 cells transfected with miR- 205 or cont-miR duplexes, as measured by qRT-PCR and normalized to nascent GAPDH mRNA transcription levels.
We also performed a luciferase reporter assay to verify that a direct interaction between miR-205 and IL24 was responsible for the increased expression of IL24. IL24 vector contained a 2.2 kb promoter sequence including the binding site for miR-205 and an empty vector that served as a negative control. These reporter vectors were co-transfected in PC3 cells with miR-205 precursor and cont-miR molecules that served as a negative control. The luciferase activity was markedly increased after miR-205 overexpression compared to negative control (Figure 5C) supporting the evidence that miR-205 induces IL24 directly by targeting promoter sequence.
MicroRNA-205 induces the in-vitro transcription in IL24 and IL32 genes
We performed nuclear run-on assay that measures the relative in situ transcription rate of specific genes in intact nuclei. Within a given experiment, the nuclear run-on assay can be used to determine the level of transcription for several different genes. The isolated nuclei contain the full transcription machinery for synthesis of mRNA. Therefore, it is regarded as the gold-standard measurement of overall transcriptional activity of a specific promoter. After the transcriptional reaction, biotinylated RNA was isolated and quantitative real-time PCR was conducted. The relative IL24 and IL32 mRNA levels reflect the transcription efficiency of both genes. As shown in Figure 5D, miR-205 transfected cells showed significantly higher mRNA expression levels of IL24 (7 fold) and IL32 (5 fold) compared to the cont-miR transfected cells suggesting that miR-205 induces the transcriptional activity of IL24 and IL32 promoters.
MicroRNA-205 causes enrichment of markers for transcriptionally active promoters
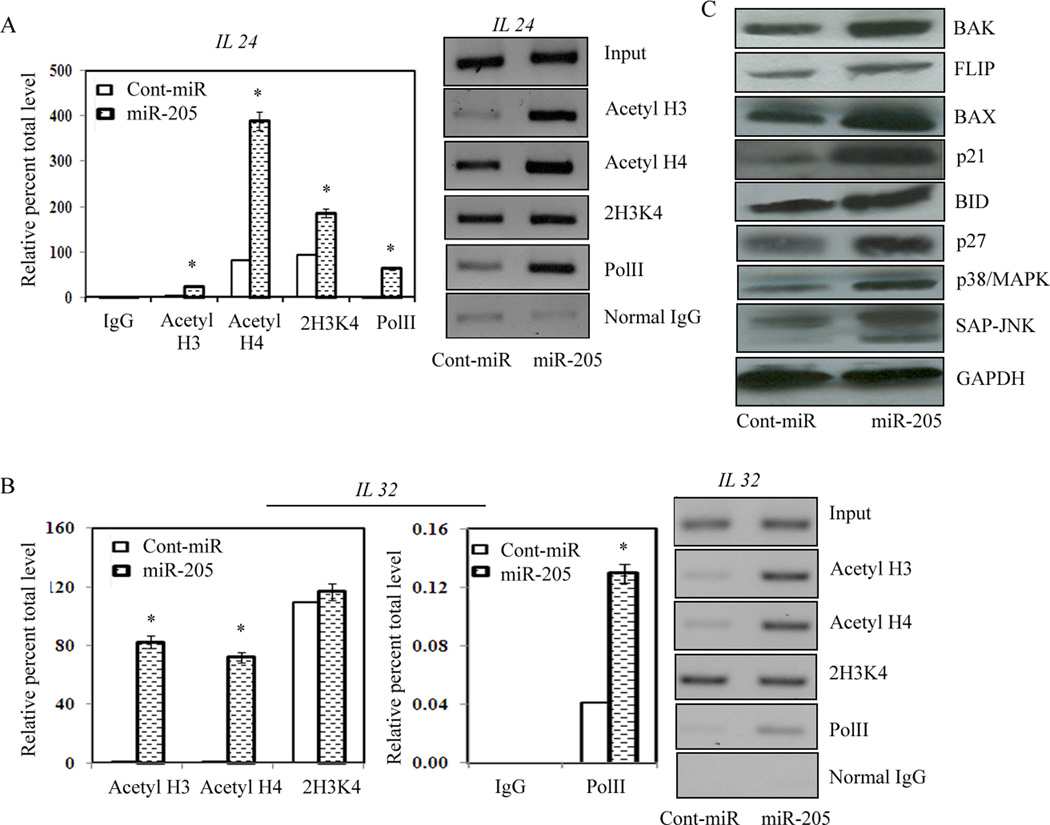
Several histone modifications, mainly acetylation of histone 3 and 4 and Di- and tri-methylation of Lys 4 of histone 3, have been established as markers for transcriptionally active promoters. To determine whether over-expression of miR-205 results in enrichment of active chromatin modifications in IL24 and IL32 promoters, we performed chromatin immunoprecipitation (ChIP) assays on miR-205 and cont-miR transfected PC3 cells. As shown in Figure 6A and B, we observed enrichment of almost all the active chromatin modifications in response to miR-205 compared to cont-miR, indicative of gene activation by miR-205. Therefore, the increased levels of mRNA and protein expression of IL24 and IL32 (Fig., 4B, C) in response to miR-205 over-expression correlated with the enrichment of markers for transcriptionally active promoters in both genes.
Figure 6.
Enrichment of markers for transcriptionally active promoters by miR-205. (A–B) ChIP assay in PC3 cells transfected with miR-205 or cont-miR. Quantification of immunoprecipitated IL24 and IL32 promoter regions were determined by quantitiative real-time PCR and normalized to input DNA. Signals were also confirmed by conventional PCR and PCR product for each sample was resolved on 2% agarose gel and visualized by staining with ethidium bromide. (C) Induction of downstream target genes by miR-205. Since miR-205 induced expression of IL24 and IL32 expression and caused cell cycle arrest and apoptosis in PC3 cells, we examined the expression of several downstream proteins involved in these pathways. Pro-apoptotic and cell cycle checkpoint proteins were induced in response to miR-205 compared to cont-miR.
Effect of miR-205 on downstream targets
MicroRNA-205 over-expression induced apoptosis, impaired cell migration and invasion and caused cell cycle arrest in metastatic PC3 cells. MicroRNA-205 also induced the expression of IL24 that is known to control cell survival and proliferation and induces apoptosis selectively in cancer cells without harming normal cells. We examined the protein expression of various downstream genes involved in these pathways. As shown in Figure 6C the protein expression of pro-apoptotic genes, cell cycle regulators and downstream targets of IL24 which are involved in inhibition of migration, were increased in PC3 cells transfected with miR-205 compared to the cont-miR (Figure 6C). Taken together these results suggest that miR-205 acts as tumor suppressor microRNA by directly targeting tumor suppressor genes IL24 and IL32 through apoptotic and cell survival pathways in prostate cancer.
Discussion
MicroRNAs play important roles in numerous cellular processes including development, proliferation, and apoptosis 2. It is currently believed that miRNAs elicit their effect by silencing the expression of target genes 14. However, in the present study we identified two genes (IL24 and IL32) that possess target sites complementary to miR-205 within their promoters and over-expression of miR-205 readily induced the expression of both the genes. Several lines of evidence indicate that gene induction was specific for miR-205 and the targeted promoter sites. (i) No modifications to miR-205 or pre-miR-205 sequences were required for gene induction. We synthesized native miR-205 molecules to ensure we were analyzing natural miR-205 sequence function. (ii) Nonspecific control molecules (cont-miR) did not impact target gene expression. (iii) Activity required complementarity with target sequences. Mutation to nucleotides involved in target recognition completely prevented gene induction by miR-205. (iv) Selective activation of IL24 was achieved by targeting binding sites in its promoter as confirmed by luciferase reporter assay and suggests that activation of this gene by miR-205 was specific for the miRNA target sites in the gene promoter. Previous reports from our lab and others 29, 30 have revealed that small dsRNAs targeting the promoters of E-cadherin, p21WAF1/CIP1, vascular endothelial growth factor (VEGF), progesterone receptor (PR) and major vault protein (MVP) readily activate gene expression. These reports indicated an unexpected ability of small RNAs to induce gene expression.
Expression of miR-205 in cancer is controversial because it has been found to be either up-regulated 10, 11 or down-regulated 12, 13 in tumor compared with normal tissues. We found that miR-205 was markedly down-regulated in PCa cell lines, irrespective of their androgen responsiveness, compared with normal cells. Reduction of miR-205 expression was also observed in carcinoma compared with BPH tissues. These findings show that reduced miR-205 expression in prostate cancer may be important for prostate cancer progression. To examine this possibility, we re-expressed miR-205 in metastatic PC-3 cells. Its re-expression induced changes in cell proliferation, cell cycle progression and cell morphology. MicroRNA-205 transfected cells showed impaired clonability, migratory and invasive capabilities. Altogether, our findings are consistent with other reports 31, 32 suggesting that miR-205 acts as a tumor suppressor microRNA. A previous report 32 describes the tumor-suppressive functions of miR-205 involving reversal of EMT and down-regulation of protein kinase-C epsilon whereas our study is distinct and identifies a new function for miR-205 to induce the tumor suppressor genes by targeting their promoters.
We also found the IL24 and IL32 promoters to be direct targets of miR-205. Luciferase activity was significantly increased after miR-205 over-expression compared to negative control indicating that miR-205 induces IL24 by directly targeting its promoter sequence. Furthermore, none of the mutated sequences of miR-205 was capable of inducing IL24 or IL32 expression suggesting that the induction of these genes was specific to the sequence of miR-205. Our results show that miR-205 induced the expression of these tumor suppressor genes by targeting their promoters.
Recent studies have identified chromatin signatures that can be used to identify active promoters in human cells 33–35. Several histone modifications, mainly di- and tri-methylation of Lys 4 of histone 3 (2H3K4, 3H3K4) and acetylation of Lys 9/14 of histone 3, have been established as markers for transcriptionally active promoters 34, 36, 37. Since IL24 and IL32 were transcriptionally activated by miR-205, we analyzed the status of local active histone modifications such as acetylation of histone H3 and H4, di-methylation of Lys 4 of histone H3 and also Pol II. We found that there was enrichment of these active histone modifications in response to miR-205 indicative of transcriptionally active promoters in both the genes. These results are consistent with the mRNA and protein expression of both IL24 and IL32 genes.
We also performed nuclear run-on assay that measures the relative in vitro transcription rate of specific genes in intact nuclei. The isolated nuclei contain the full transcription machinery for synthesis of mRNA. The IL24 and IL32 gene transcription activity, revealed by the nuclear run-on assay, was increased in the nuclear extracts from cells transfected with miR-205 compared to cont-miR cells suggesting that miR-205 induces the transcriptional activity of IL24 and IL32 promoters.
Given the functional complexity of miRNA mediated gene regulation, it is unlikely that the effects of these molecules are limited to gene silencing. A previous report from our lab 29 has shown that exogenously introduced dsRNAs activated the expression of E-cadherin, p21, or VEGF by targeting noncoding regulatory regions in gene promoters. Gene activation by dsRNA required the Argonaute 2 (Ago2) protein and was associated with a loss of lysine-9 methylation on histone 3 at dsRNA-target sites 29. Like these exogenously introduced dsRNAs, it is possible that miRNAs may also positively regulate gene expression. However, to understand the mechanism, further research is required to identify the molecular components involved in miRNA activated gene expression.
In summary, we have provided a proof of principle that miRNA-205 function as a tumor suppressor by up-regulating tumor suppressor genes IL24 and IL32 by targeting specific sites in their promoters. Enrichment of the active histone modifications in response to miR-205, indicative of transcriptionally active promoters in both genes was also observed. The identification of such a phenomenon where cancer cells can be targeted with microRNA to turn-on silenced tumor suppressor genes may have significant therapeutic potential. The use of RNAi is currently being implemented as a gene-specific approach for molecular medicine. By the same principle, the specific activation of tumor suppressor genes (e.g., IL24, IL32) or other dysregulated genes by micro-RNA may contribute to the novel therapeutic approach in the treatment of prostate cancer.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by Grants RO1CA 111470, T32DK007790 (NIH), VA Research Enhancement Award Program (REAP) and Merit Review grants.
References
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18955434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12869753. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17011485. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15944708. [DOI] [PubMed] [Google Scholar]
- 5.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16461460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16557279. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16557279. [DOI] [PubMed] [Google Scholar]
- 8.Mimeault M, Batra SK. Recent advances on multiple tumorigenic cascades involved in prostatic cancer progression and targeting therapies. Carcinogenesis. 2006;27(1):1–22. doi: 10.1093/carcin/bgi229. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16195239. [DOI] [PubMed] [Google Scholar]
- 9.Shi XB, Tepper CG, White RW. MicroRNAs and prostate cancer. J Cell Mol Med. 2008;12(5A):1456–1465. doi: 10.1111/j.1582-4934.2008.00420.x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18624768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25(5):387–392. doi: 10.1016/j.urolonc.2007.01.019. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17826655. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17875710. [DOI] [PubMed] [Google Scholar]
- 12.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–260. doi: 10.1016/j.jtcvs.2007.08.055. discussion 60. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18242245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67(24):11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18089790. [DOI] [PubMed] [Google Scholar]
- 14.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15211354. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB. Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995;11(12):2477–2486. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8545104. [PubMed] [Google Scholar]
- 16.Saeki T, Mhashilkar A, Chada S, Branch C, Roth JA, Ramesh R. Tumor-suppressive effects by adenovirus-mediated mda-7 gene transfer in non-small cell lung cancer cell in vitro. Gene Ther. 2000;7(23):2051–2057. doi: 10.1038/sj.gt.3301330. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11175318. [DOI] [PubMed] [Google Scholar]
- 17.Su ZZ, Madireddi MT, Lin JJ, Young CS, Kitada S, Reed JC, et al. The cancer growth suppressor gene mda-7 selectively induces apoptosis in human breast cancer cells and inhibits tumor growth in nude mice. Proc Natl Acad Sci U S A. 1998;95(24):14400–14405. doi: 10.1073/pnas.95.24.14400. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9826712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mhashilkar AM, Schrock RD, Hindi M, Liao J, Sieger K, Kourouma F, et al. Melanoma differentiation associated gene-7 (mda-7): a novel anti-tumor gene for cancer gene therapy. Mol Med. 2001;7(4):271–282. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9826712. [PMC free article] [PubMed] [Google Scholar]
- 19.Chada S, Sutton RB, Ekmekcioglu S, Ellerhorst J, Mumm JB, Leitner WW, et al. MDA-7/IL-24 is a unique cytokine--tumor suppressor in the IL-10 family. Int Immunopharmacol. 2004;4(5):649–667. doi: 10.1016/j.intimp.2004.01.017. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15120650. [DOI] [PubMed] [Google Scholar]
- 20.Su Z, Emdad L, Sauane M, Lebedeva IV, Sarkar D, Gupta P, et al. Unique aspects of mda-7/IL-24 antitumor bystander activity: establishing a role for secretion of MDA-7/IL-24 protein by normal cells. Oncogene. 2005;24(51):7552–7566. doi: 10.1038/sj.onc.1208911. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16044151. [DOI] [PubMed] [Google Scholar]
- 21.Su ZZ, Lebedeva IV, Sarkar D, Gopalkrishnan RV, Sauane M, Sigmon C, et al. Melanoma differentiation associated gene-7, mda-7/IL-24, selectively induces growth suppression, apoptosis and radiosensitization in malignant gliomas in a p53-independent manner. Oncogene. 2003;22(8):1164–1180. doi: 10.1038/sj.onc.1206062. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12606943. [DOI] [PubMed] [Google Scholar]
- 22.Ramesh R, Mhashilkar AM, Tanaka F, Saito Y, Branch CD, Sieger K, et al. Melanoma differentiation-associated gene 7/interleukin (IL)-24 is a novel ligand that regulates angiogenesis via the IL-22 receptor. Cancer Res. 2003;63(16):5105–5113. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12941841. [PubMed] [Google Scholar]
- 23.Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity. 2005;22(1):131–142. doi: 10.1016/j.immuni.2004.12.003. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15664165. [DOI] [PubMed] [Google Scholar]
- 24.Goda C, Kanaji T, Kanaji S, Tanaka G, Arima K, Ohno S, et al. Involvement of IL-32 in activation-induced cell death in T cells. Int Immunol. 2006;18(2):233–240. doi: 10.1093/intimm/dxh339. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16410314. [DOI] [PubMed] [Google Scholar]
- 25.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 116(1):66–76. doi: 10.1002/cncr.24662. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19885928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68(8):2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18413741. [DOI] [PubMed] [Google Scholar]
- 27.Torabian SZ, de Semir D, Nosrati M, Bagheri S, Dar AA, Fong S, et al. Ribozyme-mediated targeting of IkappaBgamma inhibits melanoma invasion and metastasis. Am J Pathol. 2009;174(3):1009–1016. doi: 10.2353/ajpath.2009.080207. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19179607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18852463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17085592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17259978. [DOI] [PubMed] [Google Scholar]
- 31.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17616669. [DOI] [PubMed] [Google Scholar]
- 32.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69(6):2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19244118. [DOI] [PubMed] [Google Scholar]
- 33.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17277777. [DOI] [PubMed] [Google Scholar]
- 34.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17632057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17604727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17320508. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15680324. [DOI] [PubMed] [Google Scholar]