Abstract
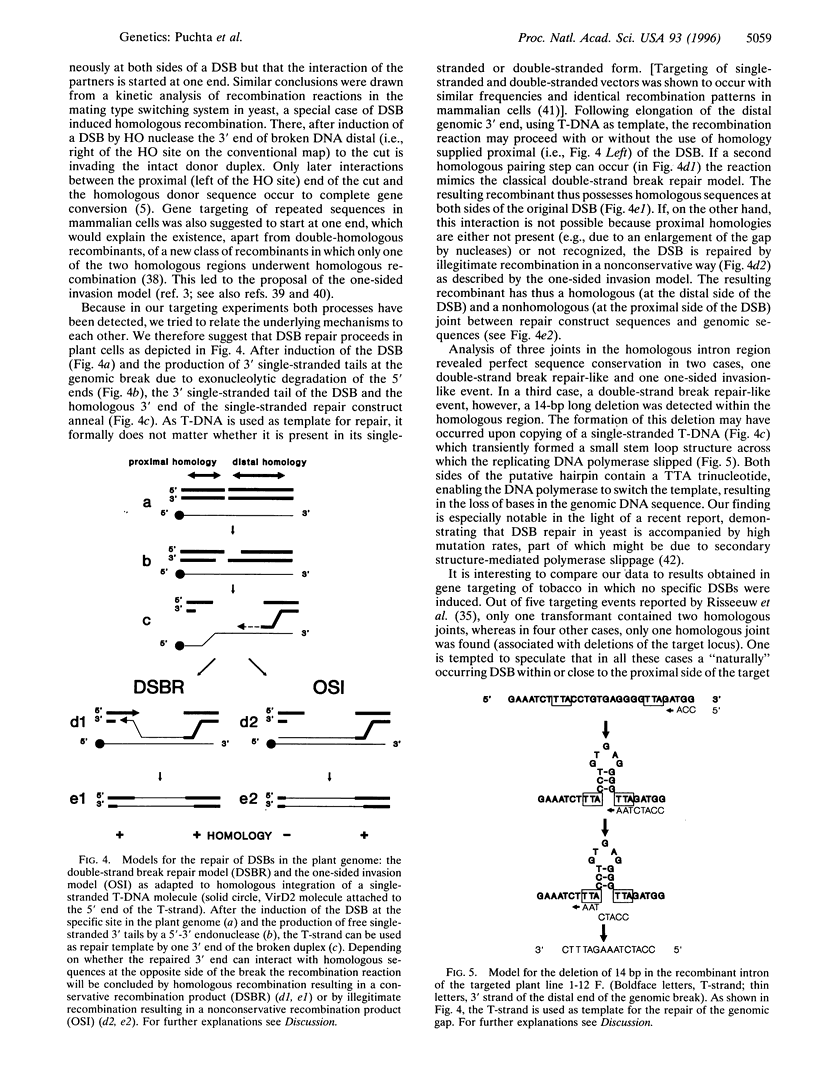
Genomic double-strand breaks (DSBs) are key intermediates in recombination reactions of living organisms. We studied the repair of genomic DSBs by homologous sequences in plants. Tobacco plants containing a site for the highly specific restriction enzyme I-Sce I were cotransformed with Agrobacterium strains carrying sequences homologous to the transgene locus and, separately, containing the gene coding for the enzyme. We show that the induction of a DSB can increase the frequency of homologous recombination at a specific locus by up to two orders of magnitude. Analysis of the recombination products demonstrates that a DSB can be repaired via homologous recombination by at least two different but related pathways. In the major pathway, homologies on both sides of the DSB are used, analogous to the conservative DSB repair model originally proposed for meiotic recombination in yeast. Homologous recombination of the minor pathway is restricted to one side of the DSB as described by the nonconservative one-sided invasion model. The sequence of the recombination partners was absolutely conserved in two cases, whereas in a third case, a deletion of 14 bp had occurred, probably due to DNA polymerase slippage during the copy process. The induction of DSB breaks to enhance homologous recombination can be applied for a variety of approaches of plant genome manipulation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albert H., Dale E. C., Lee E., Ow D. W. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995 Apr;7(4):649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Ashraf M., Altschuler M., Galasinski S., Griffiths T. D. Alteration of gene expression by restriction enzymes electroporated into plant cells. Mutat Res. 1993 Jun;302(2):75–82. doi: 10.1016/0165-7992(93)90007-i. [DOI] [PubMed] [Google Scholar]
- Athma P., Peterson T. Ac induces homologous recombination at the maize P locus. Genetics. 1991 May;128(1):163–173. doi: 10.1093/genetics/128.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmaaza A., Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutat Res. 1994 May;314(3):199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Belmaaza A., Milot E., Villemure J. F., Chartrand P. Interference of DNA sequence divergence with precise recombinational DNA repair in mammalian cells. EMBO J. 1994 Nov 15;13(22):5355–5360. doi: 10.1002/j.1460-2075.1994.tb06870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berinstein N., Pennell N., Ottaway C. A., Shulman M. J. Gene replacement with one-sided homologous recombination. Mol Cell Biol. 1992 Jan;12(1):360–367. doi: 10.1128/mcb.12.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984 Nov 26;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. Altering the genome by homologous recombination. Science. 1989 Jun 16;244(4910):1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Choulika A., Perrin A., Dujon B., Nicolas J. F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995 Apr;15(4):1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka K., Aratani Y., Kusano K., Koyama H. Targeted recombination with single-stranded DNA vectors in mammalian cells. Nucleic Acids Res. 1993 Feb 11;21(3):407–412. doi: 10.1093/nar/21.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995 Jul;17(7):609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- Haber J. E. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 1992 Dec;8(12):446–452. doi: 10.1016/0168-9525(92)90329-3. [DOI] [PubMed] [Google Scholar]
- Halfter U., Morris P. C., Willmitzer L. Gene targeting in Arabidopsis thaliana. Mol Gen Genet. 1992 Jan;231(2):186–193. doi: 10.1007/BF00279790. [DOI] [PubMed] [Google Scholar]
- Hrouda M., Paszkowski J. High fidelity extrachromosomal recombination and gene targeting in plants. Mol Gen Genet. 1994 Apr;243(1):106–111. doi: 10.1007/BF00283882. [DOI] [PubMed] [Google Scholar]
- Lebel E. G., Masson J., Bogucki A., Paszkowski J. Stress-induced intrachromosomal recombination in plant somatic cells. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):422–426. doi: 10.1073/pnas.90.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Lund P., Lowe K., Dunsmuir P. Homologous recombination in plant cells after Agrobacterium-mediated transformation. Plant Cell. 1990 May;2(5):415–425. doi: 10.1105/tpc.2.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe B., Mathern J., Hake S. Active Mutator elements suppress the knotted phenotype and increase recombination at the Kn1-O tandem duplication. Genetics. 1992 Nov;132(3):813–822. doi: 10.1093/genetics/132.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich T., Yang D., Waldman A. S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 1994 Dec 25;22(25):5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K. E., Summerfelt K. R. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990 Feb;14(2):269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Medberry S. L., Dale E., Qin M., Ow D. W. Intra-chromosomal rearrangements generated by Cre-lox site-specific recombination. Nucleic Acids Res. 1995 Feb 11;23(3):485–490. doi: 10.1093/nar/23.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z. H., Lam E. Targeted disruption of the TGA3 locus in Arabidopsis thaliana. Plant J. 1995 Feb;7(2):359–365. doi: 10.1046/j.1365-313x.1995.7020359.x. [DOI] [PubMed] [Google Scholar]
- Offringa R., Franke-van Dijk M. E., De Groot M. J., van den Elzen P. J., Hooykaas P. J. Nonreciprocal homologous recombination between Agrobacterium transferred DNA and a plant chromosomal locus. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7346–7350. doi: 10.1073/pnas.90.15.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offringa R., de Groot M. J., Haagsman H. J., Does M. P., van den Elzen P. J., Hooykaas P. J. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 1990 Oct;9(10):3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Wirtz U., Baker B. A system for insertional mutagenesis and chromosomal rearrangement using the Ds transposon and Cre-lox. Plant J. 1995 Apr;7(4):687–701. doi: 10.1046/j.1365-313x.1995.7040687.x. [DOI] [PubMed] [Google Scholar]
- Paszkowski J., Baur M., Bogucki A., Potrykus I. Gene targeting in plants. EMBO J. 1988 Dec 20;7(13):4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J., Peterhans A., Bilang R., Filipowicz W. Expression in transgenic tobacco of the bacterial neomycin phosphotransferase gene modified by intron insertions of various sizes. Plant Mol Biol. 1992 Aug;19(5):825–836. doi: 10.1007/BF00027078. [DOI] [PubMed] [Google Scholar]
- Perrin A., Buckle M., Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO J. 1993 Jul;12(7):2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A., Perrin A., Haber J. E., Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992 Mar;130(3):451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
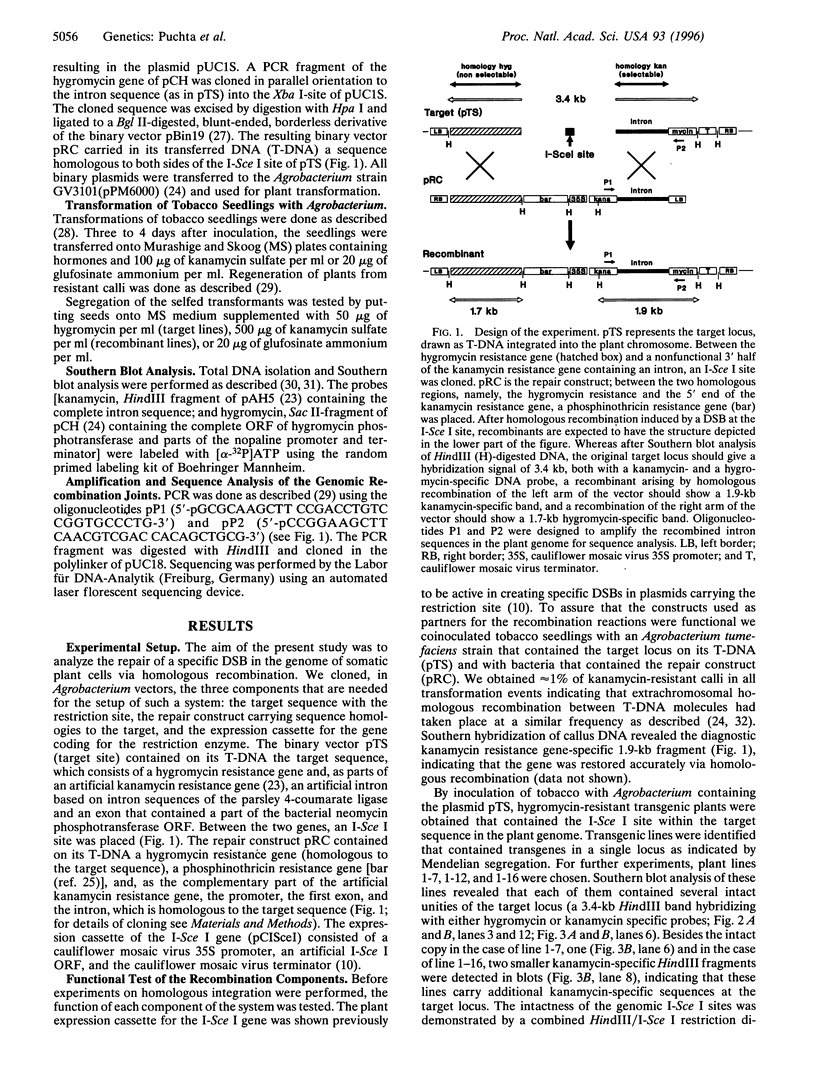
- Puchta H., Dujon B., Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993 Nov 11;21(22):5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H., Kocher S., Hohn B. Extrachromosomal homologous DNA recombination in plant cells is fast and is not affected by CpG methylation. Mol Cell Biol. 1992 Aug;12(8):3372–3379. doi: 10.1128/mcb.12.8.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M., Bayley C., Stockton T., Ow D. W. Cre recombinase-mediated site-specific recombination between plant chromosomes. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1706–1710. doi: 10.1073/pnas.91.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw E., Offringa R., Franke-van Dijk M. E., Hooykaas P. J. Targeted recombination in plants using Agrobacterium coincides with additional rearrangements at the target locus. Plant J. 1995 Jan;7(1):109–119. doi: 10.1046/j.1365-313x.1995.07010109.x. [DOI] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994 Dec;14(12):8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami K., Tokinaga Y., Yoshikura H., Kobayashi I. Homology-associated nonhomologous recombination in mammalian gene targeting. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8527–8531. doi: 10.1073/pnas.91.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal D. J., Carroll D. Endonuclease-induced, targeted homologous extrachromosomal recombination in Xenopus oocytes. Proc Natl Acad Sci U S A. 1995 Jan 31;92(3):806–810. doi: 10.1073/pnas.92.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Shafer B. K., McGill C. B. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995 Jul;140(3):965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. Intrachromosomal homologous recombination in whole plants. EMBO J. 1994 Jan 15;13(2):484–489. doi: 10.1002/j.1460-2075.1994.tb06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
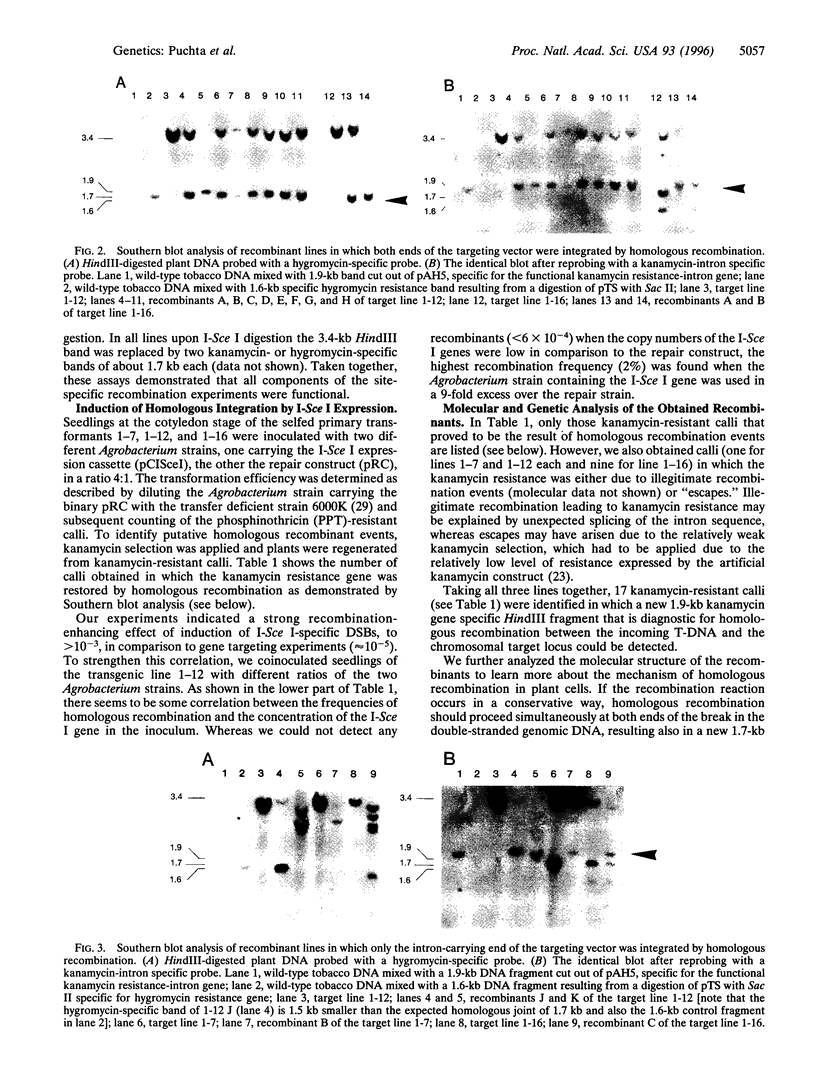
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thierry A., Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992 Nov 11;20(21):5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A., Fairhead C., Dujon B. The complete sequence of the 8.2 kb segment left of MAT on chromosome III reveals five ORFs, including a gene for a yeast ribokinase. Yeast. 1990 Nov-Dec;6(6):521–534. doi: 10.1002/yea.320060609. [DOI] [PubMed] [Google Scholar]
- Tinland B., Hohn B., Puchta H. Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8000–8004. doi: 10.1073/pnas.91.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B., Hohn B. Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet Eng (N Y) 1995;17:209–229. [PubMed] [Google Scholar]
- Tinland B., Schoumacher F., Gloeckler V., Bravo-Angel A. M., Hohn B. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995 Jul 17;14(14):3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar J., Lichtenstein C. Somatic and Meiotic Chromosomal Recombination between Inverted Duplications in Transgenic Tobacco Plants. Plant Cell. 1992 Mar;4(3):319–332. doi: 10.1105/tpc.4.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994 Jan 28;263(5146):515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]